Abstract
During the study of multimodal brain tumor MR image segmentation, the large differences in the image distributions make the assumption that the conditional probabilities are similar when the edge distributions of the target and source domains are similar, and that the edge distributions are similar when the conditional probabilities are similar, not valid. In addition, the training network is usually trained on single domain data, which creates a tendency for the network to represent the image towards the source domain when the target domain is not labeled. Based on the aforementioned reasons, a new multimodal brain tumor MR segmentation strategy based on domain adaptation is proposed in this study. First, the source domain targets for each modality are derived through the clustering methods in the pre-training stage to select the target domain images with the strongest complementarity in the source domain and further produce the pseudo labels. Second, feature adapters are proposed to improve the feature alignment, and a network sensitive to both source and target domain images is designed to comprehensively leverage the multimodal image information. These measures mitigate the domain shift problem and improve the generalization ability of the model, enhancing the accuracy of multimodal brain tumor MR image segmentation.
1. Introduction
The brain is the center of control of the human body, processing the information collected by sensory organs through a neural network. Controls many functions such as reasoning, language, and movement. However, uncontrolled cell growth in the brain can cause brain tumors that affect the health of individuals and even lead to death. To date, approximately 120 types of brain tumors have been identified [1]. It is proposed that the early detection and treatment of brain tumors will not only increase the survival rate of patients but also reduce the incidence of brain tumor-related complications. Therefore, the precise segmentation of brain tumors is crucial.
In medical image analysis, images acquired by X-ray, computed tomography, and magnetic resonance imaging usually need to be pixelated manually, which is highly tedious and time consuming and involves a large investment in human resources. To improve network performance, traditional segmentation methods usually assume that the training dataset (source domain) and the test dataset (target domain) have a similar feature space or are derived from the same domain (assuming a close data distribution between the labeled training samples and test samples) and use a large amount of labeled source domain data for training.
In multimodal brain tumor medical image segmentation tasks, data sets usually contain multiple domains (i.e., the data in the training set and the test set come from different hospitals, scanning devices, or scanning parameters), while the data lack a lot of manual labeling, which makes the assumptions mentioned above too strict and idealistic. In addition, trained networks are affected by the domain shift problem, that is, they perform well on the source domain data but perform poorly. in the target domain. To address these problems, previous studies have focused on domain-adaptive-based approaches. Next, the concepts related to domains, tasks, transfer learning, and domain adaptation are described in detail.
Definition 1.
(Domain ): The domain (data source) comprises the input feature space , the label space , and the associated joint probability distribution , where: X indicates a set of samples, each sample being a feature representation in the feature space , that is, is one of the two or more types of label space; and is the joint probability distribution on the feature label space . Therefore, the domain can be expressed by the formula . If the domain is different, its feature space or joint probability distribution will also change.
Definition 2.
(Task ): For a given domain , a task (an operation to be performed by a learned model) consists of two parts, the label space and the target category detection function . Given the characteristic vectors of an instance , the target category detection function can predict the class labels corresponding to the instance, and from a probabilistic point of view, the target category detection function can be displayed through a marginal probability distribution , so that a task for a given domain can be represented as is the set of all labels. The target category detection function cannot be intuitively predicted, but can be derived by learning the training samples.
The definitions of the source/reference and target domains are presented as follows. The source dataset in the source domain consists of the sample–label pairs , where denotes the observation samples in the source domain denotes the corresponding class labels of the observation samples in the source domain . The target domain comprises the target samples and their labels, expressed as , where represents The input samples of the target domain and is the label / prediction of the target samples , that is, the output of the corresponding target class detection function . The unsupervised domain is unknown. In general, the data set for the source domain and the data set for the target domain exhibit the following relationship: . The source domain contains a large amount of sample data, and the corresponding class labels can be easily derived, whereas the target domain has fewer sample data and usually has no class labels or only a small number of class labels. Generally, when the data in the source and target domains are derived from the same task but are distributed differently, considering the basic knowledge contained in one domain to be migrated to the other domain may exert a negative effect on the performance of the learned model in the target domain, i.e., a common negative migration.
Definition 3.
(Transfer Learning): Given a source domain and a source task , a target domain and a target task , the data of the source and target domains exhibit different distributions and feature spaces ( or ), that is, and under different settings are aimed at improving the prediction performance of the detection function of the target category learning the knowledge of and [2]. In particular, both the domain and the task can change during the transfer learning process. Therefore, domain adaptation ensues.
Definition 4.
(Domain Adaptation): DA generally assumes that the feature space and tasks are constant in all domains, while the distribution of edges (features) varies between the source and target domains. The main objective is to map data from the same task, but with diverse distributions of labels in the source domain and data that are unlabeled or with few labels in the target domain, into a common feature space so that the data are mapped as closely as possible in this feature space. Consequently, the learned target category detection function utilizes the source domain in this space and can perform well in the target domain, thus improving the accuracy of the model’s prediction in the target domain, which can be mathematically described as follows:
Define as the common feature space and the corresponding labeling space. Define the source domain and the target domain in this space with the different distributions and . There are m samples with labels in the source domain , , and there are n samples (with or without labels) in the target domain . The purpose of DA is to transfer the knowledge learned in the source domain to the target domain and perform a specific task in the target domain with good performance shared between the source domain and the target domain .
Domain adaptation can be adopted in different scenarios and settings through distinct methods such as instance weighting, feature transformation, and supervised, semi-supervised, and unsupervised learning. Therefore, DA is expected to be widely applied in medical image segmentation. The main strategies for this study are as follows.
To improve the generalization ability of the multimodal brain tumor MR image segmentation task and address the domain shift problem, we propose a new adaptive multimodal brain tumor MR segmentation strategy based on three principles, outlined below:
(1) There are large visual differences and similarities between different modal brain tumor MR images, suggesting that the different modal images possess unique and common features.
(2) Multimodal images can cause a large difference in data distribution between training samples and test samples, making the idealization assumption invalid.
(3) Training data from only one domain can lead to a tendency for the network to still have a representation on the target domain that lacks labeling towards the source-domain image.
Taking into account the above principles, the specifics of the new adaptive multimodal brain tumor MRI segmentation strategy are as follows:
- Targets with different modal features of the source domain are derived using clustering methods in the pre-training stage.
- For each target, samples with the strongest complementarity to the source domain are identified in the target domain data, and the corresponding pseudo labels are produced and applied to train the network.
- Feature alignment is performed using a feature adapter; the feature adapter is a modular structure that learns to align features from source- and target-domain images.
- To improve the generalization ability of the network, a network structure is designed that is sensitive to the source- and target-domain images, making it better adapted to the data distribution in different domains.
2. Related Work
2.1. Relevant Research
Domain adaptation is a special case of transfer learning that primarily aims to improve the performance of the model in the target domain in the absence of a large amount of labeled data in that domain, using the knowledge of the relevant auxiliary domains and a large amount of labeled training data. Pan et al. [2] conducted the first systematic analysis of the development of transfer learning, and proposed several definitions and a classification of transfer learning techniques, in which domain adaptation (DA) as a subfield was formally coined. In principle, DA aims to utilize a model trained in one domain in another domain. The primary aim of this technique is to adapt the model by analyzing and understanding the differences between the source and target domains to improve its performance in the target domain. Since then, domain-adaptive methods have been extensively utilized in various fields. In the past few years, many approaches based on domain adaptation have been designed to address different scenarios arising under different application settings. For example, studies [3,4] applied DA to cross-language text classification; studies [5,6,7] applied DA to pose estimation; studies [8,9,10] summarized the application of domain adaptation in target detection; and in study [11], the boosting algorithm, a linear fusion algorithm, and a concept-association model were proposed, all of which are based on DA, to resolve the various challenges affecting the feature layers, multi-feature fusion, and semantic associations among the concepts of temporal evaluation in video concept detection. In addition, domain adaptation is often applied to recommender systems, sentiment analysis, machine translation, and other directions.
In recent years, data mining-based segmentation techniques have been developed, improving the performance of cross-domain image analysis. Therefore, the application of domain-based adaptive methods in medical image analysis is expected to become one of the hot spots for research.
Currently, DA is mainly applied in medical image analysis tasks such as image segmentation, image classification, and image reconstruction. Wachinger et al. [12] proposed a supervised domain-adaptive method that leverages the instance weighting to prevent the effect of distributional differences between source- and target-domain datasets, thereby enhancing the accuracy in Alzheimer’s disease classification. The method weights the sample data from the source domain by maximizing the similarity between the source and target domains, allowing the effective adaptation of the model to the data distribution in the target domain. Goetz et al. [13] proposed an adaptive supervised brain tumor segmentation method based on instance weighting, with the aim of solving the challenges of incorrect sample selection seen in supervised learning due to limited annotation. This method first trains a domain classifier using paired samples from the source and target domains, then adjusts the weights of the source samples based on the output (computed probabilities) of the domain classifier, and finally performs segmentation through the training of a random forest classifier. Chen et al. [14] developed a novel unsupervised domain adaptive framework that is leveraged on collaborative images and feature matching. The method allows the synergistic fusion of feature transformation and image migration into a unified network for the severe domain shift problem prevalent in cross-modal medical images. Han et al. [15] proposed a new symmetric unsupervised domain-adaptive structure that utilizes a symmetric migration subnetwork to map the source domain features into the target-domain feature space. It learns the feature mapping between the source and target images, and thus aligns the data distribution between the two domains. To optimize the segmentation performance, the method uses a complementary information-extraction mechanism, i.e., the feature information of both the source and target domains in the segmentation subnetwork, to increase the segmentation accuracy. Vesal et al. [16] proposed a supervised domain-adaptive method based on CNNs and the task of multisequence cardiac MRI image segmentation, which effectively addressed the reliance of the traditional CNN-based multisequence cardiac MRI image on pixel-level annotations, making it unsuitable for images of different domains. When segmenting MR images, the variability from different image domains may introduce model reproducibility problems. To perform the same operation in different image domains, traditional methods are retrained on the network. Kushibar et al. [17] proposed domain-adaptive strategies by combining spatial and deep convolutional features to address this problem. For small target-domain data samples, it cannot fine-tune the model effectively. Therefore, several studies have proposed the use of auxiliary domains as a strategy to enhance the performance on target domain data. Gu et al. [18] developed a multi-step domain-adaptation algorithm for the skin cancer classification problem by fine-tuning and training a ResNet model on a larger dataset of medical images of skin cancer and applying the model to train the target domain dataset. In addition, Kaur et al. [19] proposed to first pre-train a 3D model on large-scale source domain data, which was optimized using small labeled target domain data. It was observed that this strategy achieved superior performance compared to retraining the network. Kamphenkel et al. [20] proposed end-to-end unsupervised domain-adaptive models that overcame the dependence of common models on specific image input channels for breast cancer classification on diffusion-weighted MR images. The technique leverages a diffusion crag imaging algorithm to convert the target domain data into source domain data, which alleviates the need to use labeled target domain data. Gao et al. [21] proposed an unsupervised approach to adapt to data discrepancies between domains by employing the central moment difference as a loss of domain regularization to maintain the consistency of the features for more effective adaptation. Bateson et al. [22] proposed an unsupervised approach to adapt to data discrepancies between domains by employing the central moment difference as a loss of domain regularization to maintain the consistency of the features for more effective adaptation.
As presented in Table 1, DA improves the precision and accuracy of image segmentation while solving the domain shift problem in medical image collars.

Table 1.
Overview of research status.
2.2. Relevant Knowledge
- Domain shiftDomain shift stems from the machine learning model, trained on a dataset, displaying reduced performance in different datasets. This phenomenon is derived from variations in the underlying data distributions, such as differences in features, labels, or sample distributions.The domain shift problem occurs even when deep convolutional neural networks are trained on large image datasets, which is a challenge in the era of the rapid development of deep learning technology. In domains such as medical images, the domain shift problem needs to be solved. The DA technique is used to solve the domain shift problem during data migration.
- Domain GeneralizationDomain generalization investigates the problems in which, in the case of there being only source domain data, multiple source domain data with different distributions can be used to train and learn a model with strong generalizability, which achieves better performance on unknown target domains. The main difference between the domain generalization problem and the domain adaptation problem is that in domain generalization the target domain is unknown and only the data of the source domain are trained. In contrast, in domain adaptation, the target domain and source domain data are utilized in the training. The advantage of domain generalization is that models that require a single training can be applied to diverse scenarios. Using MR and CT as examples, a great difference is observed in the appearance between the images acquired by these two methods. Training two respective segmentation models on CT or MR alone and followed by the adoption of these two models to segment a common set of CT images, called ‘CT to CT’ and ‘MR to CT’, respectively, leads to the appearance of the segmentation model trained on MR images alone. The model trained on MR images alone does not generalize well to CT images directly, or the model trained on CT images alone does not generalize well to MR images directly.
- Negative migrationThe main focus of DA is to learn from the knowledge of the source domain to perform well on the target task. However, in some cases, DA may face the following challenges: (1) the tasks in the source and target domains are not related or similar; (2) the feature space or data distribution of the data in the source and target domains are different; (3) the trained model cannot be applied to the source and target domains, leading to negative migration. In medical imaging, data from different modalities exhibit specific distributions, and sometimes it is necessary to encode the appearance, shape, and context simultaneously to eliminate distributional differences. Negative migration is a major challenge during the application of domain adaptation in medical tasks.
3. Proposed Method
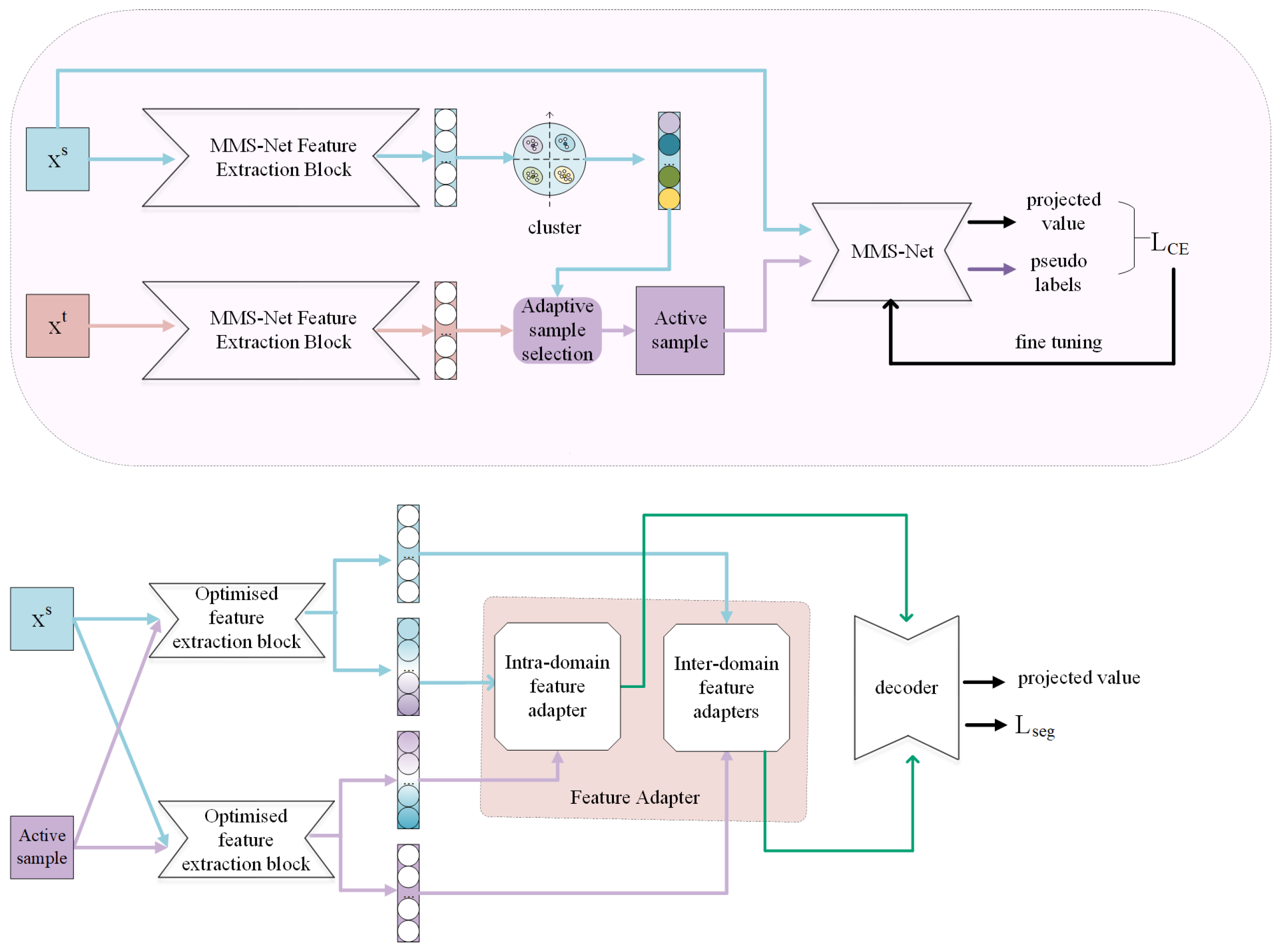
In this section, we provide an overview of the proposed network (Figure 1), and discuss the key modules in the proposed method (Figure 2 and Figure 3).
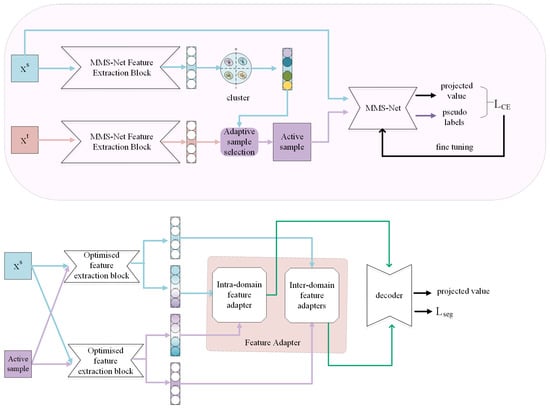
Figure 1.
Overall architecture.
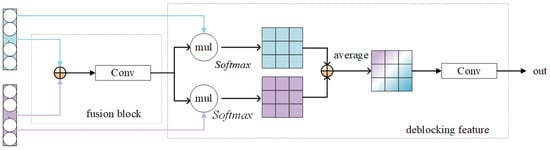
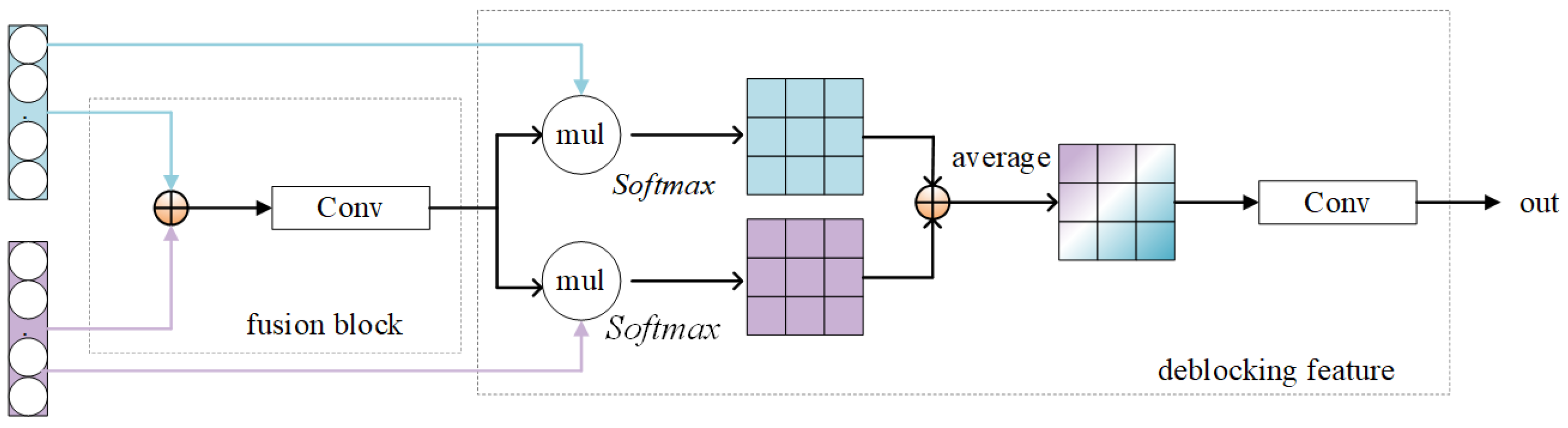
Figure 2.
Inter-domain feature adapters.
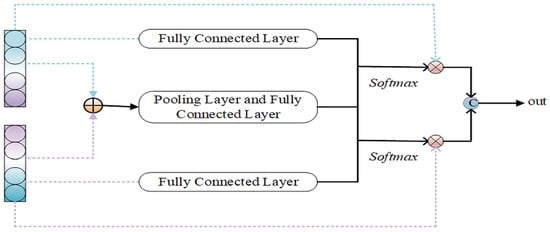
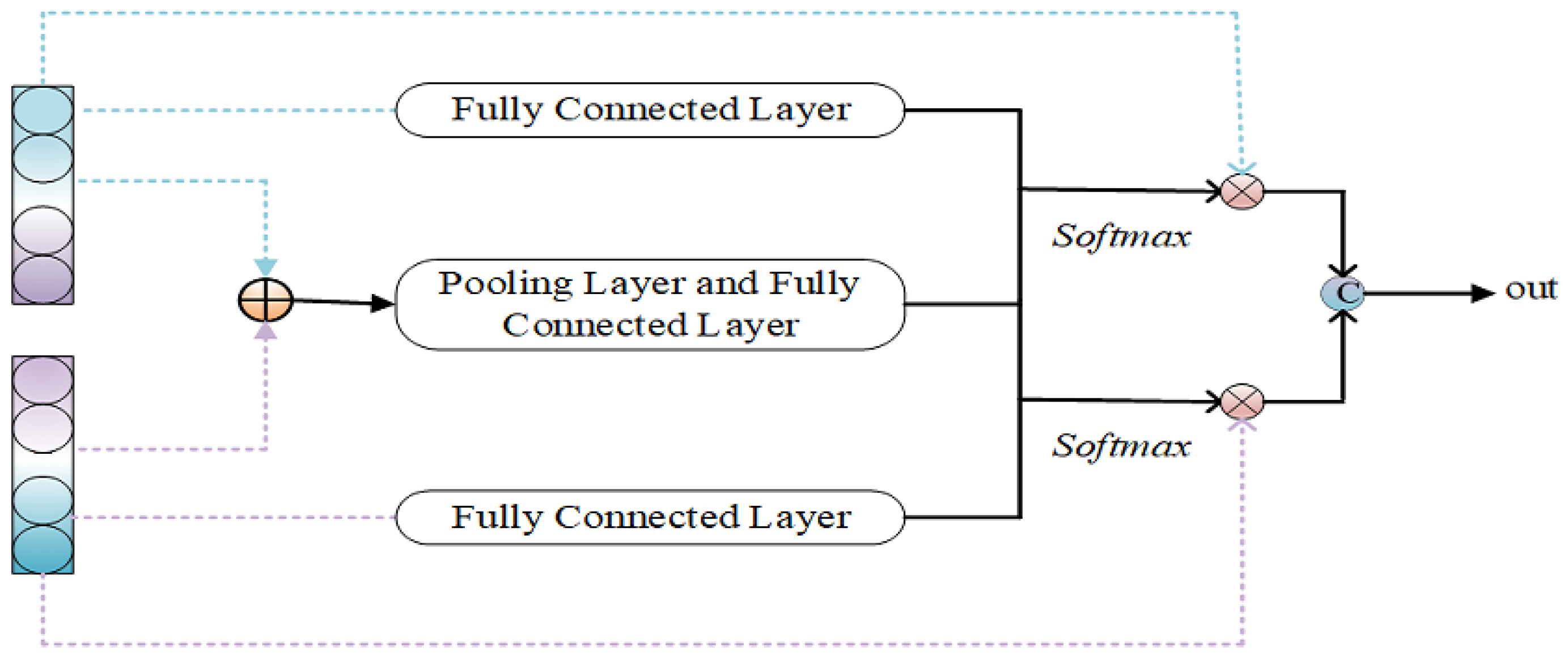
Figure 3.
Intra-domain feature adapters.
3.1. Network Framework
The overall structure is presented in Figure 1. Based on the first principle, this paper uses the MMS-Net encoder [23] as a feature extractor. This encoder can capture different modal features separately, as well as the links between different modal features. Based on the second and third principles, Firstly, use the target points in the source domain for adaptive sample selection during the pre-training stage (Figure 1). Next, the selected adaptive samples are trained together with the source domain samples, and pseudo-labels are generated for the target samples to improve the complementary ability between the source and target domains. In addition, the network is optimized using the cross-entropy loss function to make it adaptable to both the source and target domains. Subsequently, feature extraction is performed on the source domain and activity samples to obtain source-domain source-style features, source-domain target-style features, target-domain target-style features, and target-domain source-domain-style features. The extracted features are then extracted using feature adapters, so that the features are aligned to some extent, including unique features and common features. Finally, the decoder stage is entered.
3.2. Adaptive Generation of Pseudo-Labels
In multimodal image segmentation, two training methods are usually applied: one is to use only the data of the source domain for training, but this method is associated with the degradation of the performance of the network in the target domain. The other is to align the feature distributions of the source and target domains; however, this method is not applicable to multimodal images. Considering that the feature distributions of different modalities vary, if we force the alignment of the features of the source and target domains of each modality, it can destroy its internal structure, causing a degradation of the segmentation performance.
Therefore, to better characterize the source and target domains, we propose a multi-target strategy, as shown in Figure 1. The method uses the feature distributions of the source and target domains to identify the target domain samples that are most complementary to the source domain and generate pseudo labels, allowing the further calibration of the model using the cross-entropy loss function. Even when the target domain samples are small or unlabeled, the method can effectively prevent the distortion of the target domain feature distribution produced when the two domains are feature-aligned, ensuring that the model achieves excellent segmentation performance.
Specifically, the approach first employs the K-means algorithm to obtain the most representative feature targets between different modalities in the source domain, and then selects the target domain samples that are most complementary to the source domain by calculating the maximum distance between the target domain features and the targets to annotate and generate pseudo-labels. Consequently, the fusion of the features of the source and target domains is achieved without destroying the internal structure, which improves the performance of multimodal image segmentation.
3.2.1. Generate Source Domain Targets
Assuming that there are pairs of image-label pairs in the source domain and pairs of unlabeled images in the target domain, the encoder is . Initially, it passes the source-domain images through to obtain the source style features of the source domain, and calculates the source-domain image feature vectors ; the operation is presented as Equation (1). The feature vectors of all the source images are divided into K clusters using the K-means method [24], to minimize the error, and the operation is as in Equation (2).
where denotes the source domain data of all modalities, denotes the source domain data of the i-th modality among them, represents the corresponding label mapping, indicates the corresponding network output, denotes the number of its pixels, c denotes the total number of modalities, L represents the operation of spreading the feature mapping, C indicates the operation of joining the feature mappings of several modalities into a single long vector, denotes the source domain target, , and ∣∣ represents the number of images of the cluster in which the target is located.
3.2.2. Adaptive Sample Selection
In adaptive sample selection, the distance between the samples in the target domain and the targets in the source domain is first calculated, and the samples that are the farthest away and contain only information specific to the target domain are selected. This strategy helps identify samples that provide important information about the target domain and may not be well represented in the source data. This is because the greater the dissimilarity between the target samples and the source domain, the more they complement the segmentation network. Therefore, we learn the unique features of the target domain from these samples. To determine the dissimilarity between the target domain samples and the target domain of the source, the distance between them is selected as a measure. As shown in Equations (3) and (4), the pseudo code is given in Algorithm 1:
where denotes the target domain data for all modalities, is the target domain data for the i-th modality among them, is its network output, and the target-domain image feature vector is the distance.
3.2.3. Generation of Pseudo-Labels
The processes of generating pseudo-labels involve the adaptive selection of active target-domain samples and source-domain samples , which are used to train the model. In this process, a cross-entropy loss function is applied for model tuning purposes. The principal elements are as follows: (1) the adaptive selection of active target domain samples and source domain samples. To select the active target domain samples, the distance between the target domain samples and the source domain targets is calculated, and a subset of the samples exhibiting the greatest distance is chosen. The aforementioned samples are utilized to achieve the pseudo labels and subsequent training. (2) The selected target domain and source domain samples are employed to train the model and generate the corresponding pseudo-labels. The selected samples and source domain samples are then fed into the training network, where the model is optimized using the cross-entropy loss function (Equation (5)). In addition, the cross-entropy loss function is employed to calculate the losses associated with the generated pseudo-labels and to augment the total loss function. (3) The execution of (1) and (2) is repeated until the performance of the model is optimal. In this process, the samples selected each time will be different and, therefore, the generated pseudo-labels also vary. This facilitates the adaptation of the model to the distinctive characteristics of the target domain. The pseudocode algorithm is described in Algorithm 2.
| Algorithm 1 Adaptive sample selection |
|
| Algorithm 2 Pseudo-Label Generation Algorithm |
|
3.3. Feature Adapter
To make the best use of the rich information between different domains, networks that are suitable for both the source and target domains are identified. Dual-domain features are deeply mined using an encoder to derive source-domain source-style features, source-domain target-style features, target-domain source-style features, and target-domain target-style features. The domain shift problem is successfully eliminated through the use of feature-level adversarial learning of these features, while generic and unique features can be extracted from cross-domain style features. The feature adapters proposed are composed of two parts: inter-domain feature adapters and intra-domain feature adapters. The inter-domain feature adapters are applied to extract domain-specific features from different domains, whereas the intra-domain feature adapters are employed to extract common feature representations for all domains.
3.3.1. Inter-Domain Feature Adapters
Initially, the generated source-style features of the source domain and the target-style features of the target domain are fused to generate a fused feature ; next, a decoupled feature block is presented to decouple unique and invariant features, which mainly serves for feature extraction. This is because although the input is a single-domain object, its output features still contains hybrid-style features. Therefore, the inter-domain feature adapter can be regarded as the purification of the fusion features into style features. The specific structure is shown in Figure 2. The specific operation is shown in Equations (6) and (7).
3.3.2. Intra-Domain Feature Adapters
The method is primarily employed to extract common features from source-domain target-style features and target-domain source-style features. In this chapter, the common feature extraction means are used, with the specific structure illustrated in Figure 3 and the specific operation described in Equations (8)–(11).
4. Experiment
4.1. Dataset
The network models for this experiment were all implemented based on PyTorch, using the Adam optimizer, the model was an MMS-Net network, the initial learning rate was set to 0.001 and the number of iterations was decreased, the initial weight was 1, the momentum was 0.1, the batch size was 4, and the maximum training period was 2000. The publicly available BraTs18 training set dataset was used as the source-domain data, BraTs19 and BraTs21 were used as the target-domain data, and the detailed information is presented in Table 2. The BraTs21 training set includes the results of the MR scan of 1,251 patients with a data size of .

Table 2.
The details of the dataset.
4.2. K-Value Setting
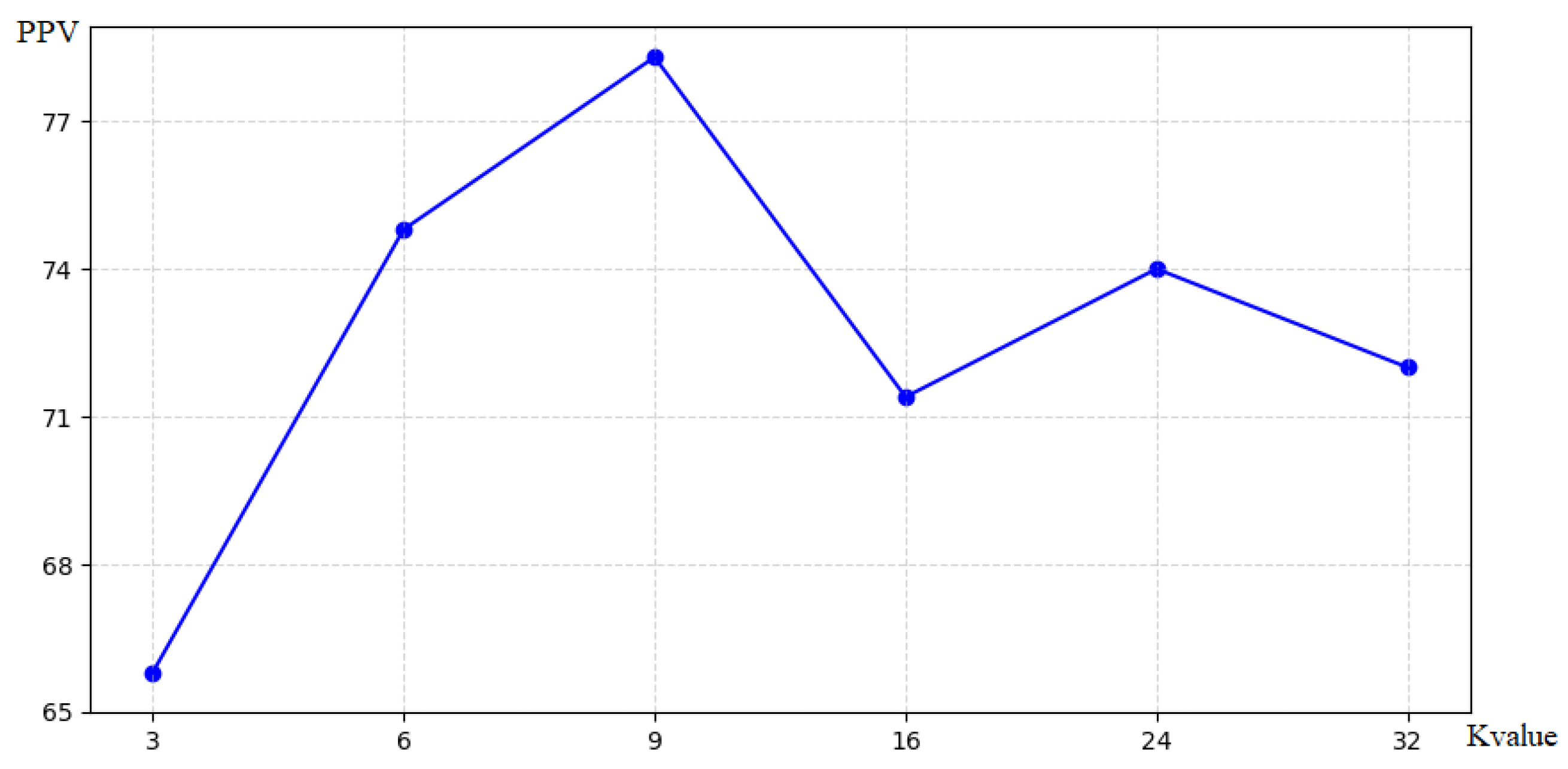
In this chapter, we evaluate the effect of different K values on the modeling of the source and target domains. For both domains, using multiple K values was consistently better than using a single center-of-mass K value, and the use of six to nine targets. achieved better performance as presented in Figure 4. Considering the best performance for both domains, eight clusters are used for averaging in this chapter.
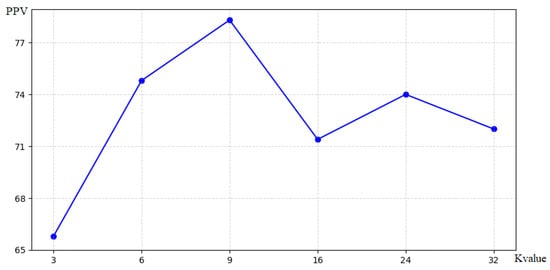
Figure 4.
Number of target spots in the source domain.
4.3. Loss Function
To achieve the alignment of the characteristic of the adapter, the maximum mean difference was selected as an estimate of the difference between the two domains. MMD is a kernel sample test based on the rejection or acceptance of the original hypothesis by a sample of observations. The principle of MMD is that all statistics are the same if the resulting distributions are the same. Thus, the MMD defines the distance between two distributions, so the MMD loss is described as follows:
where FA represents the feature adapter.
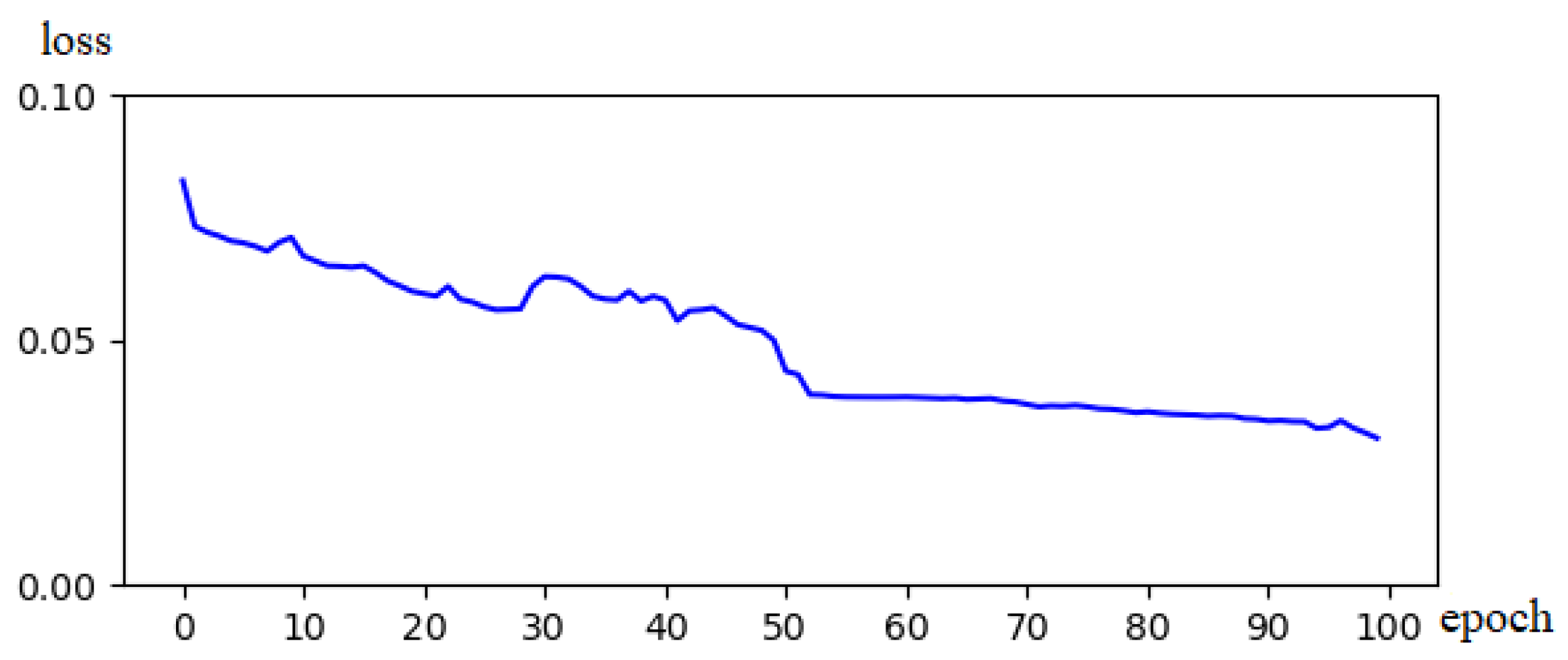
Figure 5 illustrates the variation curve of the values of the loss function during segmentation training of the BraTs18 dataset using the proposed strategy. During rounds 0–20, the network converges rapidly, and the loss decreases significantly. In round 50, the network gradually stabilizes and the loss values drop below 5%. In the experiments, several validations were conducted, and a similar trend was observed. This loss change curve confirms that the proposed strategy has a specific stability and good convergence speed.
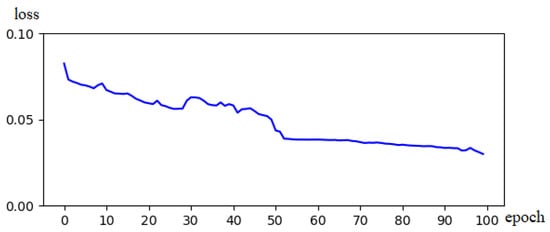
Figure 5.
Schematic diagram of the Loss of decline.
4.4. Comparison of Similar Methods
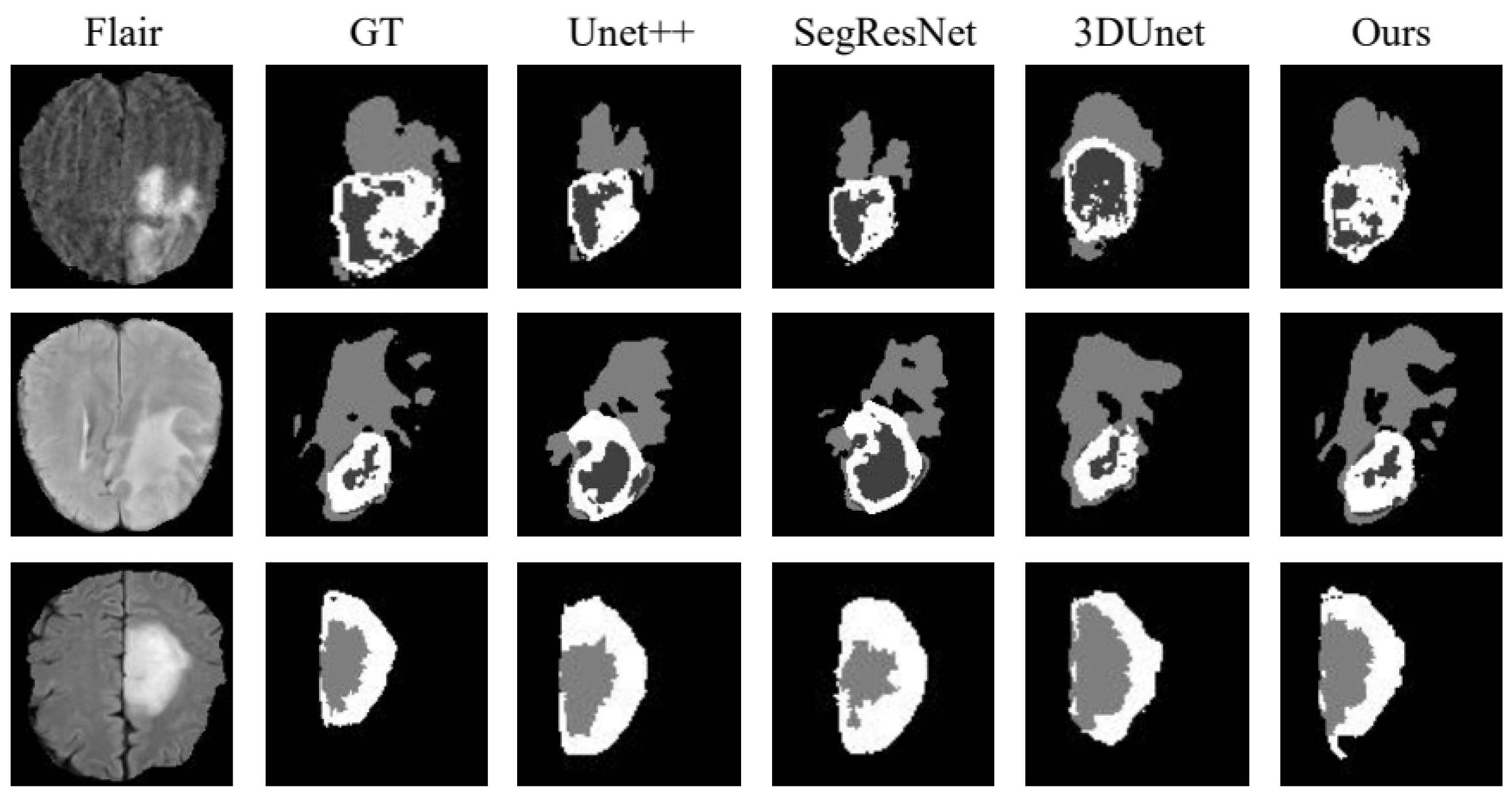
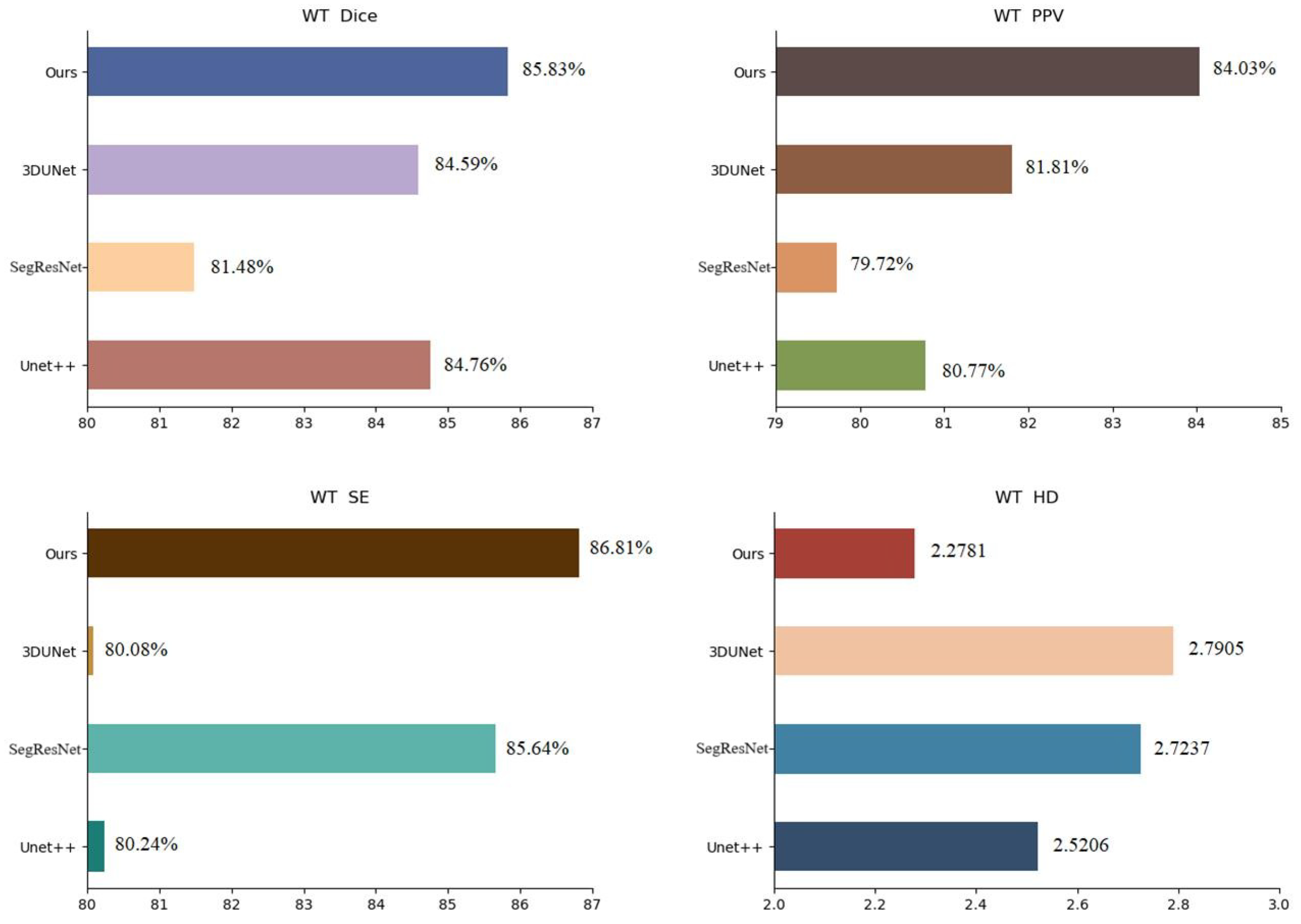
To validate the performance of the proposed method, we compared it with Unet++ [25], SegResNet [26] and 3DUnet [27] in the BraTs19 dataset. The experimental results are presented in Table 3, the visualization of the results is shown in Figure 6 and the analysis of the results is shown in Figure 7, using WT as an example. The metrics shown in Table 3 demonstrate that the segmentation strategy performs well in each metric. In terms of the WT Dice score, the method proposed in this chapter is higher than Unet++ by approximately 1.07%, SegResNet by approximately 4.35%, and 3DUnet by approximately 1.24%. The data shown in Figure 6 suggest that the proposed strategy can capture the details well in the target domain, indicating that the strategy solves the domain shift problem.

Table 3.
BraTs19 dataset division results comparison.
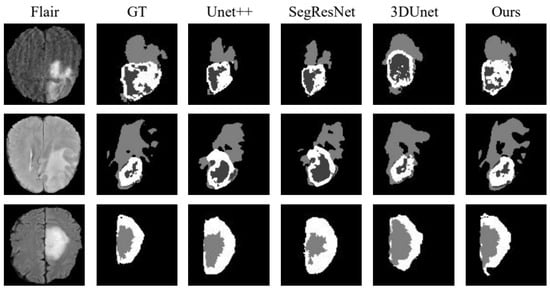
Figure 6.
BraTs19 dataset division result visualization.
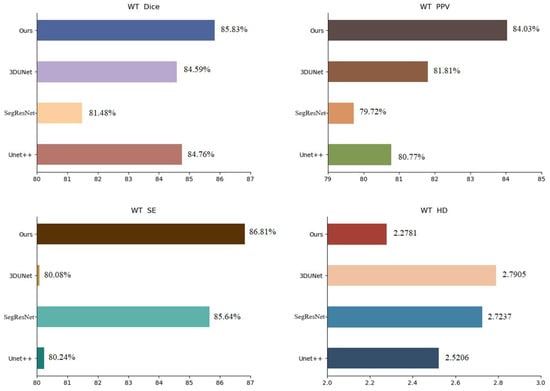
Figure 7.
BraTs19 dataset segmentation results analysis (WT).
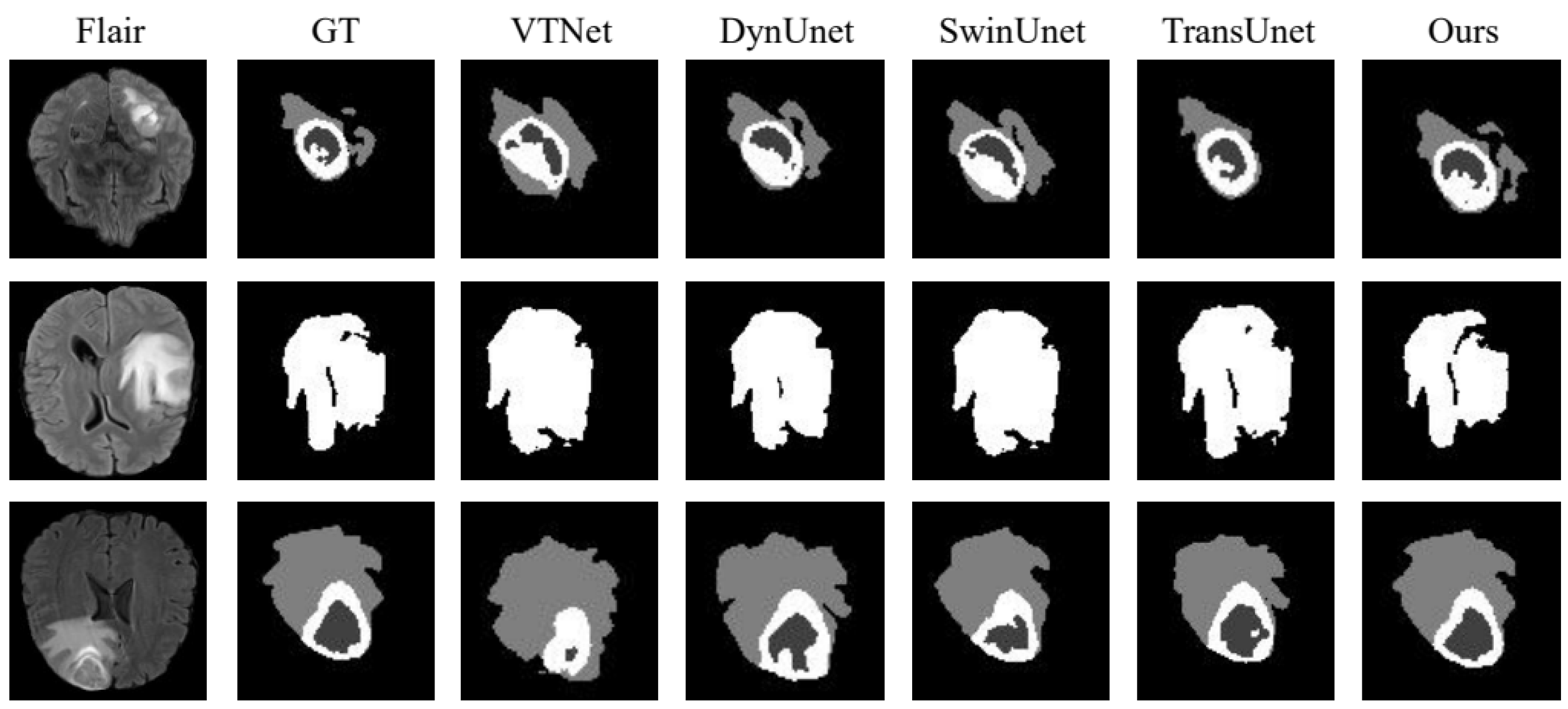
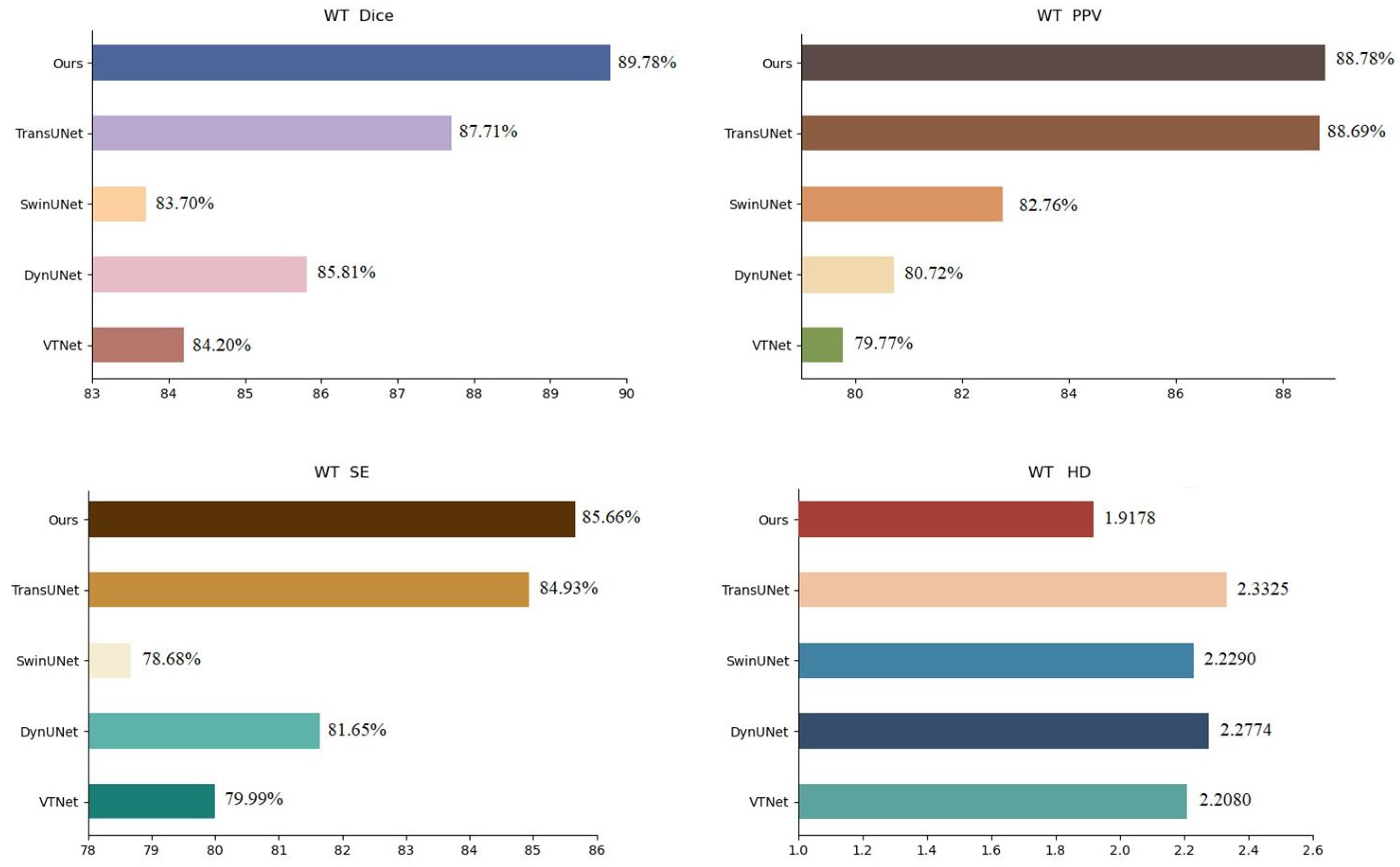
To further confirm the effectiveness of the proposed strategy, we compared it with VTNet [28], DynUNet [29], winUnet [30], and TransUnet [31] in the BraTs21 data set. The experimental results are presented in Table 4, the visualization of the results in terms of WT is shown in Figure 8 and the analysis of the results is shown in Figure 9. In Table 4, we infer that the performance of the proposed strategy in this chapter is slightly higher than that of the popular TransUnet. In Figure 8, we observe that the training of the network with both the source and active samples accurately extracted the unique features of the samples from the target domain during segmentation, which weakens the possibility of the network being skewed towards the source domain.

Table 4.
BraTs21 dataset division results comparison.
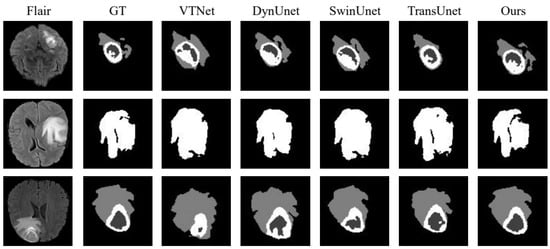
Figure 8.
BraTs21 dataset division result visualization.
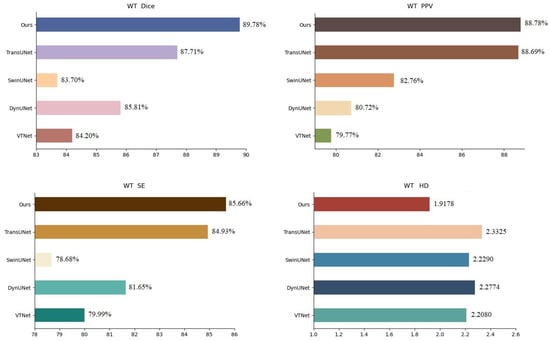
Figure 9.
BraTs21 dataset segmentation results analysis (WT).
5. Discussion
The experimental results in Section 4.4 demonstrate that the brain tumor image segmentation strategy based on domain adaptation proposed in this paper can be used to mitigate the domain shift problem and enhance the model generalization ability, thus improving the multi-modal brain tumor MR image segmentation accuracy. Experiments confirmed that adaptively selecting samples and generating pseudo-labels through the generated source domain targets can help the model to learn the unique features of different domains, and in addition, the feature adapters can improve the model’s ability to learn the common features of different domains.
A comparison with other famous methods, including Unet++, SegResNet, and 3DUnet, and VT-Net, DynUnet, swinUnet, and TransUnet on the two datasets of BraTs19 and BraTs21, respectively, showed that the proposed strategy performs better in terms of Dice scores, accuracy, precision, and HD, which proves that this network weakens the possibility of tilting towards the source domain.
In future applications, the proposed strategy is expected to provide a research direction to attenuate the effect of domain shifts on the accuracy of image segmentation.
6. Summary
In this paper, we propose a multitarget adaptive sample selection strategy based on domain adaptation to select the distribution most complementary to the source domain, and simultaneously, the most unique distribution as the active samples, mainly to avoid the distortion of the distribution of the target domain that occurs when the model is trained. In addition, feature alignment is achieved using feature adapters to prevent a strong adaptation to a single domain when training the network.
Author Contributions
Q.Y. and R.J.; methodology, Q.Y. and R.J.; software, Q.Y. and J.M.; validation, Q.Y.; investigation, Q.Y.; writing—original draft preparation, Q.Y.; writing—review and editing, J.M.; supervision, Q.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Shanxi Higher Education Institutions Science and Technology Innovation Project OF FUNDER grant number 2024L494.
Institutional Review Board Statement
Ethical review and approval were waived by the Human Research Ethics Committee of Henan Polytechnic University, as the research involved the use of existing data collections that contained only nonidentifiable data about human beings.
Informed Consent Statement
The study received a waiver of written patient consent, as all cases were anonymized, and personal identifying information was removed.
Data Availability Statement
The data used in this study are public datasets accessible at https://www.med.upenn.edu/cbica/brats-2019/ (accessed on 15 December 2022).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Biratu, E.S.; Schwenker, F.; Ayano, Y.M.; Debelee, T.G. A survey of brain tumor segmentation and classification algorithms. J. Imaging 2021, 7, 179. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Guo, H.; Pasunuru, R.; Bansal, M. Multi-source domain adaptation for text classification via distance-net bandits. AAAI Conf. Artif. Intell. 2020, 34, 7830–7838. [Google Scholar]
- Zou, H.; Yang, J.; Wu, X. Unsupervised energy-based adversarial domain adaptation for cross-domain text classification. In Proceedings of the Findings of the Association for Computational Linguistics, Online, 1–6 August 2021; pp. 1208–1218. [Google Scholar]
- Xu, L.; Gong, H.; Zhong, Y.; Wang, F.; Wang, S.; Lu, L.; Ding, J.; Zhao, C.; Tang, W.; Xu, J. Real-time monitoring of manual acupuncture stimulation parameters based on domain adaptive 3D hand pose estimation. Biomed. Signal Process. Control 2023, 83, 104681. [Google Scholar] [CrossRef]
- Zhang, X.; Ji, J.; Wang, L.; He, Z.; Liu, S. Review of video-based identification and detection methods of abnormal human behavior. Control Decis. Mak. 2022, 37, 14–27. [Google Scholar]
- Chen, Y.; Liu, Q.; Peng, D. A component-aware adaptive algorithm for pose estimation. Comput. Eng. 2018, 44, 263–270. [Google Scholar]
- Kiran, M.; Pedersoli, M.; Dolz, J.; Blais-Morin, L.A.; Granger, E. Incremental multi-target domain adaptation for object detection with efficient domain transfer. Pattern Recognit. 2022, 129, 108771. [Google Scholar]
- Oza, P.; Sindagi, V.A.; Sharmini, V.V.; Patel, V.M. Unsupervised domain adaptation of object detectors: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 4018–4040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ye, M.; Liu, Y.; Xiong, L.; Zhou, L. Multi-source unsupervised domain adaptation for object detection. Inf. Fusion 2022, 78, 138–148. [Google Scholar] [CrossRef]
- Jie, G. Research on Domain Adaptation and Semantic Association in Video Concept Detection; Beijing Jiaotong University: Beijing, China, 2016. [Google Scholar]
- Wachinger, C.; Reuter, M.; Initiative, A.D.N. Domain adaptation for Alzheimer’s disease diagnostics. Neuroimage 2016, 139, 470–479. [Google Scholar] [CrossRef]
- Goetz, M.; Weber, C.; Binczyk, F.; Polanska, J.; Tarnawski, R.; Bobek-Billewicz, B.; Koethe, U.; Kleesiek, J.; Stieltjes, B.; Maier-Hein, K.H. DALSA: Domain adaptation for supervised learning from sparsely annotated MR images. IEEE Trans. Med. Imaging 2015, 35, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Synergistic image and feature adaptation: Towards cross-modality domain adaptation for medical image segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 865–872. [Google Scholar]
- Han, X.; Qi, L.; Yu, Q.; Zhou, Z.; Zheng, Y.; Shi, Y.; Gao, Y. Deep symmetric adaptation network for cross-modality medical image segmentation. IEEE Trans. Med. Imaging 2021, 41, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Vesal, S.; Ravikumar, N.; Maier, A. Automated multi-sequence cardiac MRI segmentation using supervised domain adaptation. In Proceedings of the Statistical Atlases and Computational Models of the Heart, Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges, Shenzhen, China, 13 October 2019; Springer: Berlin/Heidelberg, Germany, 2020; pp. 300–308. [Google Scholar]
- Kushibar, K.; Valverde, S.; Gonzalez-Villa, S.; Bernal, J.; Cabezas, M.; Oliver, A.; Llado, X. Supervised domain adaptation for automatic sub-cortical brain structure segmentation with minimal user interaction. Sci. Rep. 2019, 9, 6742. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Ge, Z.; Bonnington, C.P.; Zhou, J. Progressive transfer learning and adversarial domain adaptation for cross-domain skin disease classification. IEEE J. Biomed. Health Inform. 2019, 24, 1379–1393. [Google Scholar] [CrossRef]
- Kaur, B.; Lemaître, P.; Mehta, R.; Sepahvand, N.M.; Precup, D.; Arnold, D.; Arbel, T. Improving pathological structure segmentation via transfer learning across diseases. In Proceedings of the Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data, Shenzhen, China, 13 and 17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 90–98. [Google Scholar]
- Kamphenkel, J.; Jäger, P.F.; Bickelhaupt, S.; Laun, F.B.; Lederer, W.; Daniel, H.; Kuder, T.A.; Delorme, S.; Schlemmer, H.P.; König, F.; et al. Domain adaptation for deviating acquisition protocols in CNN-based lesion classification on diffusion-weighted MR images. In Image Analysis for Moving Organ, Breast, and Thoracic Images; Springer: Berlin/Heidelberg, Germany, 2018; pp. 73–80. [Google Scholar]
- Gao, Y.; Zhang, Y.; Cao, Z.; Guo, X.; Zhang, J. Decoding brain states from fMRI signals by using unsupervised domain adaptation. IEEE J. Biomed. Health Inform. 2019, 24, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Bateson, M.; Kervadec, H.; Dolz, J.; Lombaert, H.; Ayed, I.B. Constrained domain adaptation for segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 326–334. [Google Scholar]
- Sheng, X.; Zhang, Y. Multimodal brain tumour MR image segmentation by incorporating attention mechanism. J.-Comput.-Aided Des. Graph. 2023, 35, 1429–1438. [Google Scholar]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the 5th Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 21 June–18 July 1965; p. 281. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Granada, Spain, 20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Myronenko, A. 3D MRI brain tumor segmentation using autoencoder regularization. In Brain-Lesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Berlin/Heidelberg, Germany, 2019; pp. 311–320. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; Proceedings, Part II 19. Springer: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 565–571. [Google Scholar]
- Futrega, M.; Milesi, A.; Marcinkiewicz, M.; Ribalta, P. Optimized U-Net for brain tumor segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Berlin/Heidelberg, Germany, 2022; pp. 15–29. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the Computer Vision—ECCV 2022 Workshops, Online, 23 October 2022; Springer: Berlin/Heidelberg, Germany, 2023; pp. 205–218. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).