Machine Learning Insight: Unveiling Overlooked Risk Factors for Postoperative Complications in Gastric Cancer
Simple Summary
Abstract
1. Introduction
2. Material and Methods
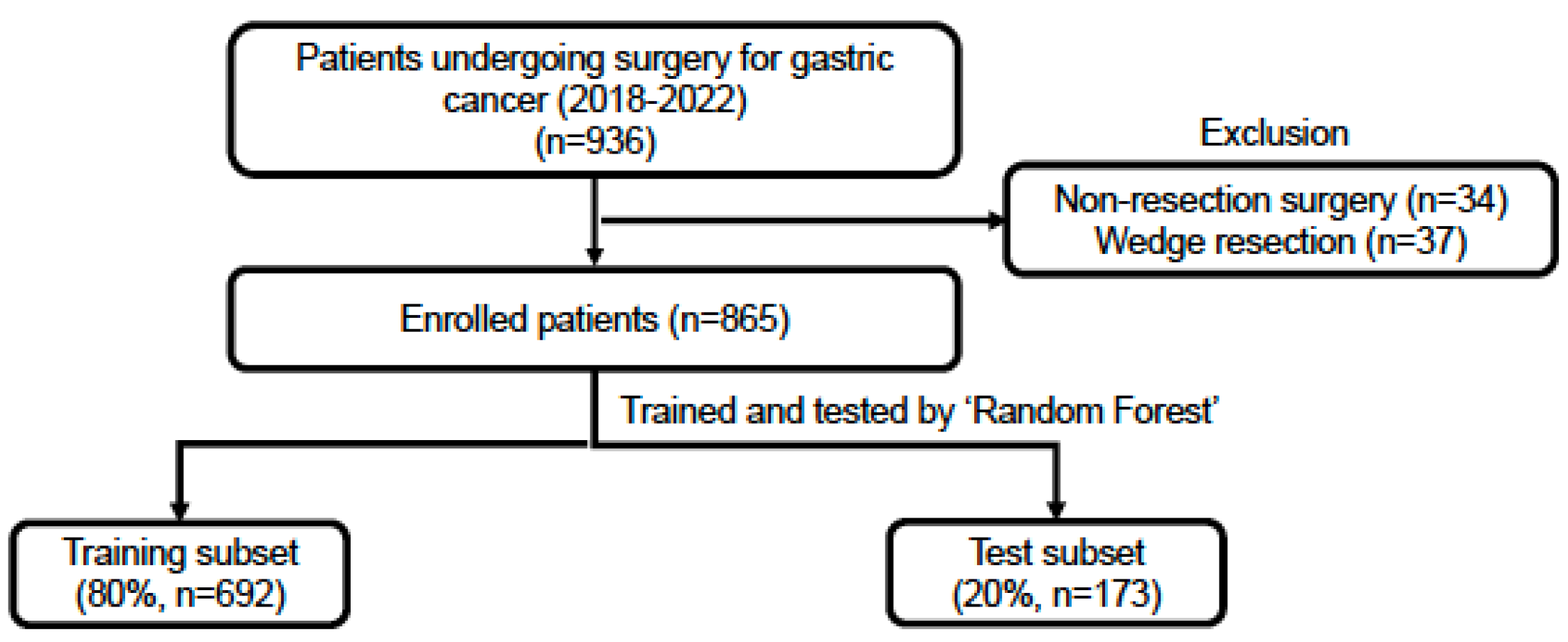
2.1. Patient Selection
2.2. Data Collection and Completeness
- Over 30 missing values—Neutrophil percentage, Lymphocyte percentage, and 21 others, totaling 23 variables
- Between 10 and 30 missing values—Total protein, Carcinoembryonic antigen, and Carbohydrate antigen 19-9, totaling 3 variables
- Less than 10 missing values—Red blood cell distribution, Plateletcrit, Mean platelet volume (MPV), Platelet distribution width (PDW), Alkaline phosphatase, and Phosphorus, totaling 6 variables.
2.3. Machine Learning Methodology
- Complete Case Strategy: All variables with any missing values were omitted, leaving us with data from all 865 patients but only 53 variables.
- Moderate Missing Data Strategy: We removed variables with more than 10 missing values and then excluded patients with any missing data. This process left us with data from 847 patients and 59 variables.
- Liberal Missing Data Strategy: We discarded variables with over 30 missing values and then excluded patients with any missing data. As a result, we retained the most variables, 62 in total, but were left with data from 799 patients.
2.4. Tools and Software
3. Results
3.1. Postoperative Complications
3.2. Optimizing AUC in Model Development
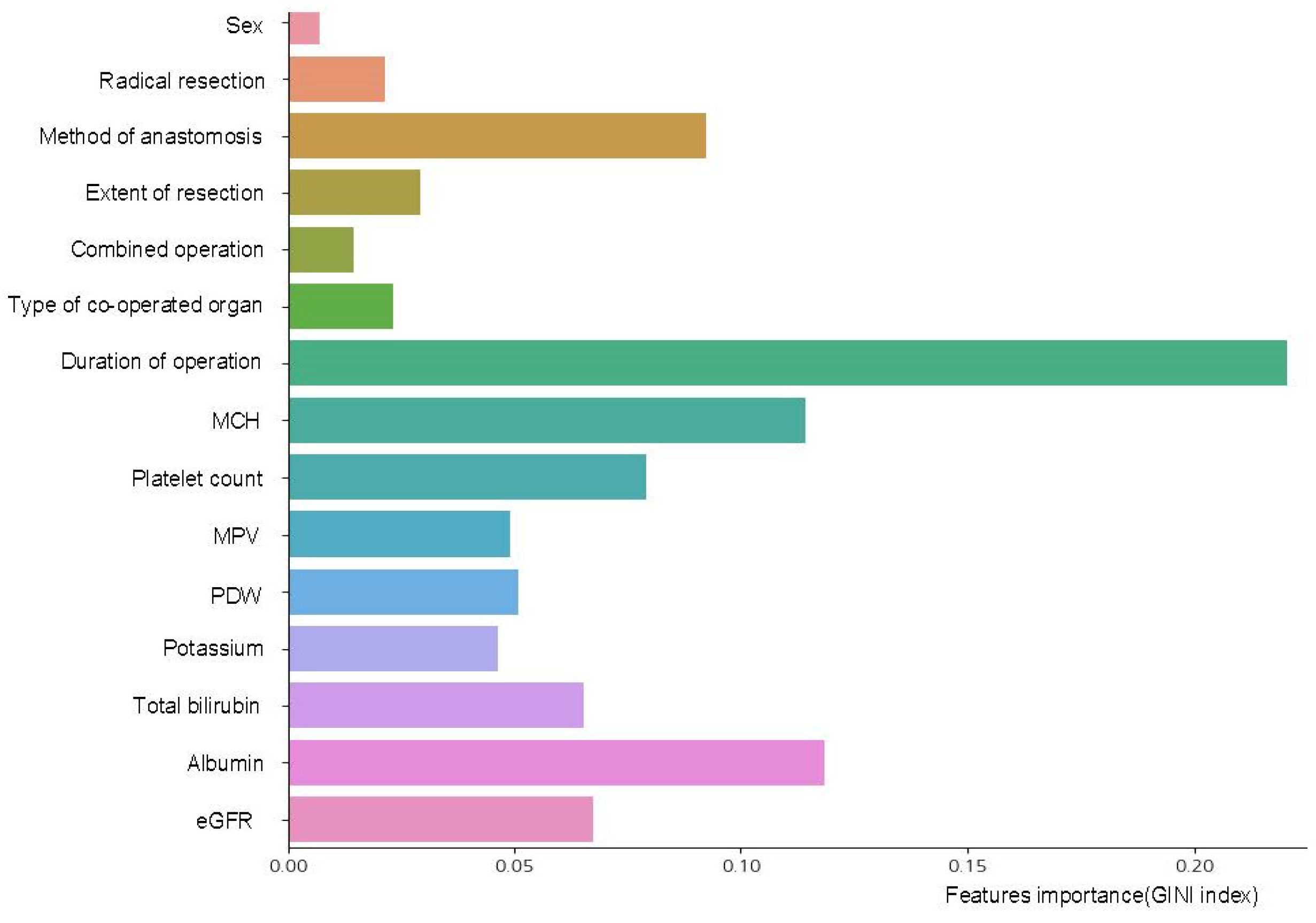
3.3. Interpretable Variables Selection
3.4. Performance Comparison of Machine Learning Models with Conventional Multiple Linear Regression
4. Discussion
- Given that our research is preliminary, the significance of variables derived from our modeling cannot be fully ascertained with confidence.
- As a result, validation through multi-institutional big data, particularly from high-volume tertiary centers [41], is imperative to reaffirm our findings.
- The data are sourced from a single institution, and although comprehensive, they might not be representative of a broader patient population.
- While we employed three strategies to handle missing values in the quest for an optimal predictive model, this approach may have inadvertently excluded variables that could be deemed significant based on clinical experience.
- While we included essential model comparisons and variable selection strategies, more comprehensive ablation analyses—such as comparing the impact of different feature subsets or evaluating confusion matrices—were not fully explored. These analyses are crucial for evaluating model robustness and methodological innovation. To address this, a follow-up study is currently underway. This upcoming research will compare feature sets selected by AI-based backward elimination with those chosen by experienced surgeons using clinical expertise. We expect this future work to provide deeper insight into the clinical applicability and interpretability of machine learning models in surgical outcomes.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, T.D.; Mahar, A.L.; Haas, B.; Beyfuss, K.; Law, C.H.L.; Karanicolas, P.J.; Coburn, N.G.; Hallet, J. The impact of advanced age on short-term outcomes following gastric cancer resection: An acs-nsqip analysis. Gastric Cancer 2018, 21, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Hyder, J.A.; Reznor, G.; Wakeam, E.; Nguyen, L.L.; Lipsitz, S.R.; Havens, J.M. Risk prediction accuracy differs for emergency versus elective cases in the acs-nsqip. Ann. Surg. 2016, 264, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Protopapa, K.L.; Simpson, J.C.; Smith, N.C.; Moonesinghe, S.R. Development and validation of the surgical outcome risk tool (sort). Br. J. Surg. 2014, 101, 1774–1783. [Google Scholar] [CrossRef]
- Copeland, G.P.; Jones, D.; Walters, M. Possum: A scoring system for surgical audit. Br. J. Surg. 1991, 78, 355–360. [Google Scholar] [CrossRef]
- Beal, E.W.; Saunders, N.D.; Kearney, J.F.; Lyon, E.; Wei, L.; Squires, M.H.; Jin, L.X.; Worhunsky, D.J.; Votanopoulos, K.I.; Ejaz, A.; et al. Accuracy of the acs nsqip online risk calculator depends on how you look at it: Results from the united states gastric cancer collaborative. Am. Surg. 2018, 84, 358–364. [Google Scholar] [CrossRef]
- Cengil, E.; Çınar, A.; Yıldırım, M. A hybrid approach for efficient multi-classification of white blood cells based on transfer learning techniques and traditional machine learning methods. Concurrency Comput. Pract. Exp. 2022, 34, e6789. [Google Scholar] [CrossRef]
- Mutlu, H.B.; Durmaz, F.; Yücel, N.; Cengil, E.; Yıldırım, M. Prediction of maternal health risk with traditional machine learning methods. Naturengs 2023, 4, 18–25. [Google Scholar] [CrossRef]
- FitzHenry, F.; Murff, H.J.; Matheny, M.E.; Gentry, N.; Fielstein, E.M.; Brown, S.H.; Reeves, R.M.; Aronsky, D.; Elkin, P.L.; Messina, V.P.; et al. Exploring the frontier of electronic health record surveillance: The case of postoperative complications. Med. Care 2013, 51, 509–516. [Google Scholar] [CrossRef]
- Hofer, I.S.; Lee, C.; Gabel, E.; Baldi, P.; Cannesson, M. Development and validation of a deep neural network model to predict postoperative mortality, acute kidney injury, and reintubation using a single feature set. NPJ Digit. Med. 2020, 3, 58. [Google Scholar] [CrossRef]
- Weller, G.B.; Lovely, J.; Larson, D.W.; Earnshaw, B.A.; Huebner, M. Leveraging electronic health records for predictive modeling of post-surgical complications. Stat. Methods Med. Res. 2018, 27, 3271–3285. [Google Scholar] [CrossRef] [PubMed]
- Fritz, B.A.; Cui, Z.; Zhang, M.; He, Y.; Chen, Y.; Kronzer, A.; Ben Abdallah, A.; King, C.R.; Avidan, M.S. Deep-learning model for predicting 30-day postoperative mortality. Br. J. Anaesth. 2019, 123, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Waljee, A.K.; Higgins, P.D. Machine learning in medicine: A primer for physicians. Am. J. Gastroenterol. 2010, 105, 1224–1226. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Sohil, F.; Sohali, M.U.; Shabbir, J. An introduction to statistical learning with applications in r. Stat. Theory Relat. Fields 2022, 6, 87. [Google Scholar] [CrossRef]
- Goldstein, B.A.; Navar, A.M.; Carter, R.E. Moving beyond regression techniques in cardiovascular risk prediction: Applying machine learning to address analytic challenges. Eur. Heart J. 2017, 38, 1805–1814. [Google Scholar] [CrossRef]
- Chen, J.H.; Asch, S.M. Machine learning and prediction in medicine—Beyond the peak of inflated expectations. N. Engl. J. Med. 2017, 376, 2507–2509. [Google Scholar] [CrossRef]
- Velmahos, C.S.; Paschalidis, A.; Paranjape, C.N. The not-so-distant future or just hype? Utilizing machine learning to predict 30-day post-operative complications in laparoscopic colectomy patients. Am. Surg. 2023, 89, 5648–5654. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the performance of prediction models a framework for traditional and novel measures. Epidemiology 2010, 21, 128–138. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Emanuel, E.J. Predicting the future—Big data, machine learning, and clinical medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Rendon, E.; Alejo, R.; Castorena, C.; Isidro-Ortega, F.J.; Granda-Gutierrez, E.E. Data sampling methods to deal with the big data multi-class imbalance problem. Appl. Sci. 2020, 10, 1276. [Google Scholar] [CrossRef]
- Jung, J.O.; Pisula, J.I.; Bozek, K.; Popp, F.; Biebl, M.; Raab, H.R.; Schiffmann, L. Prediction of postoperative complications after oesophagectomy using machine-learning methods. Br. J. Surg. 2023, 110, 1362–1370. [Google Scholar]
- Zhou, C.M.; Wang, Y.; Yang, J.J.; Zhu, Y. Predicting postoperative gastric cancer prognosis based on inflammatory factors and machine learning technology. BMC Med. Inform. Decis. Mak. 2023, 23, 144. [Google Scholar]
- Guideline Committee of the Korean Gastric Cancer Association (KGCA), Development Working Group & Review Panel. Korean practice guideline for gastric cancer 2018: An evidence-based, multi-disciplinary approach. J. Gastric Cancer 2019, 19, 1–48. [Google Scholar]
- Mathew, G.; Agha, R.; Albrecht, J.; Goel, P.; Mukherjee, I.; Pai, P.; D’Cruz, A.K.; Nixon, I.J.; Roberto, K.; Enam, S.A.; et al. Strocss 2021: Strengthening the reporting of cohort, cross-sectional and case-control studies in surgery. Int. J. Surg. 2021, 96, 106165. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition ajcc cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar]
- Clavien, P.A.; Sanabria, J.R.; Strasberg, S.M. Proposed classification of complications of surgery with examples of utility in cholecystectomy. Surgery 1992, 111, 518–526. [Google Scholar]
- Bihorac, A.; Ozrazgat-Baslanti, T.; Ebadi, A.; Motaei, A.; Madkour, M.; Pardalos, P.M.; Lipori, G.; Hogan, W.R.; Efron, P.A.; Moore, F.; et al. Mysurgeryrisk: Development and validation of a machine-learning risk algorithm for major complications and death after surgery. Ann. Surg. 2019, 269, 652–662. [Google Scholar]
- Horvath, B.; Kloesel, B.; Todd, M.M.; Cole, D.J.; Prielipp, R.C. The evolution, current value, and future of the american society of anesthesiologists physical status classification system. Anesthesiology 2021, 135, 904–919. [Google Scholar] [CrossRef]
- Deo, R.C. Machine learning in medicine. Circulation 2015, 132, 1920–1930. [Google Scholar]
- Klinger, M.H.; Jelkmann, W. Role of blood platelets in infection and inflammation. J. Interferon Cytokine Res. 2002, 22, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Fowler, A.J.; Agha, R.A. Neutrophil/lymphocyte ratio is related to the severity of coronary artery disease and clinical outcome in patients undergoing angiography—The growing versatility of nlr. Atherosclerosis 2013, 228, 44–45. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, G.J.; Charles, K.A.; Roxburgh, C.S.; Horgan, P.G.; McMillan, D.C.; Clarke, S.J. The systemic inflammation-based neutrophil-lymphocyte ratio: Experience in patients with cancer. Crit. Rev. Oncol. Hematol. 2013, 88, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Gunaldi, M.; Erdem, D.; Goksu, S.; Gunduz, S.; Okuturlar, Y.; Tiken, E.; Aksoy, H.; Yildirim, M. Platelet distribution width as a predictor of metastasis in gastric cancer patients. J. Gastrointest. Cancer 2017, 48, 341–346. [Google Scholar] [CrossRef]
- Korniluk, A.; Koper-Lenkiewicz, O.M.; Kaminska, J.; Kemona, H.; Dymicka-Piekarska, V. Mean platelet volume (mpv): New perspectives for an old marker in the course and prognosis of inflammatory conditions. Mediat. Inflamm. 2019, 2019, 9213074. [Google Scholar] [CrossRef]
- Gasparyan, A.Y.; Ayvazyan, L.; Mikhailidis, D.P.; Kitas, G.D. Mean platelet volume: A link between thrombosis and inflammation? Curr. Pharm. Des. 2011, 17, 47–58. [Google Scholar] [CrossRef]
- Chae, D.; Kim, N.Y.; Kim, H.J.; Kim, T.L.; Kang, S.J.; Kim, S.Y. A risk scoring system integrating postoperative factors for predicting early mortality after major non-cardiac surgery. Clin. Transl. Sci. 2022, 15, 2230–2240. [Google Scholar] [CrossRef]
- Dai, L.; Mick, S.L.; McCrae, K.R.; Houghtaling, P.L.; Blackstone, E.H.; Koch, C.G. Prognostic value of preoperative red cell distribution width: Fine-tuning by mean corpuscular volume. Ann. Thorac. Surg. 2019, 108, 1830–1838. [Google Scholar] [CrossRef]
- Patel, K.V.; Semba, R.D.; Ferrucci, L.; Newman, A.B.; Fried, L.P.; Wallace, R.B.; Bandinelli, S.; Phillips, C.S.; Yu, B.; Connelly, S.; et al. Red cell distribution width and mortality in older adults: A meta-analysis. J. Gerontol. A Biol. Sci. Med. Sci. 2010, 65, 258–265. [Google Scholar] [CrossRef]
- Marano, L.; Verre, L.; Carbone, L.; Poto, G.E.; Marrelli, D.; Roviello, F. Current trends in volume and surgical outcomes in gastric cancer. J. Clin. Med. 2023, 12, 4238. [Google Scholar] [CrossRef]

| Variables | Value (N = 865) |
|---|---|
| Overall complication | 222 (25.7) |
| Surgical complication | |
| Anastomotic leakage | 30 (3.5) |
| Anastomotic stricture | 3 (0.3) |
| Intra-abdominal bleeding | 4 (0.5) |
| Intra-luminal bleeding | 4 (0.5) |
| Fluid collection | 9 (1.0) |
| Intra-abdominal abscess | 2 (0.2) |
| Pancreatitis | 6 (0.7) |
| Gastric stasis | 19 (2.2) |
| Ileus | 14 (1.6) |
| Intestinal obstruction | 6 (0.7) |
| Fever | 19 (2.2) |
| Wound | 19 (2.2) |
| Other surgical complication | 7 (0.8) |
| Medical complication | |
| Pulmonary | 45 (5.2) |
| Renal | 5 (0.6) |
| Urinary | 8 (0.9) |
| Gastrointestinal | 9 (1.0) |
| Hepatic | 4 (0.5) |
| Other medical complication | 9 (1.0) |
| Clavien–Dindo grade | |
| I | 23 (2.7) |
| II | 122 (14.1) |
| IIIa | 42 (4.9) |
| IIIb | 19 (2.2) |
| IV | 4 (0.5) |
| V | 12 (1.4) |
| Step of Analysis | Missing Data Tx | Sampling | Train Accuracy | Test Accuracy | Test AUC | Test Precision | Test Recall |
|---|---|---|---|---|---|---|---|
| Missing data strategy | Complete strategy | Base | 0.808 (0.783–0.824) | 0.752 (0.717–0.780) | 0.709 (0.593–0.838) | 0.586 (0.000–1.000) | 0.098 (0.000–0.182) |
| Under | 0.830 (0.792–0.871) | 0.668 (0.572–0.751) | 0.707 (0.597–0.803) | 0.400 (0.313–0.508) | 0.609 (0.364–0.864) | ||
| Over | 0.815 (0.775–0.841) | 0.712 (0.642–0.786) | 0.712 (0.558–0.797) | 0.447 (0.318–0.590) | 0.530 (0.318–0.773) | ||
| Moderate strategy | Base | 0.808 (0.793–0.829) | 0.751 (0.712–0.788) | 0.718 (0.601–0.789) | 0.620 (0.000–1.000) | 0.100 (0.000–0.205) | |
| Under | 0.841 (0.793–0.891) | 0.671 (0.582–0.735) | 0.715 (0.604–0.796) | 0.411 (0.319–0.493) | 0.618 (0.455–0.818) | ||
| Over | 0.827 (0.792–0.853) | 0.711 (0.629–0.788) | 0.718 (0.589–0.797) | 0.452 (0.327–0.611) | 0.536 (0.364–0.750) | ||
| Liberal strategy | Base | 0.810 (0.789–0.826) | 0.758 (0.725–0.800) | 0.712 (0.630–0.825) | 0.687 (0.000–1.000) | 0.110 (0.000–0.244) | |
| Under | 0.853 (0.807–0.883) | 0.679 (0.606–0.769) | 0.709 (0.611–0.824) | 0.415 (0.333–0.550) | 0.605 (0.415–0.805) | ||
| Over | 0.840 (0.792–0.870) | 0.727 (0.650–0.850) | 0.712 (0.623–0.852) | 0.472 (0.340–0.707) | 0.509 (0.293–0.756) | ||
| Backward elimination | Only moderate strategy | Base | 0.799 (0.780–0.799) | 0.747 (0.706–0.782) | 0.741 (0.665–0.820) | 0.562 (0.000–1.000) | 0.099 (0.000–0.227) |
| Under | 0.807 (0.767–0.848) | 0.675 (0.600–0.735) | 0.738 (0.666–0.822) | 0.424 (0.346–0.493) | 0.694 (0.523–0.864) | ||
| Over | 0.799 (0.759–0.828) | 0.710 (0.629–0.800) | 0.741 (0.645–0.815) | 0.459 (0.344–0.600) | 0.649 (0.477–0.818) | ||
| Hyper parameter tuning | Only moderate strategy with backward elimination | Base | 0.761 (0.747–0.783) | 0.740 (0.718–0.759) | 0.739 (0.663–0.814) | 0.377 (0.000–1.000) | 0.017 (0.000–0.091) |
| Under | 0.773 (0.741–0.813) | 0.687 (0.629–0.759) | 0.741 (0.659–0.841) | 0.435 (0.368–0.526) | 0.683(0.500–0.864) | ||
| Over | 0.767 (0.721–0.801) | 0.711 (0.624–0.776) | 0.744 (0.646–0.808) | 0.461 (0.353–0.552) | 0.657 (0.500–0.818) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Oh, H.-J.; Yoo, H.; Kim, C.-Y. Machine Learning Insight: Unveiling Overlooked Risk Factors for Postoperative Complications in Gastric Cancer. Cancers 2025, 17, 1225. https://doi.org/10.3390/cancers17071225
Lee S, Oh H-J, Yoo H, Kim C-Y. Machine Learning Insight: Unveiling Overlooked Risk Factors for Postoperative Complications in Gastric Cancer. Cancers. 2025; 17(7):1225. https://doi.org/10.3390/cancers17071225
Chicago/Turabian StyleLee, Sejin, Hyo-Jung Oh, Hosuon Yoo, and Chan-Young Kim. 2025. "Machine Learning Insight: Unveiling Overlooked Risk Factors for Postoperative Complications in Gastric Cancer" Cancers 17, no. 7: 1225. https://doi.org/10.3390/cancers17071225
APA StyleLee, S., Oh, H.-J., Yoo, H., & Kim, C.-Y. (2025). Machine Learning Insight: Unveiling Overlooked Risk Factors for Postoperative Complications in Gastric Cancer. Cancers, 17(7), 1225. https://doi.org/10.3390/cancers17071225






