Radiomic Profiling of Orthotopic Mouse Models of Glioblastoma Reveals Histopathological Correlations Associated with Tumour Response to Ionising Radiation
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Establishment of Orthotopic Glioma Tumours
2.2. Molecular Profile of Cell Lines Used in This Study
2.3. Radiation Treatment
2.4. MRI Experiment
2.5. Histopathology
2.6. Patient Data
2.7. Statistical Analyses
2.8. Radiomic Feature Selection and Statistical Evaluation
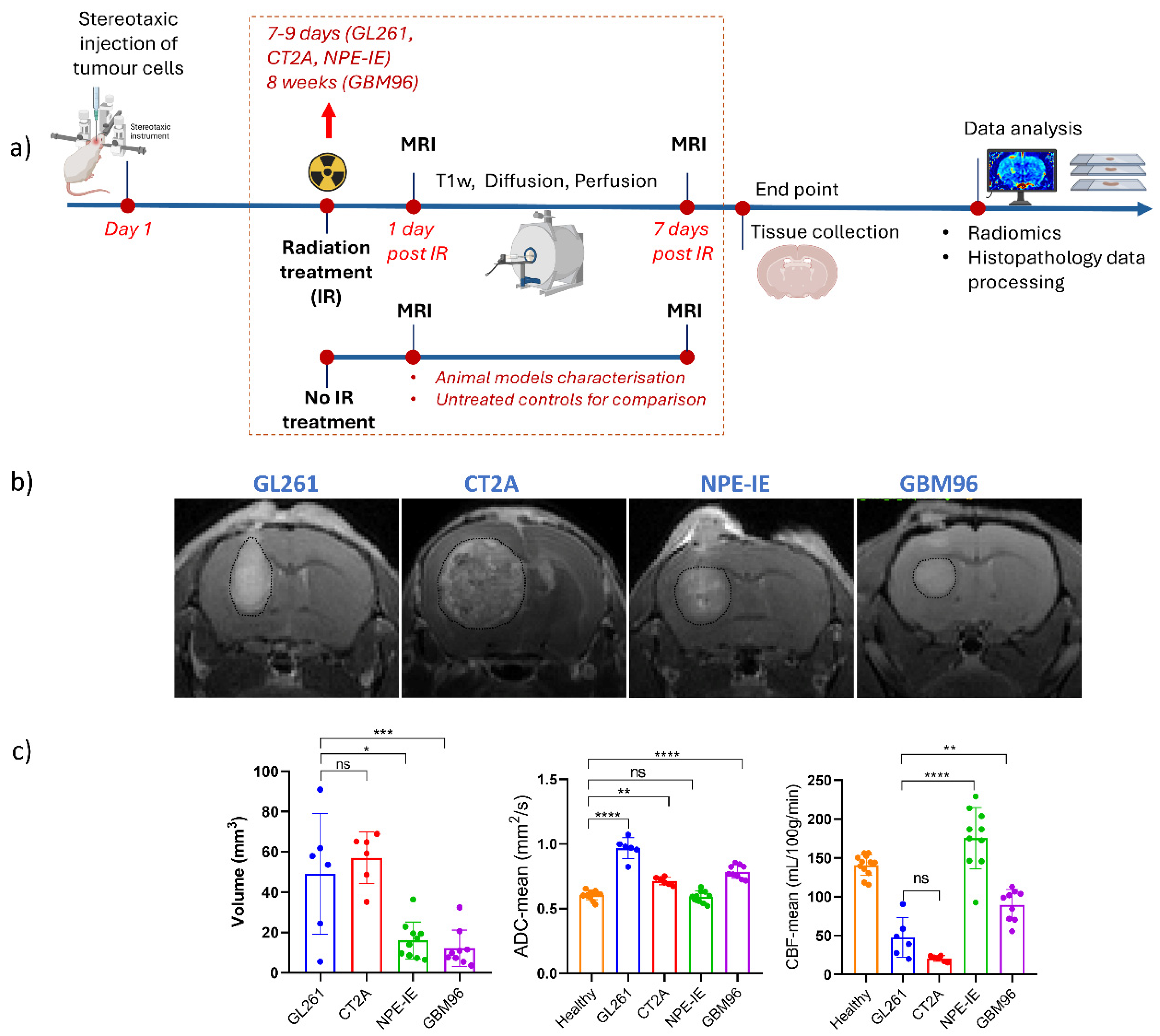
3. Results
3.1. High Diffusion Coefficients and Low Perfusion Were Found in GL261, CT-2A, and GBM96, but Not in NPE-IE Tumours
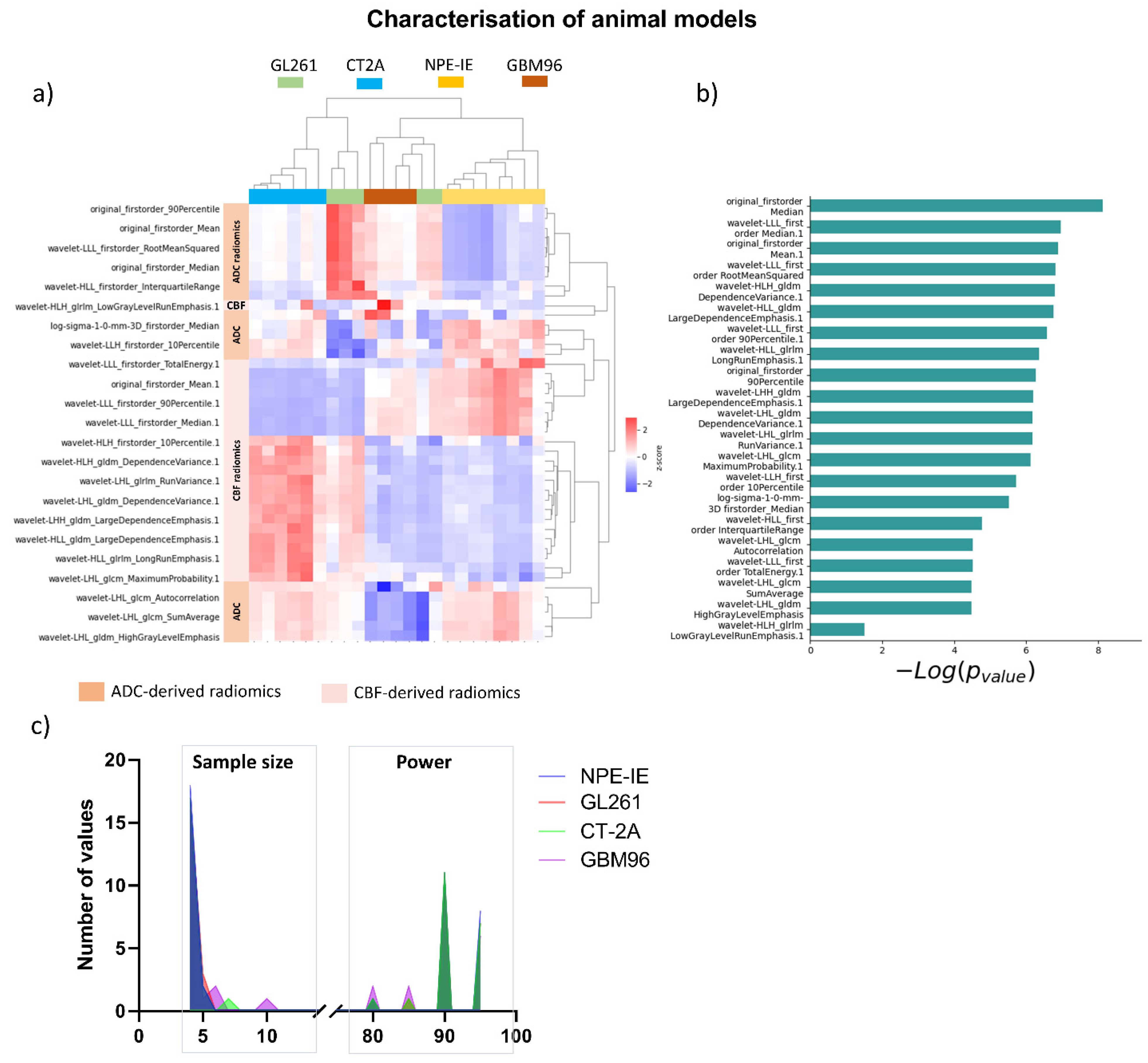
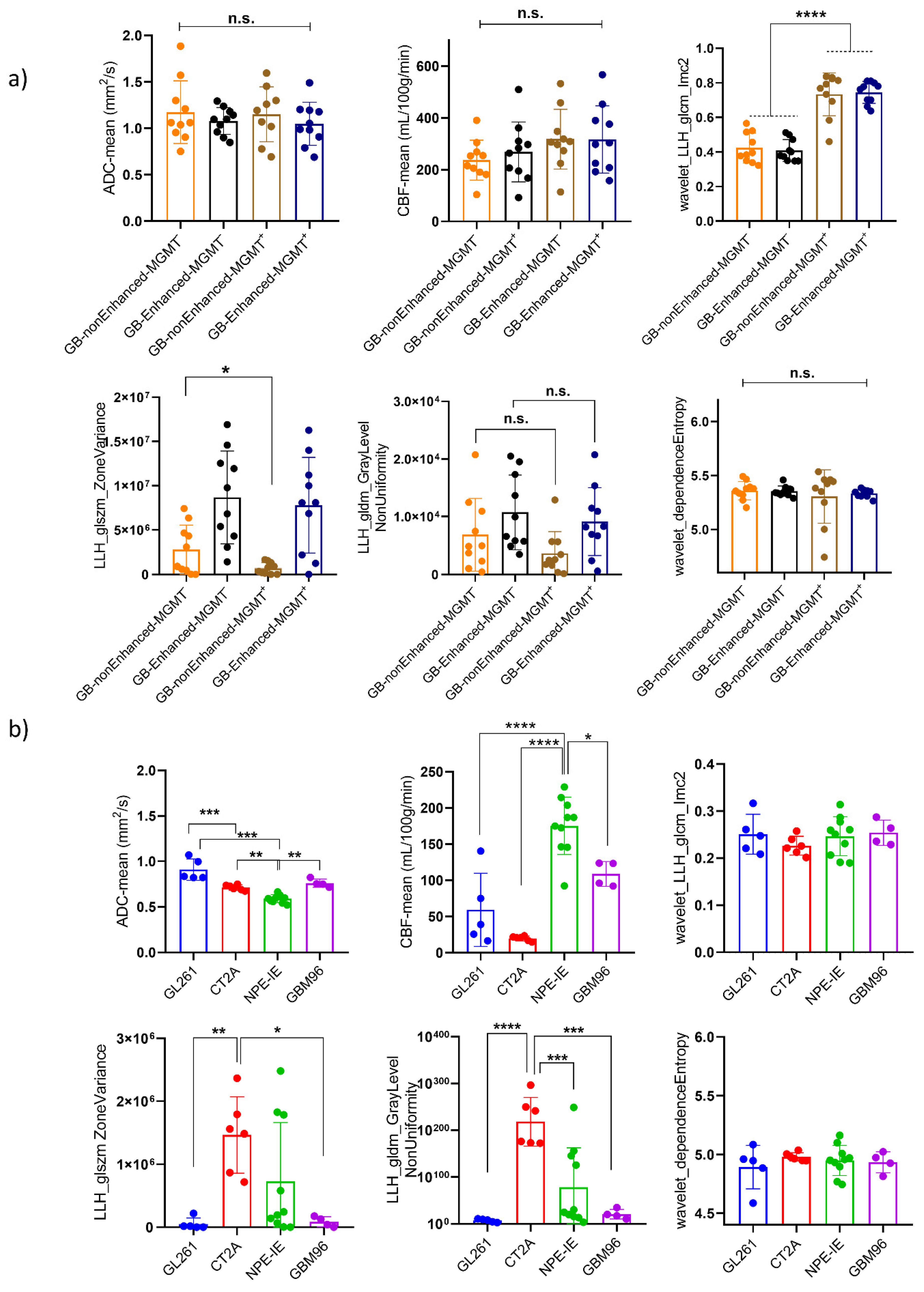
3.2. Diffusion and Perfusion Radiomic Profiling Demonstrates Notable Differences Among Orthotopic Glioma Tumours
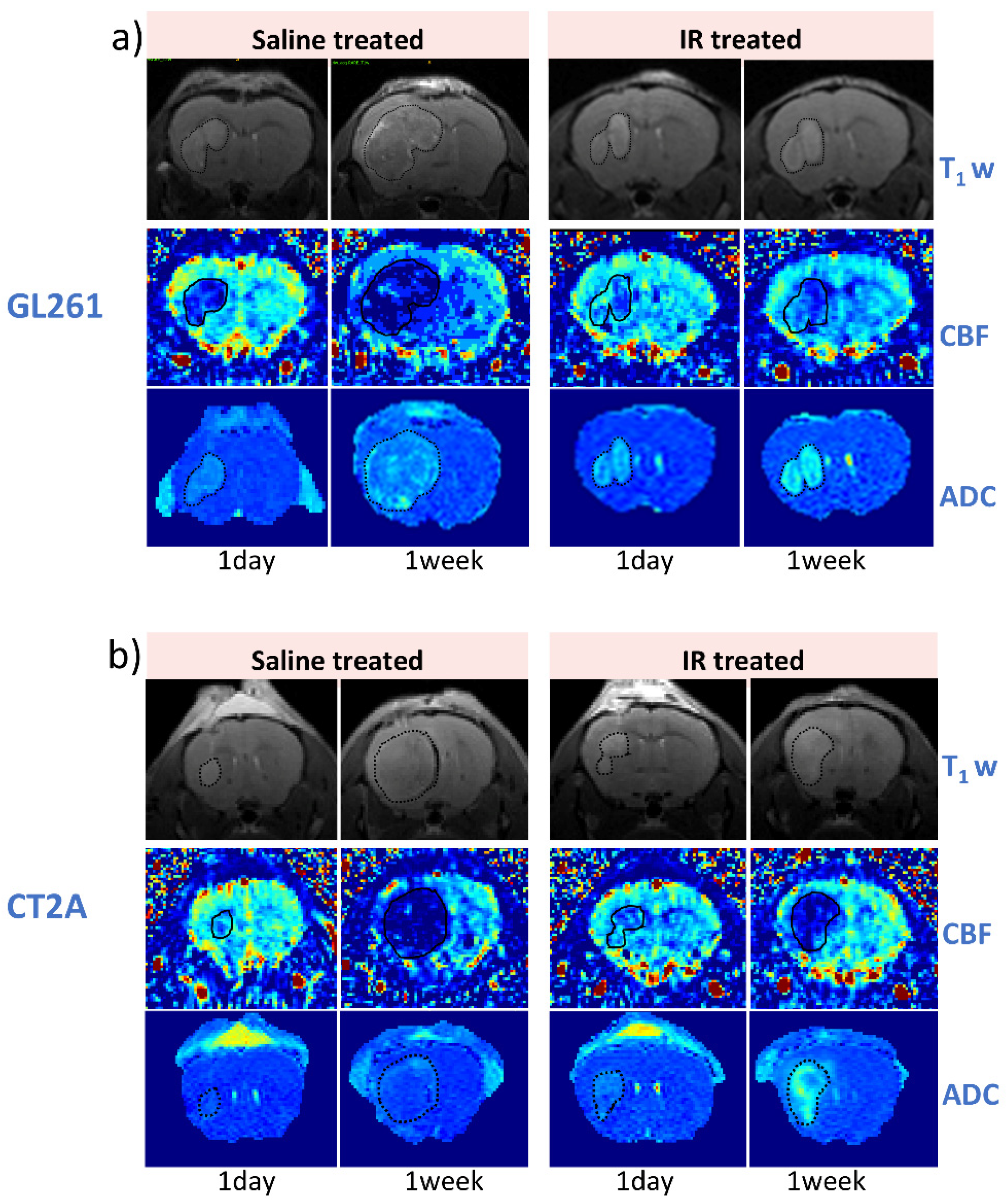
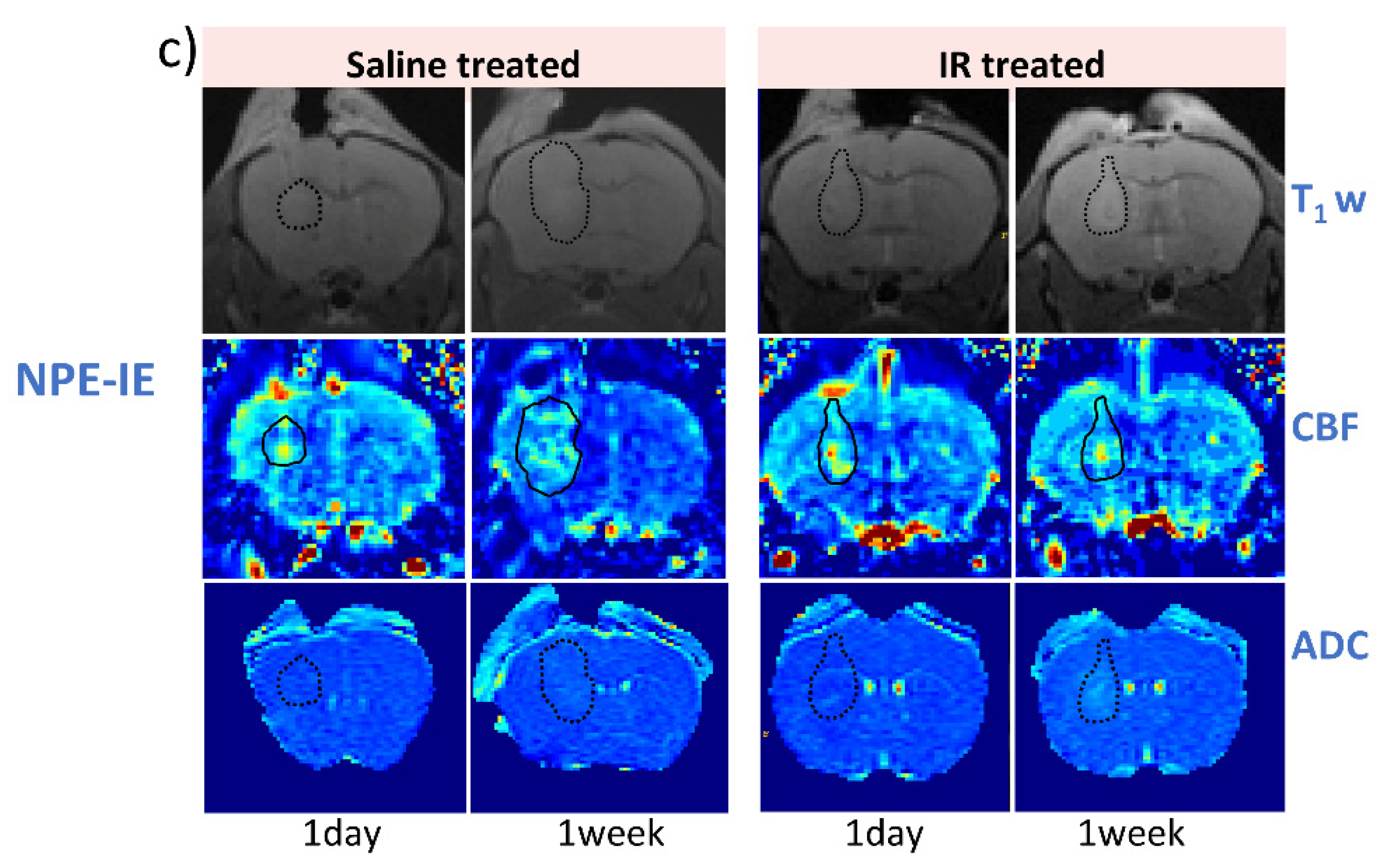
3.3. IR Therapy Leads to Increased ADC and Improved CBF in GL261 and CT-2A Tumours
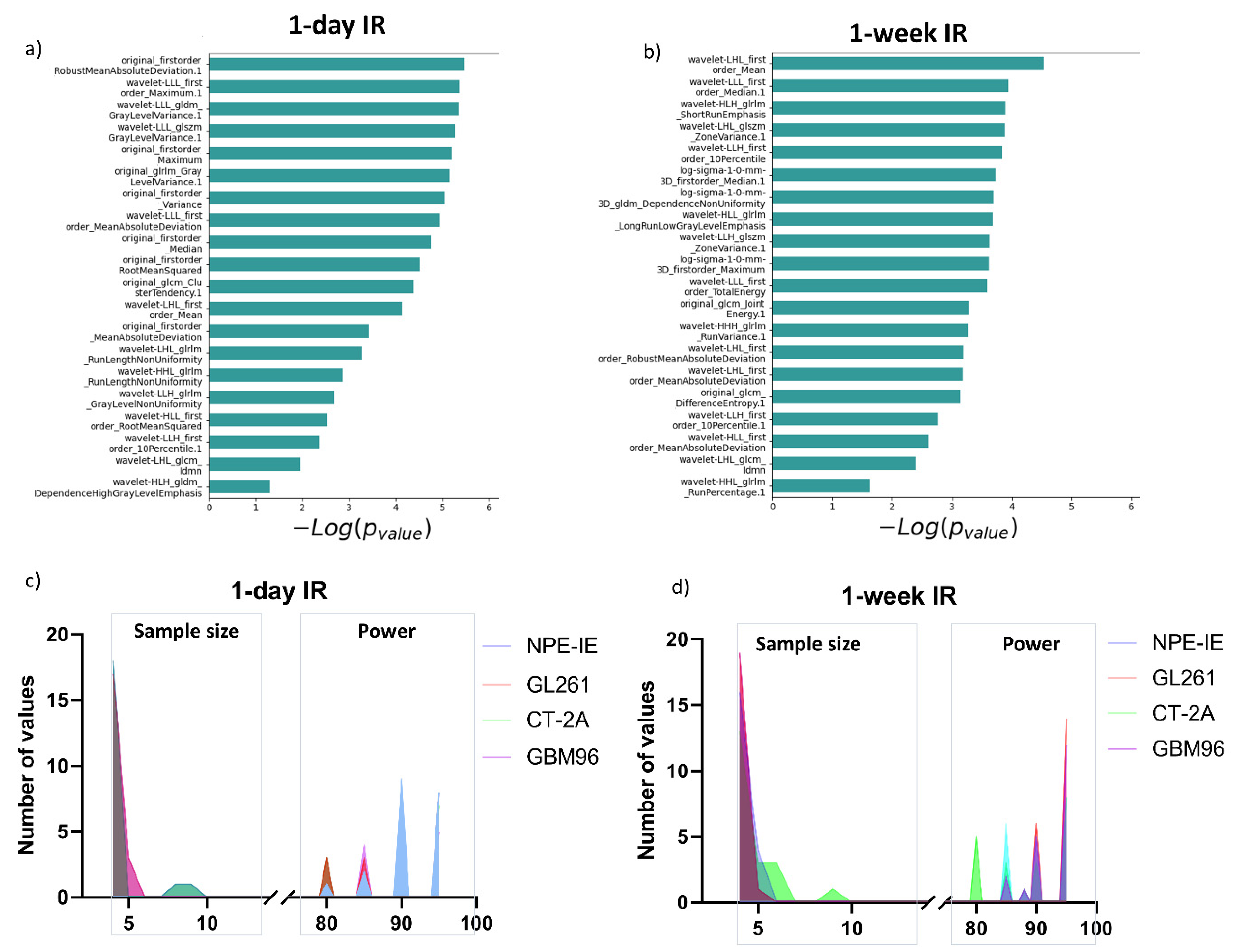
3.4. Radiomic Descriptors of MRI Heterogeneity Show Specific Changes 1 Day and 7 Days Post IR
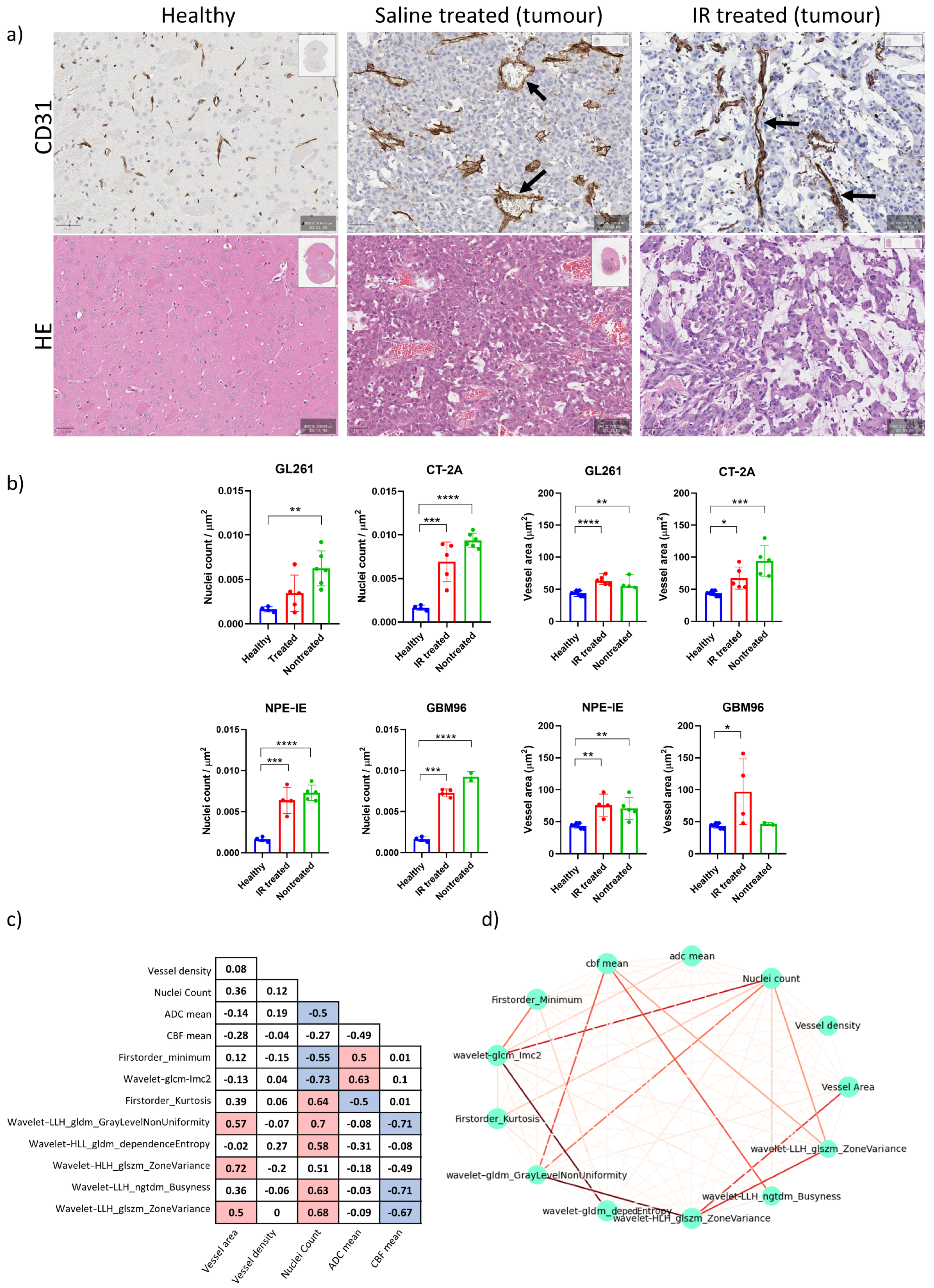
3.5. High Tumour Cellularity Is Present in All Orthotopic Tumours Along with Dilated Vessels in GL261 and CT-2A Tumours
3.6. Radiomics Yielded Higher Correlations with Histopathology than ADC and CBF Alone
3.7. Mouse Orthotopic Tumours Share Similarities with Central Non-Enhancing Patient Tumours
4. Discussion
Challenges and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nabian, N.; Ghalehtaki, R.; Zeinalizadeh, M.; Balana, C.; Jablonska, P.A. State of the neoadjuvant therapy for glioblastoma multiforme-Where do we stand? Neuro-Oncol. Adv. 2024, 6, vdae028. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Eidel, O.; Neumann, J.O.; Burth, S.; Kieslich, P.J.; Jungk, C.; Sahm, F.; Kickingereder, P.; Kiening, K.; Unterberg, A.; Wick, W.; et al. Automatic Analysis of Cellularity in Glioblastoma and Correlation with ADC Using Trajectory Analysis and Automatic Nuclei Counting. PLoS ONE 2016, 11, e0160250. [Google Scholar] [CrossRef]
- Andre, J.B.; Nagpal, S.; Hippe, D.S.; Ravanpay, A.C.; Schmiedeskamp, H.; Bammer, R.; Palagallo, G.J.; Recht, L.; Zaharchuk, G. Cerebral Blood Flow Changes in Glioblastoma Patients Undergoing Bevacizumab Treatment Are Seen in Both Tumor and Normal Brain. Neuroradiol. J. 2015, 28, 112–119. [Google Scholar] [CrossRef]
- Hu, L.S.; Eschbacher, J.M.; Heiserman, J.E.; Dueck, A.C.; Shapiro, W.R.; Liu, S.; Karis, J.P.; Smith, K.A.; Coons, S.W.; Nakaji, P.; et al. Reevaluating the imaging definition of tumor progression: Perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro-Oncol. 2012, 14, 919–930. [Google Scholar] [CrossRef]
- Hooper, G.W.; Ginat, D.T. MRI radiomics and potential applications to glioblastoma. Front. Oncol. 2023, 13, 1134109. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, R.; Gao, J.; Tan, D.; Yang, X.; Wen, M.; Wang, J.; Xie, X.; Liao, R.; Tang, Y.; et al. An Integrated Radiomics Model Incorporating Diffusion-Weighted Imaging and (18)F-FDG PET Imaging Improves the Performance of Differentiating Glioblastoma From Solitary Brain Metastases. Front. Oncol. 2021, 11, 732704. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, H.S.; Jo, Y.; Yoo, R.E.; Choi, S.H.; Nam, S.J.; Kim, J.H. Radiomics prognostication model in glioblastoma using diffusion- and perfusion-weighted MRI. Sci. Rep. 2020, 10, 4250. [Google Scholar] [CrossRef]
- Malik, N.; Geraghty, B.; Dasgupta, A.; Maralani, P.J.; Sandhu, M.; Detsky, J.; Tseng, C.L.; Soliman, H.; Myrehaug, S.; Husain, Z.; et al. MRI radiomics to differentiate between low grade glioma and glioblastoma peritumoral region. J. Neuro-Oncol. 2021, 155, 181–191. [Google Scholar] [CrossRef]
- Kobayashi, K.; Miyake, M.; Takahashi, M.; Hamamoto, R. Observing deep radiomics for the classification of glioma grades. Sci. Rep. 2021, 11, 10942. [Google Scholar] [CrossRef]
- Kickingereder, P.; Gotz, M.; Muschelli, J.; Wick, A.; Neuberger, U.; Shinohara, R.T.; Sill, M.; Nowosielski, M.; Schlemmer, H.P.; Radbruch, A.; et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin. Cancer Res. 2016, 22, 5765–5771. [Google Scholar] [CrossRef] [PubMed]
- Mammadov, O.; Akkurt, B.H.; Musigmann, M.; Ari, A.P.; Blomer, D.A.; Kasap, D.N.G.; Henssen, D.; Nacul, N.G.; Sartoretti, E.; Sartoretti, T.; et al. Radiomics for pseudoprogression prediction in high grade gliomas: Added value of MR contrast agent. Heliyon 2022, 8, e10023. [Google Scholar] [CrossRef] [PubMed]
- Kandalgaonkar, P.; Sahu, A.; Saju, A.C.; Joshi, A.; Mahajan, A.; Thakur, M.; Sahay, A.; Epari, S.; Sinha, S.; Dasgupta, A.; et al. Predicting IDH subtype of grade 4 astrocytoma and glioblastoma from tumor radiomic patterns extracted from multiparametric magnetic resonance images using a machine learning approach. Front. Oncol. 2022, 12, 879376. [Google Scholar] [CrossRef]
- Verma, N.; Cowperthwaite, M.C.; Burnett, M.G.; Markey, M.K. Differentiating tumor recurrence from treatment necrosis: A review of neuro-oncologic imaging strategies. Neuro-Oncol. 2013, 15, 515–534. [Google Scholar] [CrossRef]
- Park, Y.W.; Choi, D.; Park, J.E.; Ahn, S.S.; Kim, H.; Chang, J.H.; Kim, S.H.; Kim, H.S.; Lee, S.K. Differentiation of recurrent glioblastoma from radiation necrosis using diffusion radiomics with machine learning model development and external validation. Sci. Rep. 2021, 11, 2913. [Google Scholar] [CrossRef]
- Maes, W.; Van Gool, S.W. Experimental immunotherapy for malignant glioma: Lessons from two decades of research in the GL261 model. Cancer Immunol. Immunother. 2011, 60, 153–160. [Google Scholar] [CrossRef]
- Seyfried, T.N.; el-Abbadi, M.; Roy, M.L. Ganglioside distribution in murine neural tumors. Mol. Chem. Neuropathol. 1992, 17, 147–167. [Google Scholar] [CrossRef]
- Gangoso, E.; Southgate, B.; Bradley, L.; Rus, S.; Galvez-Cancino, F.; McGivern, N.; Guc, E.; Kapourani, C.A.; Byron, A.; Ferguson, K.M.; et al. Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell 2021, 184, 2454–2470.e26. [Google Scholar] [CrossRef]
- Przystal, J.M.; Hajji, N.; Khozoie, C.; Renziehausen, A.; Zeng, Q.; Abaitua, F.; Hajitou, A.; Suwan, K.; Want, E.; Bomalaski, J.; et al. Efficacy of arginine depletion by ADI-PEG20 in an intracranial model of GBM. Cell Death Dis. 2018, 9, 1192. [Google Scholar] [CrossRef]
- Grech-Sollars, M.; Hales, P.W.; Miyazaki, K.; Raschke, F.; Rodriguez, D.; Wilson, M.; Gill, S.K.; Banks, T.; Saunders, D.E.; Clayden, J.D.; et al. Multi-centre reproducibility of diffusion MRI parameters for clinical sequences in the brain. NMR Biomed. 2015, 28, 468–485. [Google Scholar] [CrossRef]
- Petersen, E.T.; Mouridsen, K.; Golay, X. The QUASAR reproducibility study, Part II: Results from a multi-center Arterial Spin Labeling test-retest study. Neuroimage 2010, 49, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhang, L.; Wei, Q.; Shao, A. O(6)-Methylguanine-DNA Methyltransferase (MGMT): Challenges and New Opportunities in Glioma Chemotherapy. Front. Oncol. 2019, 9, 1547. [Google Scholar] [CrossRef] [PubMed]
- Haddad, A.F.; Young, J.S.; Amara, D.; Berger, M.S.; Raleigh, D.R.; Aghi, M.K.; Butowski, N.A. Mouse models of glioblastoma for the evaluation of novel therapeutic strategies. Neuro-Oncol. Adv. 2021, 3, vdab100. [Google Scholar] [CrossRef] [PubMed]
- Iorgulescu, J.B.; Ruthen, N.; Ahn, R.; Panagioti, E.; Gokhale, P.C.; Neagu, M.; Speranza, M.C.; Eschle, B.K.; Soroko, K.M.; Piranlioglu, R.; et al. Antigen presentation deficiency, mesenchymal differentiation, and resistance to immunotherapy in the murine syngeneic CT2A tumor model. Front. Immunol. 2023, 14, 1297932. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Walker, A.J.; Ruzevick, J.; Malayeri, A.A.; Rigamonti, D.; Lim, M.; Redmond, K.J.; Kleinberg, L. Postradiation imaging changes in the CNS: How can we differentiate between treatment effect and disease progression? Future Oncol. 2014, 10, 1277–1297. [Google Scholar] [CrossRef]
- Alsop, D.C.; Detre, J.A.; Golay, X.; Gunther, M.; Hendrikse, J.; Hernandez-Garcia, L.; Lu, H.; MacIntosh, B.J.; Parkes, L.M.; Smits, M.; et al. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn. Reson. Med. 2015, 73, 102–116. [Google Scholar] [CrossRef]
- Wong, E.C.; Buxton, R.B.; Frank, L.R. Quantitative imaging of perfusion using a single subtraction (QUIPSS and QUIPSS II). Magn. Reson. Med. 1998, 39, 702–708. [Google Scholar] [CrossRef]
- Andersson, J.L.; Skare, S.; Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: Application to diffusion tensor imaging. Neuroimage 2003, 20, 870–888. [Google Scholar] [CrossRef]
- Smith, S.M.; Jenkinson, M.; Woolrich, M.W.; Beckmann, C.F.; Behrens, T.E.; Johansen-Berg, H.; Bannister, P.R.; De Luca, M.; Drobnjak, I.; Flitney, D.E.; et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004, 23 (Suppl. S1), S208–S219. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, E104–E107. [Google Scholar] [CrossRef]
- Hectors, S.J.; Lewis, S.; Besa, C.; King, M.J.; Said, D.; Putra, J.; Ward, S.; Higashi, T.; Thung, S.; Yao, S.; et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur. Radiol. 2020, 30, 3759–3769. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Calabrese, E.; Villanueva-Meyer, J.E.; Rudie, J.D.; Rauschecker, A.M.; Baid, U.; Bakas, S.; Cha, S.; Mongan, J.T.; Hess, C.P. The University of California San Francisco Preoperative Diffuse Glioma MRI Dataset. Radiol. Artif. Intell. 2022, 4, e220058. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Anaraki, J.R.; Usefi, H. A Comparative Study of Feature Selection Methods on Genomic Datasets. In Proceedings of the 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), Cordoba, Spain, 5–7 June 2019; pp. 471–476. [Google Scholar] [CrossRef]
- Radovic, M.; Ghalwash, M.; Filipovic, N.; Obradovic, Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinform. 2017, 18, 9. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso: A retrospective. J. R. Stat. Soc. B 2011, 73, 273–282. [Google Scholar] [CrossRef]
- Tanaka, T. [Fundamentals] 5. Python+scikit-learn for Machine Learning in Medical Imaging. Nihon Hoshasen Gijutsu Gakkai Zasshi 2023, 79, 1189–1193. [Google Scholar] [CrossRef]
- Martinez-Murillo, R.; Martinez, A. Standardization of an orthotopic mouse brain tumor model following transplantation of CT-2A astrocytoma cells. Histol. Histopathol. 2007, 22, 1309–1326. [Google Scholar] [CrossRef]
- Breen, W.G.; Aryal, M.P.; Cao, Y.; Kim, M.M. Integrating multi-modal imaging in radiation treatments for glioblastoma. Neuro-oncology 2024, 26, S17–S25. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Holloway, L.; Metcalfe, P.; Koh, E.S.; Brighi, C. Challenges in Glioblastoma Radiomics and the Path to Clinical Implementation. Cancers 2022, 14, 3897. [Google Scholar] [CrossRef] [PubMed]
- Sottoriva, A.; Spiteri, I.; Piccirillo, S.G.; Touloumis, A.; Collins, V.P.; Marioni, J.C.; Curtis, C.; Watts, C.; Tavare, S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 4009–4014. [Google Scholar] [CrossRef] [PubMed]
- Ravi, V.M.; Will, P.; Kueckelhaus, J.; Sun, N.; Joseph, K.; Salie, H.; Vollmer, L.; Kuliesiute, U.; von Ehr, J.; Benotmane, J.K.; et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 2022, 40, 639–655.e13. [Google Scholar] [CrossRef]
- Dextraze, K.; Saha, A.; Kim, D.; Narang, S.; Lehrer, M.; Rao, A.; Narang, S.; Rao, D.; Ahmed, S.; Madhugiri, V.; et al. Spatial habitats from multiparametric MR imaging are associated with signaling pathway activities and survival in glioblastoma. Oncotarget 2017, 8, 112992–113001. [Google Scholar] [CrossRef]
- Monsky, W.L.; Jin, B.; Molloy, C.; Canter, R.J.; Li, C.S.; Lin, T.C.; Borys, D.; Mack, W.; Kim, I.; Buonocore, M.H.; et al. Semi-automated volumetric quantification of tumor necrosis in soft tissue sarcoma using contrast-enhanced MRI. Anticancer Res. 2012, 32, 4951–4961. [Google Scholar]
- Butler, M.; Pongor, L.; Su, Y.T.; Xi, L.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 2020, 6, 380–391. [Google Scholar] [CrossRef]
- Li, L.; Xiao, F.; Wang, S.; Kuang, S.; Li, Z.; Zhong, Y.; Xu, D.; Cai, Y.; Li, S.; Chen, J.; et al. Preoperative prediction of MGMT promoter methylation in glioblastoma based on multiregional and multi-sequence MRI radiomics analysis. Sci. Rep. 2024, 14, 16031. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Muller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baxan, N.; Perryman, R.; Chatziathanasiadou, M.V.; Syed, N. Radiomic Profiling of Orthotopic Mouse Models of Glioblastoma Reveals Histopathological Correlations Associated with Tumour Response to Ionising Radiation. Cancers 2025, 17, 1258. https://doi.org/10.3390/cancers17081258
Baxan N, Perryman R, Chatziathanasiadou MV, Syed N. Radiomic Profiling of Orthotopic Mouse Models of Glioblastoma Reveals Histopathological Correlations Associated with Tumour Response to Ionising Radiation. Cancers. 2025; 17(8):1258. https://doi.org/10.3390/cancers17081258
Chicago/Turabian StyleBaxan, Nicoleta, Richard Perryman, Maria V. Chatziathanasiadou, and Nelofer Syed. 2025. "Radiomic Profiling of Orthotopic Mouse Models of Glioblastoma Reveals Histopathological Correlations Associated with Tumour Response to Ionising Radiation" Cancers 17, no. 8: 1258. https://doi.org/10.3390/cancers17081258
APA StyleBaxan, N., Perryman, R., Chatziathanasiadou, M. V., & Syed, N. (2025). Radiomic Profiling of Orthotopic Mouse Models of Glioblastoma Reveals Histopathological Correlations Associated with Tumour Response to Ionising Radiation. Cancers, 17(8), 1258. https://doi.org/10.3390/cancers17081258





