Deep Learning Models Based on Pretreatment MRI and Clinicopathological Data to Predict Responses to Neoadjuvant Systemic Therapy in Triple-Negative Breast Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Data
2.2. Imaging and Clinicopathological Data
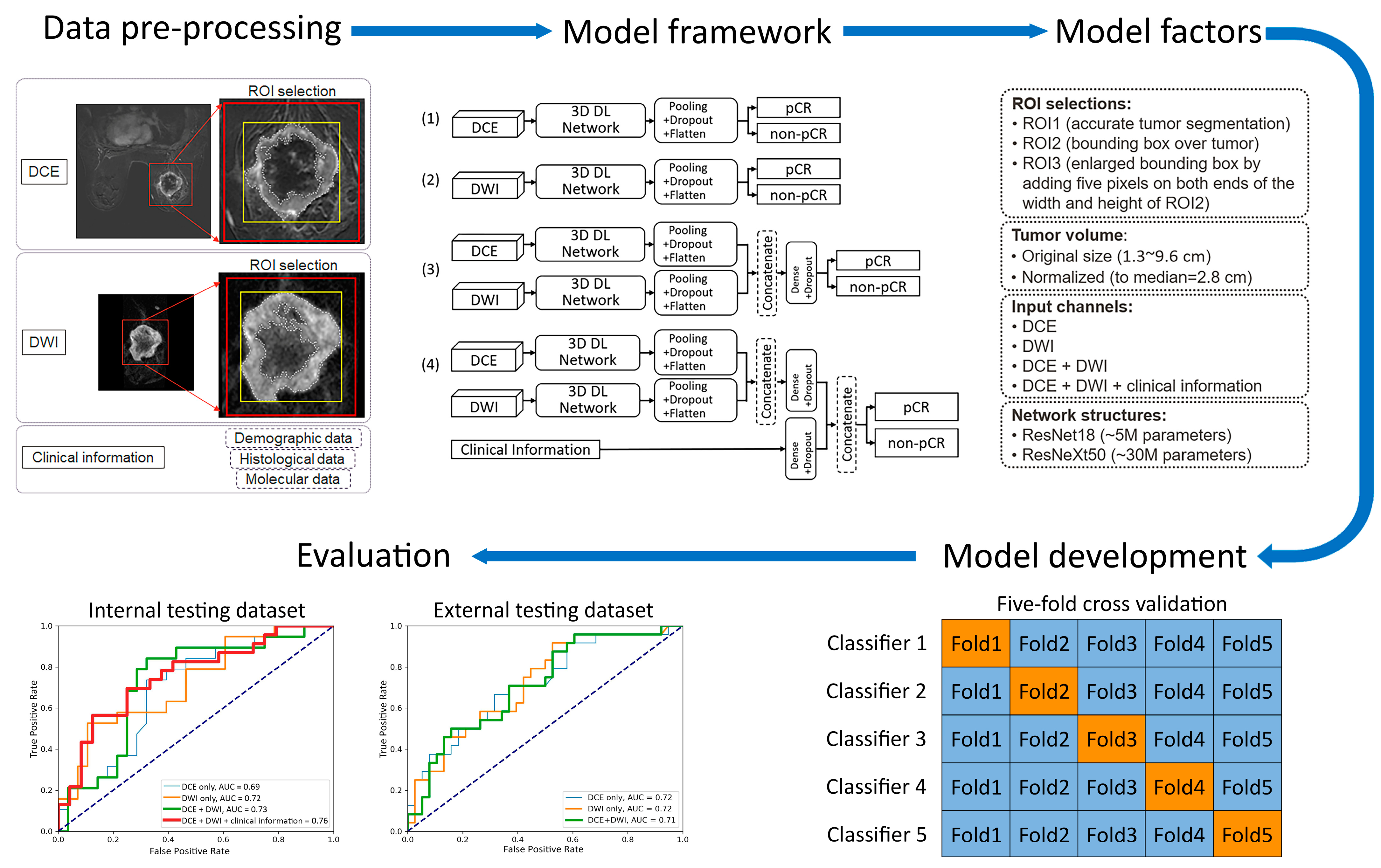
2.3. Models
- (1)
- Two 3D frameworks for transfer learning: ResNet18 [22,23,26,31], with ~5 million parameters and ~1000 trainable parameters, and ResNeXt50 [16,24], with ~30 million parameters and ~4000 trainable parameters. These two frameworks were chosen for their popularity and reported success in applications of medical-imaging-based prediction/classification.
- (2)
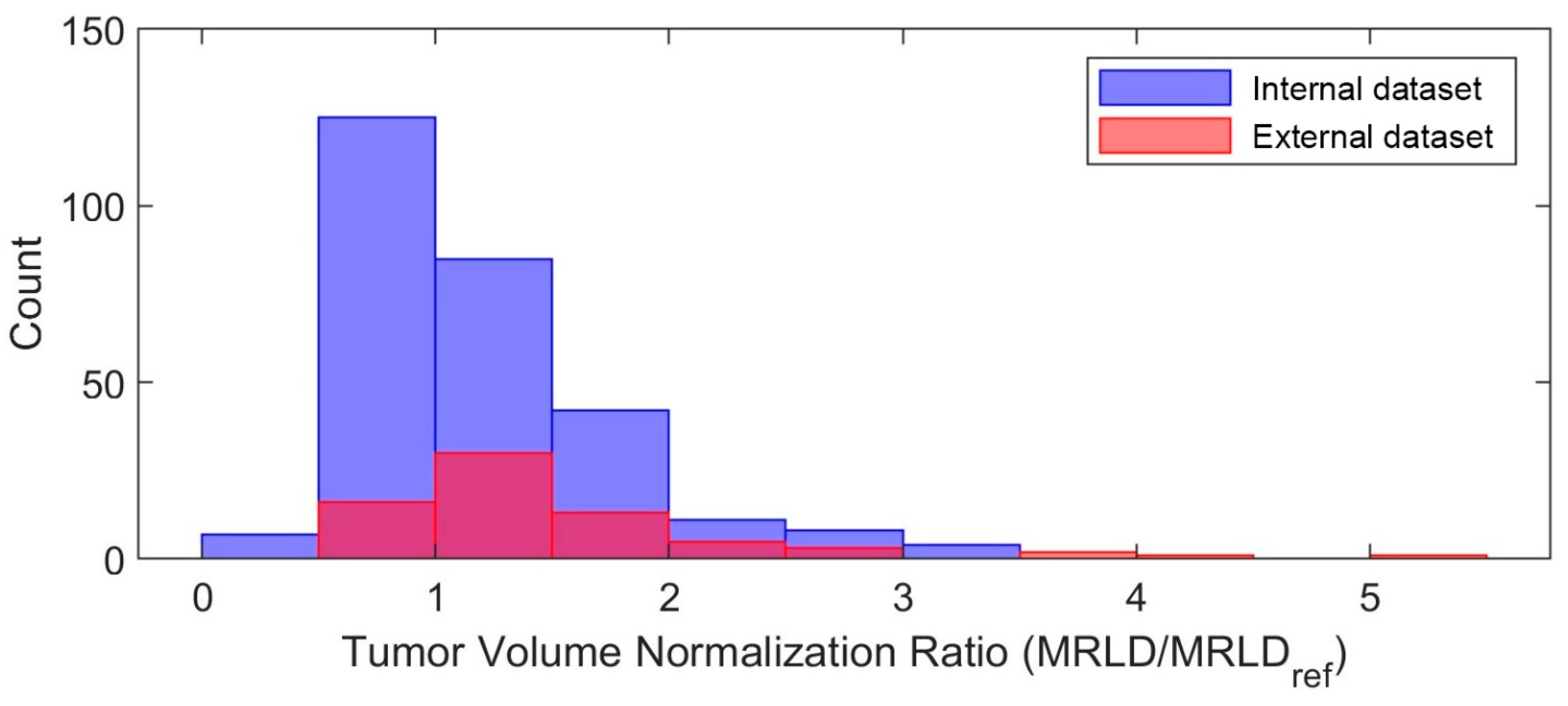
- Tumor volume preprocessing: normalized tumor volume [11,13,25] and original tumor matrix size [14,16,26,27,28,29,30]. The normalization referred to the median tumor size across all 282 subjects in the internal dataset, defined as the median MRI-measured longest diameter (cm) in the baseline study, which was 2.8 cm. The histogram of the tumor volume normalization ratio is shown in Figure A1 in Appendix B. The purpose here was to evaluate if using the original or normalizing tumor volume in preprocessing would impact the model performance, since both approaches [11,13,14,16,25,26,27,28,29,30] are reported in the literature.
- (3)
- Tumor ROI selection (Figure 2, Figure A2 in Appendix B): ROI1, which included voxels within the aforementioned reference masks of tumors [11,16,27]; ROI2, which included voxels within a tight bounding box of the reference masks [13,14,25]; and ROI3, which included voxels within an enlarged bounding box of the ROI2 (dilated by 5 mm along the top, bottom, left, and right sides) [28,29,30].
- (4)
- Data inputs: DCE images only; DWI images only; DCE and DWI images; or DCE and DWI images plus clinicopathological information. For each input channel of the image, the base layer parameters of the chosen transfer-learned 3D framework were locked and then linked to the top layer, which remained unlocked for training. In models that fused multiple inputs, the base layer features of each input were combined through concatenation and then linked to the top layer.
2.4. Statistical Analysis
3. Results
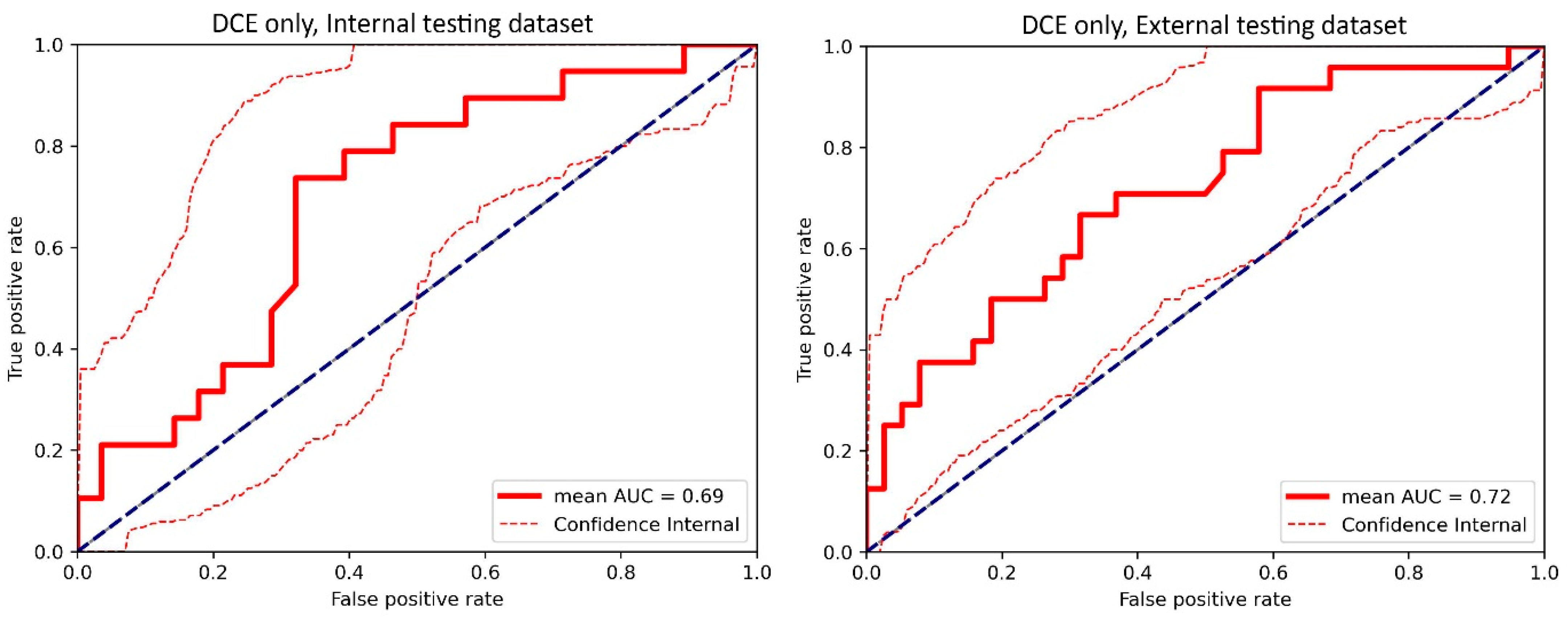
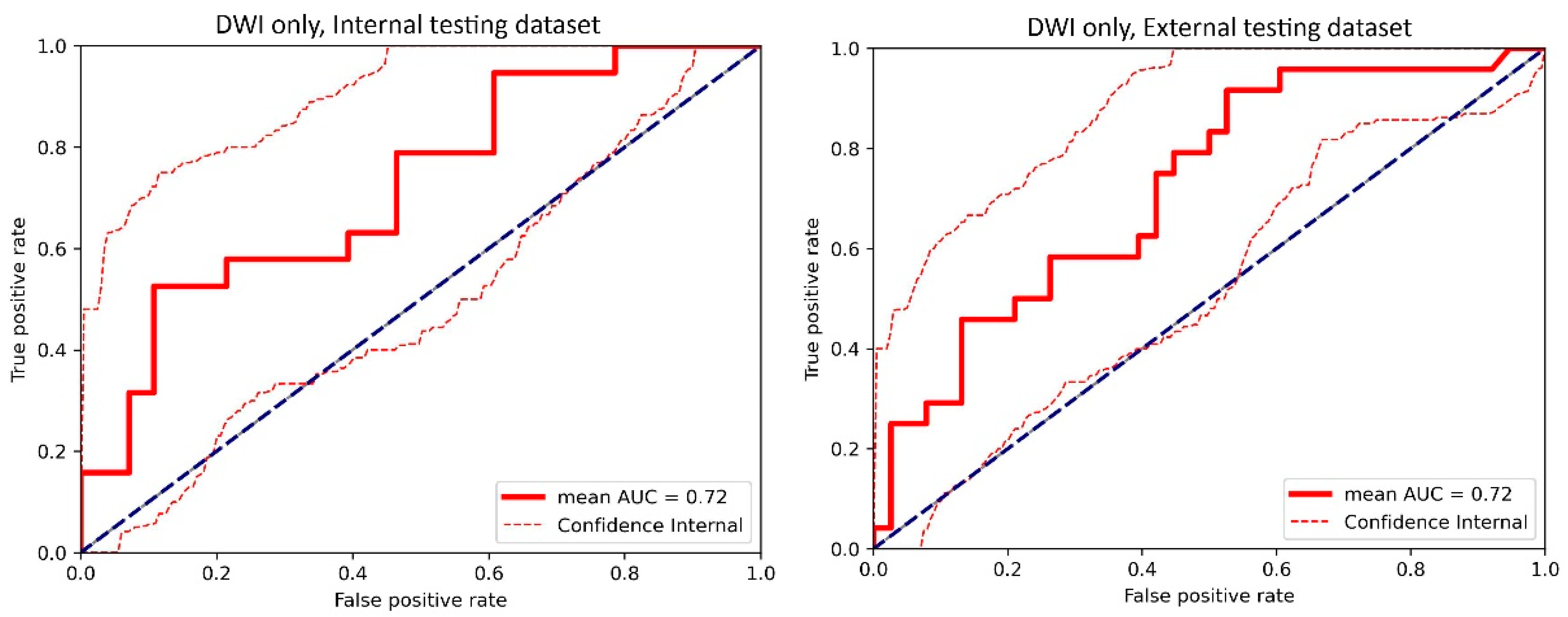
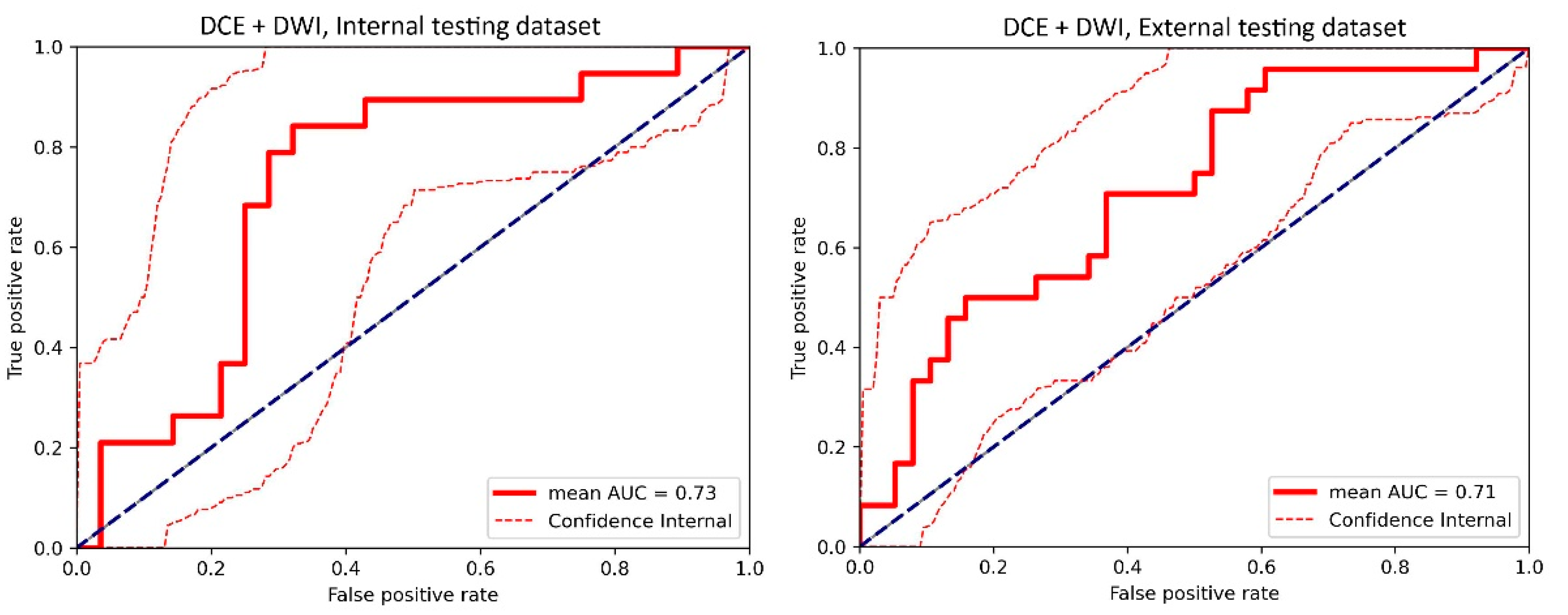
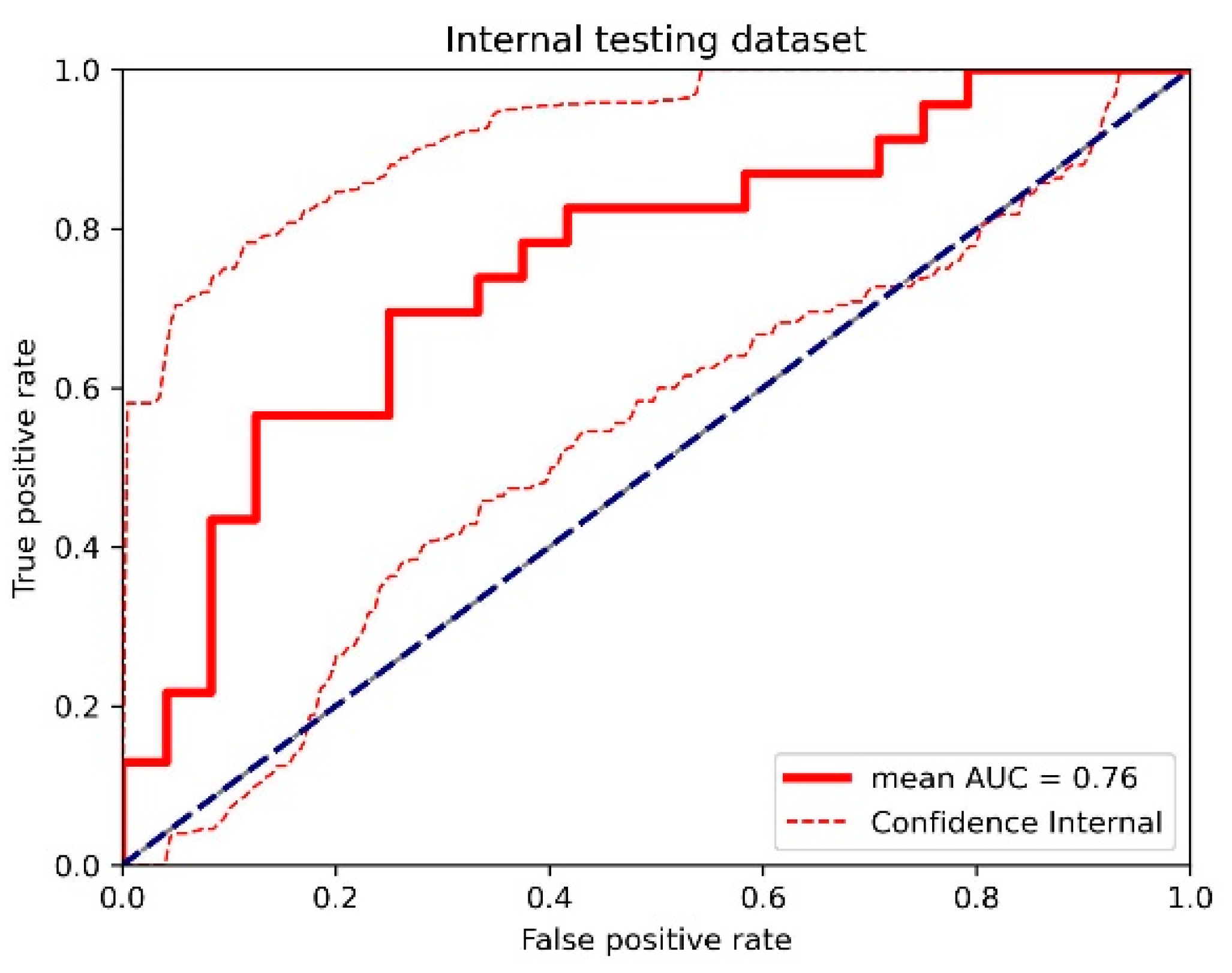
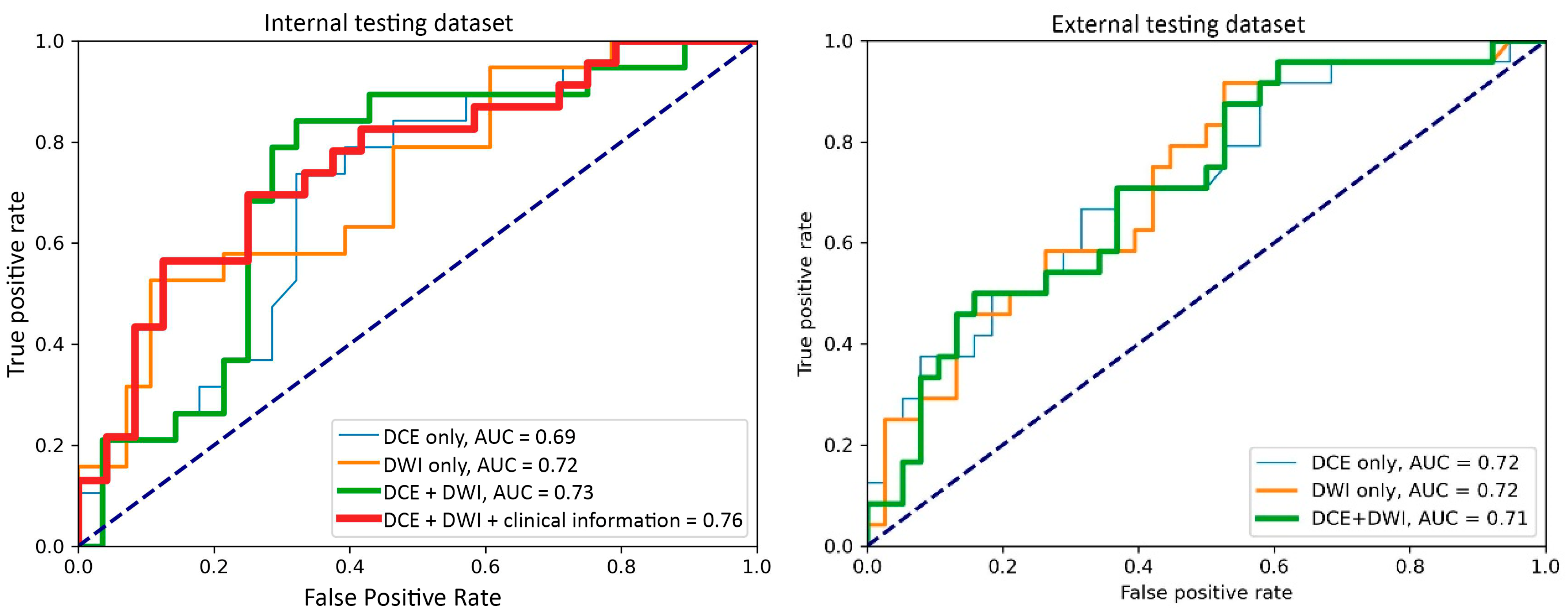
3.1. Top-Performing Models
3.2. Impact of 3D Frameworks, Tumor Volume Preprocessing, and Tumor ROI Selection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | area under the receiver operating characteristic curve |
| DCE | dynamic contrast-enhanced MRI |
| DL | deep learning |
| DWI | diffusion-weighted imaging |
| NAST | neoadjuvant systemic therapy |
| pCR | pathologic complete response |
| ROC | receiver operating characteristic |
| TNBC | triple-negative breast cancer |
| 3D | three-dimensional |
Appendix A
Technical Details of Model Construction
Appendix B
| (A) Internal Dataset | ||||
| DCE: 3D T1-Weighted DISCO | DWI: FOCUS | Clinical Information | ||
| Reconstruction matrix size | 512 × 512 | Reconstruction matrix size | 80 × 80 | Age |
| Field-of-view, mm | 300 × 300 | Field-of-view, mm | 160 × 160 | Clinical stage |
| Number of slices | 112~192 | Number of slices | 16 | T category |
| Slice thickness/gap, mm | 3.2/−1.6 | Slice thickness/gap, mm | 4/0 | N category |
| Flip angle, degrees | 12 | Flip angle, degrees | 90 | sTIL index |
| TR/TE1/TE2, ms | 6/1.1/2.3 | TR/TE, ms | 4000/70 | Ki-67 index |
| Number of temporal phases | 32~64 | b-values, s/mm2 | 100, 800 | BMI |
| (B) External Dataset | ||||
| DCE: 3D T1-Weighted | DWI: 2D SE-EPI | Clinical Information | ||
| Reconstruction matrix size | 512 × 512 | Reconstruction matrix size | 256 × 256 | Age |
| Field-of-view, mm | (260~360, 300~360) | Field-of-view, mm | (260~360, 300~360) | Clinical stage |
| Number of slices | >60 | Number of slices | variable | T category |
| Slice thickness/gap, mm | ≤2.5/0 | Slice thickness/gap, mm | 4–5/0 | |
| Flip angle, degrees | 10–20 | Flip angle, degrees | 90 | |
| TR/TE, ms | 4~10/1.4~4.8 | TR/TE, ms | 4000~10,600/50~100 | |
| Number of temporal phases | 68–256 | b-values, s/mm2 | 0, 100, 600, 800 | |

References
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-Negative Breast Cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Yost, S.E.; Yuan, Y. Neoadjuvant Treatment for Triple Negative Breast Cancer: Recent Progresses and Challenges. Cancers 2020, 12, 1404. [Google Scholar] [CrossRef] [PubMed]
- Esserman, L.J.; Berry, D.A.; DeMichele, A.; Carey, L.; Davis, S.E.; Buxton, M.; Hudis, C.; Gray, J.W.; Perou, C.; Yau, C.; et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: Results from the I-SPY 1 TRIAL—CALGB 150007/150012, ACRIN 6657. J. Clin. Oncol. 2012, 30, 3242–3249. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef]
- Gentile, D.; Martorana, F.; Karakatsanis, A.; Caruso, F.; Caruso, M.; Castiglione, G.; Di Grazia, A.; Pane, F.; Rizzo, A.; Vigneri, P.; et al. Predictors of mastectomy in breast cancer patients with complete remission of primary tumor after neoadjuvant therapy: A retrospective study. Eur. J. Surg. Oncol. 2024, 50, 108732. [Google Scholar] [CrossRef]
- Karagiannis, G.S.; Pastoriza, J.M.; Wang, Y.; Harney, A.S.; Entenberg, D.; Pignatelli, J.; Sharma, V.P.; Xue, E.A.; Cheng, E.; D’alfonso, T.M.; et al. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci. Transl. Med. 2017, 9, eaan0026. [Google Scholar] [CrossRef]
- Sharma, U.; Danishad, K.K.A.; Seenu, V.; Jagannathan, N.R. Longitudinal study of the assessment by MRI and diffusion-weighted imaging of tumor response in patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy. NMR Biomed. 2009, 22, 104–113. [Google Scholar] [CrossRef]
- Fangberget, A.; Nilsen, L.B.; Hole, K.H.; Holmen, M.M.; Engebraaten, O.; Naume, B.; Smith, H.-J.; Olsen, D.R.; Seierstad, T. Neoadjuvant chemotherapy in breast cancer-response evaluation and prediction of response to treatment using dynamic contrast-enhanced and diffusion-weighted MR imaging. Eur. Radiol. 2011, 21, 1188–1199. [Google Scholar] [CrossRef]
- Li, X.; Abramson, R.G.; Arlinghaus, L.R.; Kang, H.; Chakravarthy, A.B.; Abramson, V.G.; Farley, J.; Kelley, M.C.; Meszoely, I.M.; Means-Powell, J.; et al. Combined DCE-MRI and DW-MRI for Predicting Breast Cancer Pathological Response After the First Cycle of Neoadjuvant Chemotherapy HHS Public Access. Investig. Radiol. 2015, 50, 195–204. [Google Scholar] [CrossRef]
- Cho, N.; Im, S.-A.; Park, I.-A.; Lee, K.-H.; Li, M.; Han, W.; Noh, D.-Y.; Moon, W.K. Breast cancer: Early Prediction of Response to Neoadjuvant Chemotherapy Using Parametric Response Maps for MR Imaging. Radiology 2014, 272, 385–396. [Google Scholar] [CrossRef]
- Comes, M.C.; Fanizzi, A.; Bove, S.; Didonna, V.; Diotaiuti, S.; La Forgia, D.; Latorre, A.; Martinelli, E.; Mencattini, A.; Nardone, A.; et al. Early prediction of neoadjuvant chemotherapy response by exploiting a transfer learning approach on breast DCE-MRIs. Sci. Rep. 2021, 11, 14123. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Kim, H.-A.; Kim, W.; Lim, I.; Lee, I.; Byun, B.H.; Noh, W.C.; Seong, M.-K.; Lee, S.-S.; Kim, B.I.; et al. Early prediction of neoadjuvant chemotherapy response for advanced breast cancer using PET/MRI image deep learning. Sci. Rep. 2020, 10, 21149. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Adrada, B.E.; Candelaria, R.P.; Elshafeey, N.A.; Boge, M.; Mohamed, R.M.; Pashapoor, S.; Sun, J.; Xu, Z.; Panthi, B.; et al. Prediction of pathologic complete response to neoadjuvant systemic therapy in triple negative breast cancer using deep learning on multiparametric, MRI. Sci. Rep. 2023, 13, 1171. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, K.; Braman, N.; Janowczyk, A.; Madabhushi, A. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE-MRI. In Medical Imaging 2018: Computer-Aided Diagnosis; Mori, K., Petrick, N., Eds.; SPIE: Bellingham, WA, USA, 2018; Volume 11. [Google Scholar] [CrossRef]
- Braman, N.; Adoui, M.E.; Vulchi, M.; Turk, P.; Etesami, M.; Fu, P.; Bera, K.; Drisis, S.; Varadan, V.; Plecha, D.; et al. Deep learning-based prediction of response to HER2-targeted neoadjuvant chemotherapy from pre-treatment dynamic breast MRI: A multi-institutional validation study. arXiv 2020, arXiv:2001.08570. [Google Scholar]
- Peng, Y.; Cheng, Z.; Gong, C.; Zheng, C.; Zhang, X.; Wu, Z.; Yang, Y.; Yang, X.; Zheng, J.; Shen, J. Pretreatment DCE-MRI-Based Deep Learning Outperforms Radiomics Analysis in Predicting Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer. Front. Oncol. 2022, 12, 846775. [Google Scholar] [CrossRef]
- Musall, B.C.; Rauch, D.E.; Mohamed, R.M.; Panthi, B.; Boge, M.; Candelaria, R.P.; Chen, H.; Guirguis, M.S.; Hunt, K.K.; Huo, L.; et al. Diffusion Tensor Imaging for Characterizing Changes in Triple-Negative Breast Cancer During Neoadjuvant Systemic Therapy. J. Magn. Reson. Imaging 2024, 60, 1367–1376. [Google Scholar] [CrossRef]
- Panthi, B.; Mohamed, R.M.; Adrada, B.E.; Boge, M.; Candelaria, R.P.; Chen, H.; Hunt, K.K.; Huo, L.; Hwang, K.-P.; Korkut, A.; et al. Longitudinal dynamic contrast-enhanced MRI radiomic models for early prediction of response to neoadjuvant systemic therapy in triple-negative breast cancer. Front. Oncol. 2023, 13, 1264259. [Google Scholar] [CrossRef]
- Mohamed, R.M.; Panthi, B.; Adrada, B.E.; Boge, M.; Candelaria, R.P.; Chen, H.; Guirguis, M.S.; Hunt, K.K.; Huo, L.; Hwang, K.-P.; et al. Multiparametric MRI—Based radiomic models for early prediction of response to neoadjuvant systemic therapy in triple—Negative breast cancer. Sci. Rep. 2024, 14, 16073. [Google Scholar] [CrossRef]
- Yee, D.; DeMichele, A.M.; Yau, C.; Isaacs, C.; Symmans, W.F.; Albain, K.S.; Chen, Y.Y.; Krings, G.; Wei, S.; Harada, S.; et al. Association of Event-Free and Distant Recurrence-Free Survival with Individual-Level Pathologic Complete Response in Neoadjuvant Treatment of Stages 2 and 3 Breast Cancer: Three-Year Follow-up Analysis for the I-SPY2 Adaptively Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1355–1362. [Google Scholar] [CrossRef]
- Li, W.; Newitt, D.C.; Gibbs, J.; Wilmes, L.J.; Jones, E.F.; Arasu, V.A.; Strand, F.; Onishi, N.; Nguyen, A.A.-T.; Kornak, J.; et al. Predicting breast cancer response to neoadjuvant treatment using multi-feature MRI: Results from the I-SPY 2 TRIAL. NPJ Breast Cancer 2020, 6, 63. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Moon, J.; Kim, J.; Shin, Y.; Hwang, S. Confidence-Aware Learning for Deep Neural Networks. In Proceedings of the 37th International Conference on Machine Learning, Virtual, 13–18 July 2020. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 5987–5995. [Google Scholar] [CrossRef]
- Liu, M.Z.; Mutasa, S.; Chang, P.; Siddique, M.; Jambawalikar, S.; Ha, R. A novel CNN algorithm for pathological complete response prediction using an I-SPY TRIAL breast MRI database. Magn. Reson. Imaging 2020, 73, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Dammu, H.; Ren, T.; Duong, T.Q. Deep learning prediction of pathological complete response, residual cancer burden, and progression-free survival in breast cancer patients. PLoS ONE 2023, 18, e0280148. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Yu, H.; Ke, J.; Ding, P.; Yi, Y.; Jiang, X.; Duan, X.; Tang, J.; Chang, D.T.; Wu, X.; et al. Predicting treatment response from longitudinal images using multi-task deep learning. Nat. Commun. 2021, 12, 1851. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.; Ko, E.S.; Kwon, S.; Jeon, E.; Jung, H.; Kim, J.-Y.; Chung, M.J.; Im, Y.-H. Multimodal deep learning models for the prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Sci. Rep. 2021, 11, 18800. [Google Scholar] [CrossRef]
- Qu, Y.H.; Zhu, H.T.; Cao, K.; Li, X.T.; Ye, M.; Sun, Y.S. Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (DL) method. Thorac. Cancer 2020, 11, 651–658. [Google Scholar] [CrossRef]
- Ha, R.; Chin, C.; Karcich, J.; Liu, M.Z.; Chang, P.; Mutasa, S.; Van Sant, E.P.; Wynn, R.T.; Connolly, E.; Jambawalikar, S. Prior to Initiation of Chemotherapy, Can We Predict Breast Tumor Response? Deep Learning Convolutional Neural Networks Approach Using a Breast MRI Tumor Dataset. J. Digit. Imaging 2019, 32, 693–701. [Google Scholar] [CrossRef]
- Duanmu, H.; Bhattarai, S.; Li, H.; Shi, Z.; Wang, F.; Teodoro, G.; Gogineni, K.; Subhedar, P.; Kiraz, U.; Janssen, E.A.M.; et al. A spatial attention guided deep learning system for prediction of pathological complete response using breast cancer histopathology images. Bioinformatics 2022, 38, 4605–4612. [Google Scholar] [CrossRef]
- Abuhadra, N.; Sun, R.; Yam, C.; Rauch, G.M.; Ding, Q.; Lim, B.; Thompson, A.M.; Mittendorf, E.A.; Adrada, B.E.; Damodaran, S.; et al. Predictive Roles of Baseline Stromal Tumor-Infiltrating Lymphocytes and Ki-67 in Pathologic Complete Response in an Early-Stage Triple-Negative Breast Cancer Prospective Trial. Cancers 2023, 15, 3275. [Google Scholar] [CrossRef]
- Cain, E.H.; Saha, A.; Harowicz, M.R.; Marks, J.R.; Marcom, P.K.; Mazurowski, M.A. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: A study using an independent validation set. Breast Cancer Res. Treat. 2019, 173, 455–463. [Google Scholar] [CrossRef]
- Li, F.; Yang, Y.; Wei, Y.; He, P.; Chen, J.; Zheng, Z.; Bu, H. Deep learning-based predictive biomarker of pathological complete response to neoadjuvant chemotherapy from histological images in breast cancer. J. Transl. Med. 2021, 19, 348. [Google Scholar] [CrossRef]
- Pei, X.; Zuo, K.; Li, Y.; Pang, Z. A Review of the Application of Multi-modal Deep Learning in Medicine: Bibliometrics and Future Directions. Int. J. Comput. Intell. Syst. 2023, 16, 44. [Google Scholar] [CrossRef]
- Zahavy, T.; Krishnan, A.; Magnani, A.; Mannor, S. Is a picture worth a thousand words? A deep multi-modal architecture for product classification in e-commerce. In Proceedings of the 30th AAAI Conference on Artificial Intelligence, AAAI 2018, New Orleans, LA, USA, 2–7 February 2018; pp. 7873–7880. [Google Scholar]

| (A) | ||||
| Internal Data | All Patients | pCR | non-pCR | p Value |
| No. of patients | 282 | 132 | 150 | |
| Age, mean ± SD, y | 49.8 ± 11.4 | 49.5 ± 11.5 | 50.1 ± 11.5 | 0.36 |
| BMI, mean, kg/m2 | 28.9 | 28.5 | 29.2 | 0.15 |
| Longest tumor diameter, mean ± SD, cm | 3.4 ± 1.5 | 2.9 ± 1.3 | 3.6 ± 1.8 | <0.001 |
| Clinical stage, n (%) | 0.018 | |||
| I | 34 (12) | 19 (14) | 15 (10) | |
| II | 206 (73) | 100 (76) | 106 (71) | |
| III | 42 (15) | 13 (10) | 29 (19) | |
| T category, n (%) | <0.001 | |||
| T1 | 51 (18) | 31 (23) | 20 (13) | |
| T2 | 189 (67) | 91 (69) | 98 (65) | |
| T3 | 36 (13) | 8 (6) | 28 (19) | |
| T4 | 6 (2) | 2 (2) | 4 (3) | |
| N category, n (%) | <0.001 | |||
| N0 | 181 (64) | 92 (70) | 89 (59) | |
| N1 | 72 (26) | 30 (23) | 42 (28) | |
| N2 | 7 (2) | 3 (2) | 4 (3) | |
| N3 | 22 (8) | 7 (5) | 15 (10) | |
| Stromal tumor-infiltrating lymphocyte level, %, median (IQR) | 10 (4–20) | 20 (4–30) | 10 (4–20) | <0.001 |
| Ki-67 index, %, median (IQR) | 70 (50–90) | 75 (50–90) | 70 (51–87) | 0.012 |
| NAST regimen, n | ||||
| Doxorubicin + Paclitaxel | 233 | 121 | 112 | |
| Doxorubicin + Enzalutamide | 11 | 3 | 8 | |
| Doxorubicin + Panitumumab | 15 | 2 | 13 | |
| Doxorubicin + Everolimus | 7 | 0 | 7 | |
| Doxorubicin + Atezolizumab | 12 | 5 | 7 | |
| Doxorubicin + Alpelisib | 4 | 1 | 3 | |
| (B) | ||||
| External Data | All Patients | pCR | non-pCR | p Value |
| No. of patients | 62 | 28 | 34 | |
| Age, mean ± SD, y | 48.8 ± 10.5 | 49.4 ± 10.4 | 48.2 ± 10.9 | 0.62 |
| Longest tumor diameter, mean ± SD, cm | 4.1 ± 2.3 | 3.5 ± 1.1 | 4.7 ± 2.9 | 0.017 |
| T category, n (%) | 0.11 | |||
| T2 | 5 (8) | 1 (4) | 4 (12) | |
| T3 | 56 (90) | 26 (92) | 30 (88) | |
| N/A | 1 (2) | 1 (4) | 0 (0) | |
| NAST regimen, n | ||||
| Paclitaxel | 13 | 3 | 10 | |
| Paclitaxel + MK-2206 | 9 | 6 | 3 | |
| Paclitaxel + AMG 386 | 15 | 7 | 8 | |
| Paclitaxel + Ganetespib | 1 | 0 | 1 | |
| Paclitaxel + Ganitumab | 24 | 12 | 12 | |
| Internal Testing Dataset | ||||||
|---|---|---|---|---|---|---|
| Network | ResNet18 | ResNeXt50 | ||||
| ROI1 | ROI2 | ROI3 | ROI1 | ROI2 | ROI3 | |
| DCE-only models | ||||||
| Normalized tumor volume | 0.63 | 0.61 | 0.63 | 0.52 | 0.48 | 0.48 |
| (0.58, 0.7) | (0.59, 0.65) | (0.55, 0.68) | (0.44, 0.61) | (0.39, 0.62) | (0.45, 0.53) | |
| Original tumor volume | 0.68 | 0.69 | 0.68 | 0.66 | 0.65 * | 0.65 * |
| (0.62, 0.7) | (0.66, 0.74) | (0.66, 0.7) | (0.65, 0.67) | (0.64, 0.66) | (0.65, 0.66) | |
| DWI-only models | ||||||
| Normalized tumor volume | 0.60 | 0.59 | 0.61 | 0.61 | 0.61 | 0.57 |
| (0.55, 0.68) | (0.49, 0.67) | (0.46, 0.74) | (0.61, 0.62) | (0.59, 0.63) | (0.56, 0.59) | |
| Original tumor volume | 0.69 | 0.72 | 0.70 | 0.53 | 0.58 | 0.60 |
| (0.67, 0.72) | (0.69, 0.75) | (0.69, 0.71) | (0.51, 0.56) | (0.53, 0.73) | (0.54, 0.73) | |
| DCE + DWI models | ||||||
| Normalized tumor volume | 0.57 | 0.60 | 0.59 | 0.58 | 0.59 | 0.57 |
| (0.5, 0.63) | (0.59, 0.64) | (0.54, 0.62) | (0.54, 0.61) | (0.58, 0.6) | (0.56, 0.58) | |
| Original tumor volume | 0.71 | 0.73 | 0.72 | 0.71 | 0.70 | 0.71 |
| (0.67, 0.76) | (0.71, 0.74) | (0.68, 0.76) | (0.66, 0.78) | (0.65, 0.78) | (0.67, 0.77) | |
| DCE + DWI + clinical information models | ||||||
| Normalized tumor volume | 0.68 | 0.69 | 0.69 | 0.67 | 0.68 | 0.67 |
| (0.66, 0.72) | (0.66, 0.72) | (0.66, 0.73) | (0.64, 0.70) | (0.65, 0.69) | (0.63, 0.70) | |
| Original tumor volume | 0.74 | 0.73 | 0.71 | 0.72 | 0.76 | 0.72 |
| (0.67, 0.79) | (0.67, 0.81) | (0.65, 0.76) | (0.69, 0.75) | (0.72, 0.78) | (0.68, 0.75) | |
| External Testing Dataset | ||||||
|---|---|---|---|---|---|---|
| Network | ResNet18 | ResNeXt50 | ||||
| ROI1 | ROI2 | ROI3 | ROI1 | ROI2 | ROI3 | |
| DCE-only models | ||||||
| Normalized tumor volume | 0.41 | 0.46 | 0.53 | 0.43 | 0.44 | 0.44 |
| (0.33, 0.48) | (0.43, 0.57) | (0.49, 0.54) | (0.40, 0.53) | (0.40, 0.58) | (0.42, 0.53) | |
| Original tumor volume | 0.69 * | 0.67 * | 0.67 * | 0.71 * | 0.71 * | 0.72 * |
| (0.63, 0.69) | (0.64, 0.69) | (0.64, 0.69) | (0.71, 0.72) | (0.71, 0.72) | (0.71, 0.72) | |
| DWI-only models | ||||||
| Normalized tumor volume | 0.61 | 0.57 | 0.68 | 0.53 | 0.62 | 0.61 |
| (0.52, 0.58) | (0.45, 0.63) | (0.59, 0.76) | (0.51, 0.54) | (0.60, 0.68) | (0.57, 0.71) | |
| Original tumor volume | 0.66 | 0.70 | 0.72 | 0.66 | 0.68 | 0.69 |
| (0.65, 0.67) | (0.68, 0.73) | (0.71, 0.72) | (0.64, 0.66) | (0.60, 0.72) | (0.68, 0.69) | |
| DCE + DWI models | ||||||
| Normalized tumor volume | 0.62 | 0.61 | 0.68 | 0.59 | 0.63 | 0.64 |
| (0.49, 0.60) | (0.38, 0.62) | (0.57, 0.68) | (0.52, 0.65) | (0.63, 0.65) | (0.62, 0.65) | |
| Original tumor volume | 0.67 | 0.70 | 0.71 | 0.66 | 0.67 | 0.71 |
| (0.64, 0.70) | (0.69, 0.72) | (0.67, 0.73) | (0.55, 0.68) | (0.50, 0.70) | (0.60, 0.72) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Zhou, Z.; Son, J.B.; Feng, H.; Adrada, B.E.; Moseley, T.W.; Candelaria, R.P.; Guirguis, M.S.; Patel, M.M.; Whitman, G.J.; et al. Deep Learning Models Based on Pretreatment MRI and Clinicopathological Data to Predict Responses to Neoadjuvant Systemic Therapy in Triple-Negative Breast Cancer. Cancers 2025, 17, 966. https://doi.org/10.3390/cancers17060966
Xu Z, Zhou Z, Son JB, Feng H, Adrada BE, Moseley TW, Candelaria RP, Guirguis MS, Patel MM, Whitman GJ, et al. Deep Learning Models Based on Pretreatment MRI and Clinicopathological Data to Predict Responses to Neoadjuvant Systemic Therapy in Triple-Negative Breast Cancer. Cancers. 2025; 17(6):966. https://doi.org/10.3390/cancers17060966
Chicago/Turabian StyleXu, Zhan, Zijian Zhou, Jong Bum Son, Haonan Feng, Beatriz E. Adrada, Tanya W. Moseley, Rosalind P. Candelaria, Mary S. Guirguis, Miral M. Patel, Gary J. Whitman, and et al. 2025. "Deep Learning Models Based on Pretreatment MRI and Clinicopathological Data to Predict Responses to Neoadjuvant Systemic Therapy in Triple-Negative Breast Cancer" Cancers 17, no. 6: 966. https://doi.org/10.3390/cancers17060966
APA StyleXu, Z., Zhou, Z., Son, J. B., Feng, H., Adrada, B. E., Moseley, T. W., Candelaria, R. P., Guirguis, M. S., Patel, M. M., Whitman, G. J., Leung, J. W. T., Le-Petross, H. T. C., Mohamed, R. M., Panthi, B., Lane, D. L., Chen, H., Wei, P., Tripathy, D., Litton, J. K., ... Ma, J. (2025). Deep Learning Models Based on Pretreatment MRI and Clinicopathological Data to Predict Responses to Neoadjuvant Systemic Therapy in Triple-Negative Breast Cancer. Cancers, 17(6), 966. https://doi.org/10.3390/cancers17060966






