Genetic Insights into Breast Cancer in Northeastern Mexico: Unveiling Gene–Environment Interactions and Their Links to Obesity and Metabolic Diseases
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Scientific and Ethics Committees Approval
2.2. Study Design and Population
2.3. Patients Group
2.4. Controls Group
2.5. Nucleic Acid Extraction
2.6. SNP Selection
2.7. Genotype Analysis
2.8. Statistical Methods
2.9. In Silico Variant Effect Predictor Analysis
2.10. Protein–RNA Interaction Prediction Using RNAct
2.11. Pathway and Gene Ontology Enrichment with ToppGene Suite
3. Results
3.1. Patients Data
3.2. Genotype Analysis Results
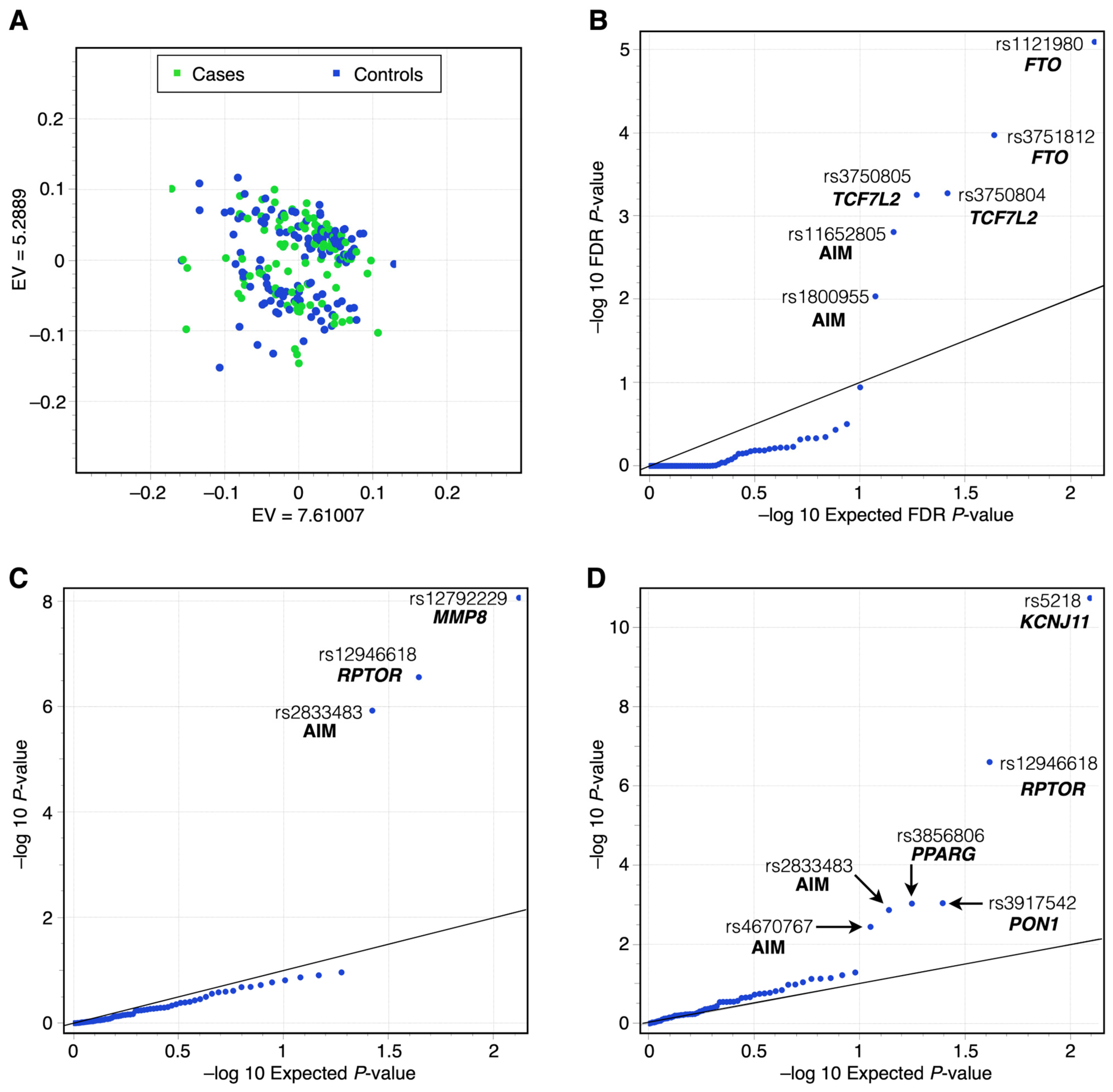
3.3. MLMM Testing
4. Discussion
4.1. The rs11652805 (AMZ2P1; GNA13 Intergenic Variant, Regulatory Region Variant)
4.1.1. AMZ2P1 (Archaelysin Family Metallopeptidase 2 Pseudogene 1)
4.1.2. DRD4 (Dopamine Receptor D4)
4.2. FTO (Fat Mass and Obesity-Associated Protein)
4.3. KCNJ11 (Potassium Voltage-Gated Channel Subfamily J Member)
4.4. MMP8 (Matrix Metalloproteinase 8)
4.5. PON1 (Human Serum Paraoxonase 1 Enzyme)
4.6. PPARG (Peroxisome Proliferator-Activated Receptor Gamma)
4.7. RPTOR (Regulatory Associated Protein of MTOR Complex 1, RPTOR, Also Named RAPTOR)
4.8. KCNJ11 Pathway and RPTOR
4.9. SCAF4 (SR-Related C-Terminal Domain-Associated Factor 4)
4.10. TCF7L2 (Transcription Factor 7 Like 2)
4.11. GNA13 and RAPTOR, TCF7L2, SCAF4, KCNJ11, and FTO
4.12. GNA13 (Gα13) and RPTOR (mTORC1) Pathway Interaction
5. Limitations of This Study
5.1. Sample Size and Study Design
5.2. Geographical and Ethnic Specificity
5.3. Restricted Genetic Scope
5.4. Limited Lifestyle and Environmental Data
5.5. BC Treatment and Subtype Representation
5.6. Functional Validation
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; Global Cancer Observatory: Cancer Today; IARC; WHO: Lyon, France, 2022. [Google Scholar]
- Makowski, L.; Sundaram, S.; Johnson, A. Obesity, Metabolism and the Microenvironment: Links to Cancer. J. Carcinog. 2013, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Wu, W.Y.; Yen, A.M.; Fann, J.C.; Chen, S.L.; Chiu, S.Y.; Chen, H.H.; Chiou, S.T. Body Mass Index and Breast Cancer: Analysis of a Nation-Wide Population-Based Prospective Cohort Study on 1,393,985 Taiwanese Women. Int. J. Obes. 2016, 40, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; Tomotaki, A.; Miyata, H.; Iwamoto, T.; Niikura, N.; Anan, K.; Hayashi, N.; Aogi, K.; Ishida, T.; Masuoka, H.; et al. Body Mass Index and Survival after Diagnosis of Invasive Breast Cancer: A Study Based on the Japanese National Clinical Database-Breast Cancer Registry. Cancer Med. 2016, 5, 1328–1340. [Google Scholar] [CrossRef]
- Samavat, H.; Kurzer, M.S. Estrogen Metabolism and Breast Cancer. Cancer Lett. 2015, 356, 231–243. [Google Scholar] [CrossRef]
- Zhang, X.; Eliassen, A.H.; Tamimi, R.M.; Hazra, A.; Beck, A.H.; Brown, M.; Collins, L.C.; Rosner, B.; Hankinson, S.E. Adult Body Size and Physical Activity in Relation to Risk of Breast Cancer According to Tumor Androgen Receptor Status. Cancer Epidemiol. Biomark. Prev. 2015, 24, 962–968. [Google Scholar] [CrossRef]
- Surakasula, A.; Nagarjunapu, G.; Raghavaiah, K. A Comparative Study of Pre- and Post-Menopausal Breast Cancer: Risk Factors, Presentation, Characteristics and Management. J. Res. Pharm. Pract. 2014, 3, 12. [Google Scholar] [CrossRef]
- Chlebowski, R.T.; Anderson, G.L. Menopausal Hormone Therapy and Breast Cancer Mortality: Clinical Implications. Ther. Adv. Drug Saf. 2015, 6, 45–56. [Google Scholar] [CrossRef]
- Millikan, R.C.; Newman, B.; Tse, C.-K.; Moorman, P.G.; Conway, K.; Smith, L.V.; Labbok, M.H.; Geradts, J.; Bensen, J.T.; Jackson, S.; et al. Epidemiology of Basal-like Breast Cancer. Breast Cancer Res. Treat. 2008, 109, 123–139. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Freedman, M.L. The Genetics of Cancer Risk. Cancer J. 2011, 17, 416–422. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to Accessing Data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2024, 52, D891–D899. [Google Scholar] [CrossRef] [PubMed]
- Shieh, Y.; Fejerman, L.; Lott, P.C.; Marker, K.; Sawyer, S.D.; Hu, D.; Huntsman, S.; Torres, J.; Echeverry, M.; Bohórquez, M.E.; et al. A Polygenic Risk Score for Breast Cancer in US Latinas and Latin American Women. JNCI J. Natl. Cancer Inst. 2020, 112, 590–598. [Google Scholar] [CrossRef] [PubMed]
- COLUMBUS Consortium; Hoffman, J.; Fejerman, L.; Hu, D.; Huntsman, S.; Li, M.; John, E.M.; Torres-Mejia, G.; Kushi, L.; Ding, Y.C.; et al. Identification of Novel Common Breast Cancer Risk Variants at the 6q25 Locus among Latinas. Breast Cancer Res. 2019, 21, 3. [Google Scholar] [CrossRef]
- Fejerman, L.; Stern, M.C.; John, E.M.; Torres-Mejía, G.; Hines, L.M.; Wolff, R.K.; Baumgartner, K.B.; Giuliano, A.R.; Ziv, E.; Pérez-Stable, E.J.; et al. Interaction between Common Breast Cancer Susceptibility Variants, Genetic Ancestry, and Nongenetic Risk Factors in Hispanic Women. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1731–1738. [Google Scholar] [CrossRef]
- Roberts, E.; Howell, S.; Evans, D.G. Polygenic Risk Scores and Breast Cancer Risk Prediction. Breast 2023, 67, 71–77. [Google Scholar] [CrossRef]
- Gallardo-Blanco, H.L.; Villarreal-Perez, J.Z.; Cerda-Flores, R.M.; Figueroa, A.; Sanchez-Dominguez, C.N.; Gutierrez-Valverde, J.M.; Torres-Muñoz, I.C.; Lavalle-Gonzalez, F.J.; Gallegos-Cabriales, E.C.; Martinez-Garza, L.E. Genetic Variants in KCNJ11, TCF7L2 and HNF4A Are Associated with Type 2 Diabetes, BMI and Dyslipidemia in Families of Northeastern Mexico: A Pilot Study. Exp. Ther. Med. 2017, 13, 523–529. [Google Scholar] [CrossRef]
- Kosoy, R.; Nassir, R.; Tian, C.; White, P.A.; Butler, L.M.; Silva, G.; Kittles, R.; Alarcon-Riquelme, M.E.; Gregersen, P.K.; Belmont, J.W.; et al. Ancestry Informative Marker Sets for Determining Continental Origin and Admixture Proportions in Common Populations in America. Hum. Mutat. 2009, 30, 69–78. [Google Scholar] [CrossRef]
- Nassir, R.; Kosoy, R.; Tian, C.; White, P.A.; Butler, L.M.; Silva, G.; Kittles, R.; Alarcon-Riquelme, M.E.; Gregersen, P.K.; Belmont, J.W.; et al. An Ancestry Informative Marker Set for Determining Continental Origin: Validation and Extension Using Human Genome Diversity Panels. BMC Genet. 2009, 10, 39. [Google Scholar] [CrossRef]
- Dandine-Roulland, C.; Bellenguez, C.; Debette, S.; Amouyel, P.; Génin, E.; Perdry, H. Accuracy of Heritability Estimations in Presence of Hidden Population Stratification. Sci. Rep. 2016, 6, 26471. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-B.; Feng, J.-Y.; Ren, W.-L.; Huang, B.; Zhou, L.; Wen, Y.-J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.-M. Improving Power and Accuracy of Genome-Wide Association Studies via a Multi-Locus Mixed Linear Model Methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef] [PubMed]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An Efficient Multi-Locus Mixed-Model Approach for Genome-Wide Association Studies in Structured Populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Lang, B.; Armaos, A.; Tartaglia, G.G. RNAct: Protein–RNA Interaction Predictions for Model Organisms with Supporting Experimental Data. Nucleic Acids Res. 2019, 47, D601–D606. [Google Scholar] [CrossRef]
- Van Den Berg, S.; Vandenplas, J.; Van Eeuwijk, F.A.; Lopes, M.S.; Veerkamp, R.F. Significance Testing and Genomic Inflation Factor Using High-Density Genotypes or Whole-genome Sequence Data. J. Anim. Breed. Genet. 2019, 136, 418–429. [Google Scholar] [CrossRef]
- Reyes, A.; Huber, W. Alternative Start and Termination Sites of Transcription Drive Most Transcript Isoform Differences Across Human Tissues. Nucleic Acids Res. 2018, 46, 582–592. [Google Scholar] [CrossRef]
- Wang, S.; Qian, F.; Zheng, Y.; Ogundiran, T.; Ojengbede, O.; Zheng, W.; Blot, W.; Nathanson, K.L.; Hennis, A.; Nemesure, B.; et al. Genetic Variants Demonstrating Flip-Flop Phenomenon and Breast Cancer Risk Prediction among Women of African Ancestry. Breast Cancer Res. Treat. 2018, 168, 703–712. [Google Scholar] [CrossRef]
- Yuzhalin, A.E.; Kutikhin, A.G. Common Genetic Variants in the Myeloperoxidase and Paraoxonase Genes and the Related Cancer Risk: A Review. J. Environ. Sci. Health Part C 2012, 30, 287–322. [Google Scholar] [CrossRef]
- Tang, W.; Chen, Y.; Wang, Y.; Gu, H.; Chen, S.; Kang, M. Peroxisome Proliferator-Activated Receptor Gamma (PPARG) Polymorphisms and Breast Cancer Susceptibility: A Meta-Analysis. Int. J. Clin. Exp. Med. 2015, 8, 12226–12238. [Google Scholar]
- Stevens, V.L.; Rodriguez, C.; Pavluck, A.L.; Thun, M.J.; Calle, E.E. Association of Polymorphisms in the Paraoxonase 1 Gene with Breast Cancer Incidence in the CPS-II Nutrition Cohort. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1226–1228. [Google Scholar] [CrossRef] [PubMed]
- Connor, A.E.; Baumgartner, R.N.; Baumgartner, K.B.; Kerber, R.A.; Pinkston, C.; John, E.M.; Torres-Mejia, G.; Hines, L.; Giuliano, A.; Wolff, R.K.; et al. Associations between TCF7L2 Polymorphisms and Risk of Breast Cancer among Hispanic and Non-Hispanic White Women: The Breast Cancer Health Disparities Study. Breast Cancer Res. Treat. 2012, 136, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Lan, N.; Lu, Y.; Zhang, Y.; Pu, S.; Xi, H.; Nie, X.; Liu, J.; Yuan, W. FTO—A Common Genetic Basis for Obesity and Cancer. Front. Genet. 2020, 11, 559138. [Google Scholar] [CrossRef] [PubMed]
- Hancock, A.M.; Witonsky, D.B.; Gordon, A.S.; Eshel, G.; Pritchard, J.K.; Coop, G.; Di Rienzo, A. Adaptations to Climate in Candidate Genes for Common Metabolic Disorders. PLoS Genet. 2008, 4, e32. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Villaseñor-Navarro, Y.; Mohar-Betancourt, A.; Ocejo-Martínez, A.; Aguilar-Cortázar, L.O.; Pérez-Badillo, M.P.; Pérez-Sénchez, V.M.; Cruz-Morales, R.A.; Pérez-Zuñiga, I.; Pavón-Hernández, C.M. Detección de Cáncer de Mama. Un Compromiso Con México (Reporte Preliminar). Gac. Mex. Oncol. 2012, 11, 220–227. [Google Scholar]
- Endogenous Hormones Breast Cancer Collaborative Group. Body Mass Index, Serum Sex Hormones, and Breast Cancer Risk in Postmenopausal Women. JNCI J. Natl. Cancer Inst. 2003, 95, 1218–1226. [Google Scholar] [CrossRef]
- Kamińska, M.; Ciszewski, T.; Łopacka-Szatan, K.; Miotła, P.; Starosławska, E. Breast Cancer Risk Factors. Przegląd Menopauzalny 2015, 3, 196–202. [Google Scholar] [CrossRef]
- Shveid Gerson, D.; Gerson-Cwilich, R.; Lara Torres, C.O.; Chousleb De Kalach, A.; Ventura Gallegos, J.L.; Badillo-Garcia, L.E.; Bargalló Rocha, J.E.; Maffuz-Aziz, A.; Sánchez Forgach, E.R.; Castorena Roji, G.; et al. Establishment of Triple-Negative Breast Cancer Cells Based on BMI: A Novel Model in the Correlation between Obesity and Breast Cancer. Front. Oncol. 2022, 12, 988968. [Google Scholar] [CrossRef]
- De Almeida, L.M.; Cortés, S.; Vilensky, M.; Valenzuela, O.; Cortes-Sanabria, L.; De Souza, M.; Barbeito, R.A.; Abdelhay, E.; Artagaveytia, N.; Daneri-Navarro, A.; et al. Socioeconomic, Clinical, and Molecular Features of Breast Cancer Influence Overall Survival of Latin American Women. Front. Oncol. 2022, 12, 845527. [Google Scholar] [CrossRef]
- Gallegos-Arreola, M.P.; Briseño-Zuno, C.J.; Figuera, L.E.; Sánchez-López, J.Y.; Zúñiga-González, G.M.; Puebla-Pérez, A.M.; Gómez-Meda, B.C.; Montoya-Fuentes, H.; Delgado-Saucedo, J.I. The Rs1008562, Rs2234671, and Rs3138060 Polymorphisms of the CXCR1 Gene Are Associated with Breast Cancer Risk in a Mexican Population. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9990–10002. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Rubio, S.A.; Quintero-Ramos, A.; Durán-Cárdenas, A.; Franco-Topete, R.A.; Castro-Cervantes, J.M.; Oceguera-Villanueva, A.; Jiménez-Pérez, L.M.; Balderas-Peña, L.M.A.; Morgan-Villela, G.; Del-Toro-Arreola, A.; et al. 1236 C/T and 3435 C/T Polymorphisms of the ABCB1 Gene in Mexican Breast Cancer Patients. Genet. Mol. Res. 2015, 14, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, G.; Aranda-Moreno, C.; Olivares-Corichi, I.M.; Garcia-Sanchez, J.R. The Association of Subtypes of Breast Cancer with Tumour Characteristics and Reproductive Factors in 1326 Mexican Women. Współczesna Onkol. 2015, 6, 462–466. [Google Scholar] [CrossRef]
- Park, S. Interaction of Polygenic Variants Specific for Abdominal Obesity Risk with Energy Metabolism in Large Korean Cohorts. Nutr. Bull. 2022, 47, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Azim, H.A.; Partridge, A.H. Biology of Breast Cancer in Young Women. Breast Cancer Res. 2014, 16, 427. [Google Scholar] [CrossRef]
- Moreno-Estrada, A.; Gignoux, C.R.; Fernandez-Lopez, J.C.; Zakharia, F.; Sikora, M.; Contreras, A.V.; Acuna-Alonzo, V.; Sandoval, K.; Eng, C.; Romero-Hidalgo, S.; et al. The Genetics of Mexico Recapitulates Native American Substructure and Affects Biomedical Traits. Science 2014, 344, 1280–1285. [Google Scholar] [CrossRef]
- Adedokun, B.; Du, Z.; Gao, G.; Ahearn, T.U.; Lunetta, K.L.; Zirpoli, G.; Figueroa, J.; John, E.M.; Bernstein, L.; Zheng, W.; et al. Cross-Ancestry GWAS Meta-Analysis Identifies Six Breast Cancer Loci in African and European Ancestry Women. Nat. Commun. 2021, 12, 4198. [Google Scholar] [CrossRef]
- Lindström, S.; Wang, L.; Feng, H.; Majumdar, A.; Huo, S.; Macdonald, J.; Harrison, T.; Turman, C.; Chen, H.; Mancuso, N.; et al. Genome-Wide Analyses Characterize Shared Heritability among Cancers and Identify Novel Cancer Susceptibility Regions. JNCI J. Natl. Cancer Inst. 2023, 115, 712–732. [Google Scholar] [CrossRef]
- Middha, P.; Wang, X.; Behrens, S.; Bolla, M.K.; Wang, Q.; Dennis, J.; Michailidou, K.; Ahearn, T.U.; Andrulis, I.L.; Anton-Culver, H.; et al. A Genome-Wide Gene-Environment Interaction Study of Breast Cancer Risk for Women of European Ancestry. Breast Cancer Res. 2023, 25, 93. [Google Scholar] [CrossRef]
- Lange, M.; Begolli, R.; Giakountis, A. Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine. ncRNA 2021, 7, 47. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for Gene List Enrichment Analysis and Candidate Gene Prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.; Stemmle, L.N.; Madden, J.F.; Fields, T.A.; Daaka, Y.; Casey, P.J. A Role for the G12 Family of Heterotrimeric G Proteins in Prostate Cancer Invasion. J. Biol. Chem. 2006, 281, 26483–26490. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.; Moeller, B.J.; Juneja, J.; Booden, M.A.; Der, C.J.; Daaka, Y.; Dewhirst, M.W.; Fields, T.A.; Casey, P.J. The G12 Family of Heterotrimeric G Proteins Promotes Breast Cancer Invasion and Metastasis. Proc. Natl. Acad. Sci. USA 2006, 103, 8173–8178. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Cui, J.; Wang, W.; Deng, K. Gα12/13 Signaling Promotes Cervical Cancer Invasion through the RhoA/ROCK-JNK Signaling Axis. Biochem. Biophys. Res. Commun. 2016, 473, 1240–1246. [Google Scholar] [CrossRef]
- Rasheed, S.A.K.; Teo, C.R.; Beillard, E.J.; Voorhoeve, P.M.; Zhou, W.; Ghosh, S.; Casey, P.J. MicroRNA-31 Controls G Protein Alpha-13 (GNA13) Expression and Cell Invasion in Breast Cancer Cells. Mol. Cancer 2015, 14, 67. [Google Scholar] [CrossRef]
- Teo, C.R.; Casey, P.J.; Rasheed, S.A.K. The GNA13-RhoA Signaling Axis Suppresses Expression of Tumor Protective Kallikreins. Cell. Signal. 2016, 28, 1479–1488. [Google Scholar] [CrossRef]
- Armendariz, D.A.; Sundarrajan, A.; Hon, G.C. Breaking Enhancers to Gain Insights into Developmental Defects. eLife 2023, 12, e88187. [Google Scholar] [CrossRef]
- Parker, S.C.J.; Stitzel, M.L.; Taylor, D.L.; Orozco, J.M.; Erdos, M.R.; Akiyama, J.A.; Van Bueren, K.L.; Chines, P.S.; Narisu, N.; NISC Comparative Sequencing Program; et al. Chromatin Stretch Enhancer States Drive Cell-Specific Gene Regulation and Harbor Human Disease Risk Variants. Proc. Natl. Acad. Sci. USA 2013, 110, 17921–17926. [Google Scholar] [CrossRef]
- Rosas-Cruz, A.; Salinas-Jazmín, N.; Valdés-Rives, A.; Velasco-Velázquez, M.A. DRD1 and DRD4 Are Differentially Expressed in Breast Tumors and Breast Cancer Stem Cells: Pharmacological Implications. Transl. Cancer Res. TCR 2022, 11, 3941–3950. [Google Scholar] [CrossRef]
- Grant, C.E.; Flis, A.L.; Ryan, B.M. Understanding the Role of Dopamine in Cancer: Past, Present and Future. Carcinogenesis 2022, 43, 517–527. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Z.-B.; Luo, C.; Mao, X.-Y.; Li, X.; Yin, J.-Y.; Zhang, W.; Zhou, H.-H.; Liu, Z.-Q. The Prospective Value of Dopamine Receptors on Bio-Behavior of Tumor. J. Cancer 2019, 10, 1622–1632. [Google Scholar] [CrossRef] [PubMed]
- Bañales-Luna, M.; Figueroa-Vega, N.; Marín-Aragón, C.I.; Perez-Luque, E.; Ibarra-Reynoso, L.; Gallardo-Blanco, H.L.; López-Aguilar, I.; Malacara, J.M. Associations of Nicotidamide-N-Methyltransferase, FTO, and IRX3 Genetic Variants with Body Mass Index and Resting Energy Expenditure in Mexican Subjects. Sci. Rep. 2020, 10, 11478. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, R.; Zhang, L.; Li, J.; Lou, K.; Shi, B. The Lipid Metabolism Gene FTO Influences Breast Cancer Cell Energy Metabolism via the PI3K/AKT Signaling Pathway. Oncol. Lett. 2017, 13, 4685–4690. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, S.; Hasanpour Ardekanizadeh, N.; Poorhosseini, S.M.; Gholamalizadeh, M.; Roumi, Z.; Goodarzi, M.O.; Doaei, S. Unraveling the Complex Interactions between the Fat Mass and Obesity-Associated (FTO) Gene, Lifestyle, and Cancer. Adv. Nutr. 2022, 13, 2406–2419. [Google Scholar] [CrossRef]
- Delahanty, R.J.; Beeghly-Fadiel, A.; Xiang, Y.-B.; Long, J.; Cai, Q.; Wen, W.; Xu, W.-H.; Cai, H.; He, J.; Gao, Y.-T.; et al. Association of Obesity-Related Genetic Variants with Endometrial Cancer Risk: A Report from the Shanghai Endometrial Cancer Genetics Study. Am. J. Epidemiol. 2011, 174, 1115–1126. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, J.; Yang, M.; Li, M.; Zheng, J. Association between FTO Gene Polymorphism (Rs9939609 T/A) and Cancer Risk: A Meta-Analysis. Eur. J. Cancer Care 2017, 26, e12464. [Google Scholar] [CrossRef]
- Kaklamani, V.; Yi, N.; Sadim, M.; Siziopikou, K.; Zhang, K.; Xu, Y.; Tofilon, S.; Agarwal, S.; Pasche, B.; Mantzoros, C. The Role of the Fat Mass and Obesity Associated Gene (FTO) in Breast Cancer Risk. BMC Med. Genet. 2011, 12, 52. [Google Scholar] [CrossRef]
- Lin, Y.; Ueda, J.; Yagyu, K.; Ishii, H.; Ueno, M.; Egawa, N.; Nakao, H.; Mori, M.; Matsuo, K.; Kikuchi, S. Association between Variations in the Fat Mass and Obesity-Associated Gene and Pancreatic Cancer Risk: A Case–Control Study in Japan. BMC Cancer 2013, 13, 337. [Google Scholar] [CrossRef]
- Zeng, X.; Ban, Z.; Cao, J.; Zhang, W.; Chu, T.; Lei, D.; Du, Y. Association of FTO Mutations with Risk and Survival of Breast Cancer in a Chinese Population. Dis. Markers 2015, 2015, 101032. [Google Scholar] [CrossRef]
- Long, J.; Zhang, B.; Signorello, L.B.; Cai, Q.; Deming-Halverson, S.; Shrubsole, M.J.; Sanderson, M.; Dennis, J.; Michailiou, K.; Easton, D.F.; et al. Evaluating Genome-Wide Association Study-Identified Breast Cancer Risk Variants in African-American Women. PLoS ONE 2013, 8, e58350. [Google Scholar] [CrossRef]
- Pan, J.; Huang, T.; Deng, Z.; Zou, C. Roles and Therapeutic Implications of m6A Modification in Cancer Immunotherapy. Front. Immunol. 2023, 14, 1132601. [Google Scholar] [CrossRef] [PubMed]
- Allen, I.C.; Hartney, J.M.; Coffman, T.M.; Penn, R.B.; Wess, J.; Koller, B.H. Thromboxane A 2 Induces Airway Constriction through an M 3 Muscarinic Acetylcholine Receptor-Dependent Mechanism. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2006, 290, L526–L533. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Maquat, L.E. Nonsense-Mediated mRNA Decay in Humans at a Glance. J. Cell Sci. 2016, 129, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Haghvirdizadeh, P.; Mohamed, Z.; Abdullah, N.A.; Haghvirdizadeh, P.; Haerian, M.S.; Haerian, B.S. KCNJ11: Genetic Polymorphisms and Risk of Diabetes Mellitus. J. Diabetes Res. 2015, 2015, 908152. [Google Scholar] [CrossRef]
- Shuhua, W.; Chenbo, S.; Yangyang, L.; Xiangqian, G.; Shuang, H.; Tangyue, L.; Dong, T. Autophagy-Related Genes Raptor, Rictor, and Beclin1 Expression and Relationship with Multidrug Resistance in Colorectal Carcinoma. Hum. Pathol. 2015, 46, 1752–1759. [Google Scholar] [CrossRef]
- Cheng, I.; Caberto, C.P.; Lum-Jones, A.; Seifried, A.; Wilkens, L.R.; Schumacher, F.R.; Monroe, K.R.; Lim, U.; Tiirikainen, M.; Kolonel, L.N.; et al. Type 2 Diabetes Risk Variants and Colorectal Cancer Risk: The Multiethnic Cohort and PAGE Studies. Gut 2011, 60, 1703–1711. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, S.; Wong, M.; Wareham, N.J.; Sha, Q. An Ensemble Learning Approach Jointly Modeling Main and Interaction Effects in Genetic Association Studies. Genet. Epidemiol. 2008, 32, 285–300. [Google Scholar] [CrossRef]
- Zhao, L.; Lian, T.; Li, J.; Wei, S.; Li, H.; Li, C.; Wang, H. NCR3LG1 (B7-H6) Is a Potential Prognostic Factor for Bladder Cancer Patients. Biomarkers 2021, 26, 260–267. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, L.; Meng, Y.; Qian, X.; Fan, Y.; Zhang, Q.; Wang, C.; Lin, F.; Chen, B.; Xu, L.; et al. Sulfonylurea Receptor 1-Expressing Cancer Cells Induce Cancer-Associated Fibroblasts to Promote Non-Small Cell Lung Cancer Progression. Cancer Lett. 2022, 536, 215611. [Google Scholar] [CrossRef]
- Davies, K.J. The Complex Interaction of Matrix Metalloproteinases in the Migration of Cancer Cells through Breast Tissue Stroma. Int. J. Breast Cancer 2014, 2014, 839094. [Google Scholar] [CrossRef] [PubMed]
- Radisky, E.S. Matrix Metalloproteinases as Drivers and Therapeutic Targets in Breast Cancer. Front. Biosci. 2015, 20, 1144–1163. [Google Scholar] [CrossRef] [PubMed]
- Walsh, L.; Roy, D. Candidate Prognostic Markers in Breast Cancer: Focus on Extracellular Proteases and Their Inhibitors. Breast Cancer Targets Ther. 2014, 6, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Juurikka, K.; Butler, S.G.; Salo, T.; Nyberg, P.; Åström, P. The Role of MMP8 in Cancer: A Systematic Review. Int. J. Mol. Sci. 2019, 20, 4506. [Google Scholar] [CrossRef]
- Decock, J.; Long, J.-R.; Laxton, R.C.; Shu, X.-O.; Hodgkinson, C.; Hendrickx, W.; Pearce, E.G.; Gao, Y.-T.; Pereira, A.C.; Paridaens, R.; et al. Association of Matrix Metalloproteinase-8 Gene Variation with Breast Cancer Prognosis. Cancer Res. 2007, 67, 10214–10221. [Google Scholar] [CrossRef]
- Decock, J.; Long, J.; Laxton, R.; Shu, X.; Hodgkinson, C.; Hendrickx, W.; Pearce, E.; Gao, Y.; Pereira, A.; Paridaens, R.; et al. Association of MMP8 Gene Variation with Breast Cancer Prognosis. Breast Cancer Res. 2008, 10, P32. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Melnichenko, A.A.; Orekhov, A.N.; Bobryshev, Y.V. Paraoxonase and Atherosclerosis-Related Cardiovascular Diseases. Biochimie 2017, 132, 19–27. [Google Scholar] [CrossRef]
- Mackness, M.; Mackness, B. Human Paraoxonase-1 (PON1): Gene Structure and Expression, Promiscuous Activities and Multiple Physiological Roles. Gene 2015, 567, 12–21. [Google Scholar] [CrossRef]
- Medina-Díaz, I.M.; Ponce-Ruíz, N.; Rojas-García, A.E.; Zambrano-Zargoza, J.F.; Bernal-Hernández, Y.Y.; González-Arias, C.A.; Barrón-Vivanco, B.S.; Herrera-Moreno, J.F. The Relationship between Cancer and Paraoxonase 1. Antioxidants 2022, 11, 697. [Google Scholar] [CrossRef]
- Kim, D.S.; Burt, A.A.; Ranchalis, J.E.; Richter, R.J.; Marshall, J.K.; Eintracht, J.F.; Rosenthal, E.A.; Furlong, C.E.; Jarvik, G.P. Additional Common Polymorphisms in the PON Gene Cluster Predict PON1 Activity but Not Vascular Disease. J. Lipids 2012, 2012, 476316. [Google Scholar] [CrossRef]
- Evans, R.M.; Barish, G.D.; Wang, Y.-X. PPARs and the Complex Journey to Obesity. Nat. Med. 2004, 10, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.W.; Park, C.-Y.; Lee, E.S.; Yoon, Y.S.; Lee, E.S.; Park, S.S.; Kim, Y.; Sung, N.J.; Yun, Y.H.; Lee, K.S.; et al. Adipokines, Insulin Resistance, Metabolic Syndrome, and Breast Cancer Recurrence: A Cohort Study. Breast Cancer Res. 2011, 13, R34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Jiang, J.; Chen, Z.; Wang, Y.; Tang, W.; Chen, Y.; Liu, L. Relationship of PPARG, PPARGC1A, and PPARGC1B Polymorphisms with Susceptibility to Hepatocellular Carcinoma in an Eastern Chinese Han Population. OncoTargets Ther. 2018, 11, 4651–4660. [Google Scholar] [CrossRef] [PubMed]
- Unal, E.; Aslan, E.I.; Ozturk, T.; Kurnaz Gomleksiz, O.; Kucukhuseyin, O.; Tuzuner, M.B.; Seyhan, M.F.; Ozturk, O.; Yilmaz Aydogan, H. Peroxisome Proliferator-Activated Receptor Gamma Pro12Ala/C161T Genotypes and Risky Haplotype Altering Risk of Breast Cancer: A Turkish Case–Control Study. Biochem. Genet. 2021, 59, 1413–1426. [Google Scholar] [CrossRef]
- Schöckel, L.; Woischke, C.; Surendran, S.A.; Michl, M.; Schiergens, T.; Hölscher, A.; Glass, F.; Kreissl, P.; Klauschen, F.; Günther, M.; et al. PPARG Activation Promotes the Proliferation of Colorectal Cancer Cell Lines and Enhances the Antiproliferative Effect of 5-Fluorouracil. BMC Cancer 2024, 24, 234. [Google Scholar] [CrossRef]
- Janku, F.; Yap, T.A.; Meric-Bernstam, F. Targeting the PI3K Pathway in Cancer: Are We Making Headway? Nat. Rev. Clin. Oncol. 2018, 15, 273–291. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, W.; Wang, Z.; Wu, Z.; Du, B.; Li, L.; Chen, Y.; Yang, X.; Hao, X.; Guo, T. RAPTOR Promotes Colorectal Cancer Proliferation by Inducing mTORC1 and Upregulating Ribosome Assembly Factor URB1. Cancer Med. 2020, 9, 1529–1543. [Google Scholar] [CrossRef]
- Shchegolev, Y.; Sorokin, D.; Scherbakov, A.; Shunaev, A.; Andreeva, O.; Mikhaevich, E.; Gudkova, M.; Bure, I.; Berstein, L.; Nemtsova, M.; et al. Upregulation of Akt/Raptor Signaling Is Associated with Rapamycin Resistance of Breast Cancer Cells. Chem.-Biol. Interact. 2020, 330, 109243. [Google Scholar] [CrossRef]
- Tang, Q.; Holland-Letz, T.; Slynko, A.; Cuk, K.; Marme, F.; Schott, S.; Heil, J.; Qu, B.; Golatta, M.; Bewerunge-Hudler, M.; et al. DNA Methylation Array Analysis Identifies Breast Cancer Associated RPTOR, MGRN1 and RAPSN Hypomethylation in Peripheral Blood DNA. Oncotarget 2016, 7, 64191–64202. [Google Scholar] [CrossRef]
- Luo, H.L.; Chiang, P.H.; Lee, N.L.; Chiang, P.H. RAPTOR Gene Polymorphism Is Independently Correlated with Urothelial Cancer Susceptibility Compared with Environmental Toxin Exposure. Urol. Sci. 2017, 28, 197–199. [Google Scholar] [CrossRef]
- Cheng, T.-Y.D.; Shankar, J.; Zirpoli, G.; Roberts, M.R.; Hong, C.-C.; Bandera, E.V.; Ambrosone, C.B.; Yao, S. Genetic Variants in the mTOR Pathway and Interaction with Body Size and Weight Gain on Breast Cancer Risk in African-American and European American Women. Cancer Causes Control 2016, 27, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qin, J.; Voruganti, S.; Nag, S.; Zhou, J.; Zhang, R. Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications. Med. Res. Rev. 2015, 35, 1220–1267. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Peterson, T.R.; Sabatini, D.M. Regulation of the mTOR Complex 1 Pathway by Nutrients, Growth Factors, and Stress. Mol. Cell 2010, 40, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Bartolome, A.; Kimura-Koyanagi, M.; Asahara, S.-I.; Guillen, C.; Inoue, H.; Teruyama, K.; Shimizu, S.; Kanno, A.; Garcia-Aguilar, A.; Koike, M.; et al. Pancreatic -Cell Failure Mediated by mTORC1 Hyperactivity and Autophagic Impairment. Diabetes 2014, 63, 2996–3008. [Google Scholar] [CrossRef]
- Makhoul, C.; Gleeson, P.A. Regulation of mTORC1 Activity by the Golgi Apparatus. Fac. Rev. 2021, 10, 50. [Google Scholar] [CrossRef]
- Yin, Q.; Ni, Q.; Wang, Y.; Zhang, H.; Li, W.; Nie, A.; Wang, S.; Gu, Y.; Wang, Q.; Ning, G. Raptor Determines β-Cell Identity and Plasticity Independent of Hyperglycemia in Mice. Nat. Commun. 2020, 11, 2538. [Google Scholar] [CrossRef]
- Le Bacquer, O.; Queniat, G.; Gmyr, V.; Kerr-Conte, J.; Lefebvre, B.; Pattou, F. mTORC1 and mTORC2 Regulate Insulin Secretion through Akt in INS-1 Cells. J. Endocrinol. 2013, 216, 21–29. [Google Scholar] [CrossRef]
- Liu, W.; Xie, L.; He, Y.-H.; Wu, Z.-Y.; Liu, L.-X.; Bai, X.-F.; Deng, D.-X.; Xu, X.-E.; Liao, L.-D.; Lin, W.; et al. Large-Scale and High-Resolution Mass Spectrometry-Based Proteomics Profiling Defines Molecular Subtypes of Esophageal Cancer for Therapeutic Targeting. Nat. Commun. 2021, 12, 4961. [Google Scholar] [CrossRef]
- Köttgen, A.; Hwang, S.-J.; Rampersaud, E.; Coresh, J.; North, K.E.; Pankow, J.S.; Meigs, J.B.; Florez, J.C.; Parsa, A.; Levy, D.; et al. TCF7L2 Variants Associate with CKD Progression and Renal Function in Population-Based Cohorts. J. Am. Soc. Nephrol. 2008, 19, 1989–1999. [Google Scholar] [CrossRef]
- Lu, X.P.; Hu, G.N.; Du, J.Q.; Li, H.Q. TCF7L2 Gene Polymorphisms and Susceptibility to Breast Cancer: A Meta-Analysis. Genet. Mol. Res. 2015, 14, 2860–2867. [Google Scholar] [CrossRef]
- Lyssenko, V. The Transcription Factor 7-like 2 Gene and Increased Risk of Type 2 Diabetes: An Update. Curr. Opin. Clin. Nutr. Metab. Care 2008, 11, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, K.Y.; Al Ahmed, H.A.; Shaaban, M.A.A.; Emam, Y.M. Study of The Association between TCF7L2 Gene Polymorphism (Rs12255372) and Breast Cancer. QJM Int. J. Med. 2021, 114, hcab091.015. [Google Scholar] [CrossRef]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Camicia, R.; Winkler, H.C.; Hassa, P.O. Novel Drug Targets for Personalized Precision Medicine in Relapsed/Refractory Diffuse Large B-Cell Lymphoma: A Comprehensive Review. Mol. Cancer 2015, 14, 207. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | Controls (n = 126) | Cases (n = 92) | p-Value 2 |
|---|---|---|---|
| Age (years) 1 | 54.93 ± 7.091 | 58.45 ± 8.811 | 0.023 |
| Height (meters) 1 | 1.578 ± 0.068 | 1.558 ± 0.080 | 0.048 |
| BMI 1 | 28.03 ± 4.950 | 29.92 ± 5.713 | 0.011 |
| Menarche (Age, years) 1 | 12.63 ± 1.543 | 12.88 ± 1.503 | 0.231 |
| Menopause (Age, years) 1 | 47.37 ± 5.630 | 45.29 ± 5.977 | 0.025 |
| Menopause confirmed 3 | 83 (65.87%) | 76 (82.61%) | 0.004 |
| Oral contraceptives 3 | 20 (15.87%) | 23 (25.00%) | 0.110 |
| Children (number) 3 | 111 (88.10%) | 82 (89.13%) | 0.187 |
| Subtype | n | Percentage (%) | Proportion | 95% CI (Proportion) | 95% CI (Percentage) |
|---|---|---|---|---|---|
| TNBC (ER–/PR–/HER2–) | 18 | 19.6% | 0.1957 | [0.1145, 0.2769] | [11.45%, 27.69%] |
| Strict HER2-Enriched (ER–/PR–/HER2+)) 1 | 7 | 7.6% | 0.0761 | [0.0220, 0.1302] | [2.20%, 13.02%] |
| Triple-Positive (ER+/PR+/HER2+) | 8 | 8.7% | 0.0870 | [0.0294, 0.1446] | [2.94%, 14.46%] |
| Luminal (HR+), HER2– | 42 | 45.7% | 0.4565 | [0.3548, 0.5582] | [35.48%, 55.82%] |
| Luminal (HR+), HER2+ (non–triple-positive) | 17 | 18.5% | 0.1848 | [0.1055, 0.2641] | [10.55%, 26.41%] |
| Total | 92 | 100% | 1.0000 | 100% |
| Chr 1 | Gene 2 | Variant | DD Frequency Cases/Controls 3 | Dd Frequency Cases/Controls 3 | dd Frequency Cases/Controls 3 | HWE p (Cases) 4 | HWE p (Controls) 5 |
|---|---|---|---|---|---|---|---|
| 3 | PPARG a | rs3856806 | TT (0.000/0.024) | TC (0.242/0.206) | CC (0.758/0.770) | 0.190 | 0.437 |
| 7 | PON1 a | rs3917542 | TT (0.088/0.064) | TC (0.385/0.376) | CC (0.527/0.560) | 0.657 | 0.977 |
| 10 | TCF7L2 d | rs3750804 | TT (0.033/0.074) | TC (0.300/0.262) | CC (0.667/0.664) | 0.986 | 0.031 |
| rs3750805 | TT (0.011/0.016) | TA (0.154/0.167) | AA (0.835/0.817) | 0.698 | 0.449 | ||
| 11 | MMP8 b | rs12792229 | TT (0.000/0.000) | TG (0.011/0.000) | GG (0.989/1.000) | 0.958 | 1.000 |
| SCT; DEAF1; DRD4 b | rs1800955 | CC (0.156/0.135) | CT (0.344/0.423) | TT (0.500/0.441) | 0.038 | 0.490 | |
| KCNJ11; ABCC8 a | rs5218 | AA (0.033/0.016) | AG (0.253/0.230) | GG (0.714/0.754) | 0.589 | 0.900 | |
| 16 | FTO a | rs1121980 | AA (0.088/0.119) | AG (0.418/0.381) | GG (0.495/0.500) | 0.996 | 0.222 |
| rs3751812 | TT (0.044/0.040) | TG (0.352/0.360) | GG (0.604/0.600) | 0.809 | 0.584 | ||
| 17 | AMZ2P1; GNA13 c | rs11652805 | CC (0.044/0.048) | CT (0.319/0.298) | TT (0.637/0.653) | 0.877 | 0.511 |
| RPTOR a | rs12946618 | AA (0.012/0.008) | AG (0.256/0.182) | GG (0.733/0.810) | 0.545 | 0.847 | |
| 21 | SCAF4 d | rs2833483 | CC (0.144/0.158) | CT (0.422/0.433) | TT (0.433/0.408) | 0.456 | 0.408 |
| Chr 1 | Gene 2 | Marker | Genetic Model 3 | p-Value 4 | FDR 5 | Regression Beta | Beta Standard Error | Prop. Var. Expl. 6 |
|---|---|---|---|---|---|---|---|---|
| 3 | PPARG a | rs3856806 | 3 | 9.39 × 10−4 | 2.84 × 10−2 | −1.014 | 0.302 | 7.59 × 10−3 |
| 7 | PON1 a | rs3917542 | 3 | 9.17 × 10−4 | 3.70 × 10−2 | 0.660 | 0.196 | 1.23 × 10−3 |
| 10 | TCF7L2 d | rs3750804 | 1 | 1.4 × 10−5 | 5.6 × 10−4 | 0.704 | 0.158 | 5.1 × 10−3 |
| rs3750805 | 1 | 1.8 × 10−5 | 5.3 × 10−4 | 0.885 | 0.201 | 5.1 × 10−3 | ||
| 11 | MMP8 b | rs12792229 | 2 | 8.56 × 10−9 | 1.04 × 10−6 | 2.002 | 0.333 | 6.81 × 10−3 |
| SCT; DEAF1; DRD4 b | rs1800955 | 1 | 4.6 × 10−4 | 9.2 × 10−3 | 0.395 | 0.111 | 5.1 × 10−3 | |
| KCNJ11; ABCC8; NCR3LG1 a | rs5218 | 3 | 1.78 × 10−11 | 2.16 × 10−9 | 1.859 | 0.261 | 1.58 × 10−3 | |
| 16 | FTO a | rs1121980 | 1 | 6.7 × 10−8 | 8.1 × 10−6 | 1.191 | 0.212 | 1.6 × 10−2 |
| rs3751812 | 1 | 1.8 × 10−6 | 1.1 × 10−4 | 1.109 | 0.225 | 1.6 × 10−2 | ||
| 17 | AMZ2P1; GNA13 c | rs11652805 | 1 | 6.4 × 10−5 | 1.6 × 10−3 | 0.650 | 0.159 | 1.6 × 10−2 |
| RPTOR a | rs12946618 | 2 | 2.75 × 10−7 | 1.66 × 10−5 | −0.898 | 0.169 | 7.97 × 10−3 | |
| 21 | SCAF4 a | rs2833483 | 2 | 1.18 × 10−6 | 4.77 × 10−5 | 0.826 | 0.165 | 7.97 × 10−3 |
| Chr 1 | Genes | Marker | HGVS Nomenclature | Consequence Details 2 |
|---|---|---|---|---|
| 3 | PPARG a | rs3856806 | NM_015869.4(PPARG):c.1431C>T (p.His477=) NM_138712.3:c.1347C>T | Synonymous variant; 3 prime UTR variant; NMDTV |
| 7 | PON1 a | rs3917542 | NC_000007.14:g.95307380C>T NM_000446.5:c.698+631G>A | RRV; NMDTV |
| 10 | TCF7L2 d | rs3750804 | NC_000010.11:g.113074091C>T | PFR, RRV |
| rs3750805 | NC_000010.10:g.114847143A>T | PFR; RRV | ||
| 11 | SCT; DEAF1; DRD4 b | rs1800955 | NC_000011.10:g.636784T>C | RRV; PFR |
| MMP8 b | rs12792229 | NC_000011.10:g.102718512G>T XM_005271556.1:c.617C>A XP_011541137.1:p.Ser206Tyr | Missense variant; SIFT: deleterious; PolyPhen: possibly damaging; NMDTV; Downstream gene variant | |
| KCNJ11; ABCC8; NCR3LG1 a | rs5218 | NC_000011.10:g.17387522G>A NP_001159762.1:p.Ala103= | Synonymous variant, Upstream gene variant Downstream gene variant, regulatory region variant; a CTCF binding site variant | |
| 16 | FTO a | rs1121980 | NC_000016.10:g.53775335G>A NM_001080432.2:c.46-34805G>A | RRV; NMDTV |
| rs3751812 | NC_000016.10:g.53784548G>T NM_001080432.2:c.46-25592G>T | RRV | ||
| 17 | AMZ2P1; GNA13 c | rs11652805 | NC_000017.11:g.64991033C>T | RRV |
| RPTOR a | rs12946618 | NC_000017.11:g.80603368G>A NM_001163034.1:c.163-22323G>A | NMDTV; upstream gene variant | |
| 21 | SCAF4 a | rs2833483 | NC_000021.9:g.31703091T>C NM_001145444.1:c.276+674A>G | Upstream gene variant |
| Marker | Chr 1 | Genes | HGVS Nomenclature | Consequence Details 2 |
|---|---|---|---|---|
| rs3856806 | 3 | PPARG a | NM_015869.4(PPARG):c.1431C>T (p.His477=) NM_138712.3:c.1347C>T | Synonymous variant 3 prime UTR variant, NMDTV |
| rs12792229 | 11 | MMP8 b | NC_000011.10:g.102718512G>T XM_005271556.1:c.617C>A XP_011541137.1:p.Ser206Tyr | Missense variant. SIFT: deleterious PolyPhen: possibly damaging. NMDTV: Downstream gene variant |
| rs5218 | 11 | KCNJ11; ABCC8; NCR3LG1 a | NC_000011.10:g.17387522G>A NP_001159762.1:p.Ala103= | Synonymous variant, upstream gene variant Downstream gene variant, regulatory region variant; a CTCF binding site variant |
| SNP | Gene | Reported Association with BC | Population/Notes | Literature Reference |
|---|---|---|---|---|
| rs3856806 | PPARG | Risk factor for BC; results are conflicting | Risk reported in European and Asian populations; flip-flop phenomenon observed in African descent | Flip-flop study [29]; Turkish study [30]; Meta-analysis [31] |
| rs3917542 | PON1 | Associated with BC risk | Potential association in post-menopausal women; identified in current study | Current study |
| rs854555 1 | PON1 | rs854555 was also related to an increased risk of BC in U.S. post-menopausal women | Potential association in post-menopausal women; ethnic variability observed | BC in U.S. [32] |
| rs3750804 | TCF7L2 | Risk factor for BC | Reported in Hispanic and European populations | Connor et al. [33] |
| rs3750805 | TCF7L2 | Risk factor for BC | Similar to rs3750804; implicated in hormone regulation | Connor et al. [33] |
| rs12792229 | MMP8 | Potential risk factor for BC | Limited prior evidence; identified in current study | Reference [28] |
| rs1800955 | DRD4/SCT/DEAF1 | Novel association with BC risk | Limited prior data; further validation required | Current study |
| rs5218 | KCNJ11-ABCC8 | Associated with metabolic disorders; unclear BC association | Reported in diabetes studies; association with BC observed in current study | Current study; see [13] |
| rs1121980 | FTO | Risk factor for obesity and BC | Widely reported in European/Asian populations; potential flip-flop effects | Numerous studies [34]; Flip-flop study [29] |
| rs3751812 | FTO | Risk factor for obesity and BC | Similar to rs1121980 | Numerous studies [34] |
| rs11652805 | AMZ2P1-GNA13 | Novel association with BC risk | Limited prior evidence; identified in current study | Current study |
| rs12946618 | RPTOR | Potential risk factor for BC | Newly identified variant; modulates mTORC1 signaling; Adaptations to Climate in Candidate Genes for Common Metabolic Disorders | Current study; [35] |
| rs2833483 | SCAF4 | Associated with BC risk | Emerging biomarker; limited prior data available | Current study; [35] |
| Gene | Function |
|---|---|
| PPARG | Nuclear receptor that regulates adipogenesis, glucose metabolism, and anti-inflammatory processes. |
| PON1 | Enzyme associated with HDL that protects against oxidative stress and inflammation. |
| TCF7L2 | Transcription factor involved in the Wnt signaling pathway and regulation of glucose metabolism; linked to diabetes and cancer. |
| MMP8 | Matrix metallopeptidase that degrades extracellular matrix components, facilitating tissue remodeling and potentially tumor invasion. |
| SCT | Gene encoding secretin, a hormone involved in regulating pancreatic secretion and water homeostasis; its direct role in BC is less defined. |
| DEAF1 | Transcription factor involved in gene expression regulation and neural development. |
| DRD4 | Dopamine receptor that modulates neuronal signaling and may influence cell proliferation and cancer-related pathways. |
| KCNJ11 | Potassium channel subunit involved in insulin secretion and glucose homeostasis. |
| ABCC8 | Encodes the sulfonylurea receptor, crucial for regulating insulin secretion. |
| NCR3LG1 | Ligand for natural cytotoxicity receptors, influencing immune responses and potentially tumor immunosurveillance. |
| FTO | Enzyme implicated in the regulation of energy balance and adipogenesis; associated with obesity and diabetes. |
| AMZ2P1 | A pseudogene related to AMZ2, possibly involved in regulatory processes via non-coding RNAs. |
| GNA13 | G-protein subunit (alpha 13) that participates in signaling pathways controlling cell migration, invasion, and proliferation. |
| RPTOR | Essential scaffolding protein for mTORC1, regulating cell growth, metabolism, and proliferation. |
| SCAF4 | Protein involved in RNA splicing and processing, potentially impacting gene expression and cancer prognosis. |
| Gene | Variant | ENSEMBL 1 | ENSEMBL 1 | RNAct 2 | RNAct 3 | TOPPGENE 4 | TOPPGENE 5 |
|---|---|---|---|---|---|---|---|
| PON1 | rs3917542 | Yes | Yes | - | - | - | - |
| TCF7L2 | rs3750804 | Yes | Yes | - | - | - | - |
| TCF7L2 | rs3750805 | Yes | Yes | - | - | - | - |
| SCT; DEAF1; DRD4 | rs1800955 | - | - | Yes | Yes | Yes | - |
| KCNJ11; ABCC8 | rs5218 | Yes | Yes | - | - | - | - |
| FTO | rs1121980 | Yes | Yes | - | - | - | - |
| FTO | rs3751812 | Yes | Yes | - | - | - | - |
| AMZ2P1; GNA13 | rs11652805 | Yes | Yes | Yes | Yes | Yes | Yes |
| RPTOR | rs12946618 | Yes | Yes | Yes | Yes | Yes | Yes |
| SCAF4 | rs2833483 | Yes | Yes | Yes | Yes | Yes | Yes |
| MMP8 | rs12792229 | - | - | - | - | - | - |
| PPARG | rs3856806 | - | - | - | - | - | - |
| SNP | Gene/Locus | Key Correlated Genes (Ensembl) | Expression Pattern in Breast Tissue | Functional Implications |
|---|---|---|---|---|
| rs3856806 | PPARG | PPARG, [additional regulatory targets] | Not significantly altered | Regulates adipogenesis, glucose metabolism, and anti-inflammatory processes |
| rs3917542 | PON1 | PON1, [related oxidative stress genes] | Downregulated | Influences oxidative stress protection and inflammation |
| rs3750804 | TCF7L2 | TCF7L2, [glucose metabolism-related genes] | Upregulated | Involved in Wnt signaling and regulation of glucose metabolism |
| rs3750805 | TCF7L2 | TCF7L2, [glucose metabolism-related genes] | Upregulated | Involved in Wnt signaling and regulation of glucose metabolism |
| rs12792229 | MMP8 | MMP8, [extracellular matrix remodeling genes] | Variable | Modulates extracellular matrix degradation and tissue remodeling |
| rs1800955 | SCT/DEAF1/DRD4 | DRD4, DEAF1, [neuronal/proliferative signaling genes] | Variable | May affect neuronal signaling and cell proliferation pathways |
| rs5218 | KCNJ11/ABCC8/NCR3LG1 | KCNJ11, ABCC8, NCR3LG1 | Not significantly altered | Related to insulin secretion and metabolic regulation |
| rs1121980 | FTO | FTO, [energy homeostasis genes] | Upregulated | Impacts energy balance and adipogenesis; associated with obesity |
| rs3751812 | FTO | FTO, [energy homeostasis genes] | Upregulated | Impacts energy balance and adipogenesis; associated with obesity |
| rs11652805 | AMZ2P1-GNA13 | AMZ2P1, GNA13 | Variable | May modulate cell migration, invasion, and proliferative signaling |
| rs12946618 | RPTOR | RPTOR and 33 correlated genes (e.g., genes involved in ubiquitin-protein ligase activity, PRC1 complex) | Downregulated | Modulates mTORC1 signaling and metabolic regulation |
| rs2833483 | SCAF4 | SCAF4, [RNA processing/splicing genes] | Variable | Involved in RNA splicing and regulation of gene expression |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallardo-Blanco, H.L.; Garza-Rodríguez, M.d.L.; Pérez-Ibave, D.C.; Burciaga-Flores, C.H.; Salinas-Torres, V.M.; González-Escamilla, M.; Piñeiro-Retif, R.; Cerda-Flores, R.M.; Vidal-Gutiérrez, O.; Sanchez-Dominguez, C.N. Genetic Insights into Breast Cancer in Northeastern Mexico: Unveiling Gene–Environment Interactions and Their Links to Obesity and Metabolic Diseases. Cancers 2025, 17, 982. https://doi.org/10.3390/cancers17060982
Gallardo-Blanco HL, Garza-Rodríguez MdL, Pérez-Ibave DC, Burciaga-Flores CH, Salinas-Torres VM, González-Escamilla M, Piñeiro-Retif R, Cerda-Flores RM, Vidal-Gutiérrez O, Sanchez-Dominguez CN. Genetic Insights into Breast Cancer in Northeastern Mexico: Unveiling Gene–Environment Interactions and Their Links to Obesity and Metabolic Diseases. Cancers. 2025; 17(6):982. https://doi.org/10.3390/cancers17060982
Chicago/Turabian StyleGallardo-Blanco, Hugo Leonid, María de Lourdes Garza-Rodríguez, Diana Cristina Pérez-Ibave, Carlos Horacio Burciaga-Flores, Víctor Michael Salinas-Torres, Moisés González-Escamilla, Rafael Piñeiro-Retif, Ricardo M. Cerda-Flores, Oscar Vidal-Gutiérrez, and Celia N. Sanchez-Dominguez. 2025. "Genetic Insights into Breast Cancer in Northeastern Mexico: Unveiling Gene–Environment Interactions and Their Links to Obesity and Metabolic Diseases" Cancers 17, no. 6: 982. https://doi.org/10.3390/cancers17060982
APA StyleGallardo-Blanco, H. L., Garza-Rodríguez, M. d. L., Pérez-Ibave, D. C., Burciaga-Flores, C. H., Salinas-Torres, V. M., González-Escamilla, M., Piñeiro-Retif, R., Cerda-Flores, R. M., Vidal-Gutiérrez, O., & Sanchez-Dominguez, C. N. (2025). Genetic Insights into Breast Cancer in Northeastern Mexico: Unveiling Gene–Environment Interactions and Their Links to Obesity and Metabolic Diseases. Cancers, 17(6), 982. https://doi.org/10.3390/cancers17060982










