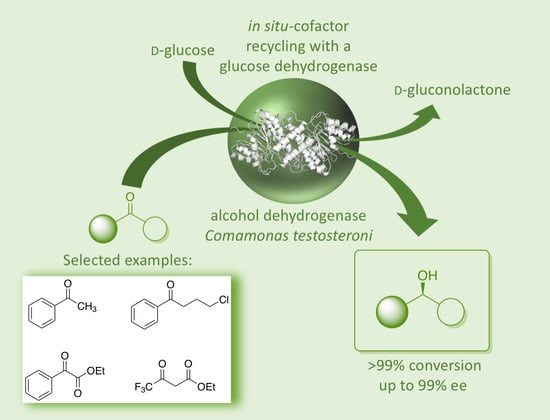
Expanding the Application Range of Microbial Oxidoreductases by an Alcohol Dehydrogenase from Comamonas testosteroni with a Broad Substrate Spectrum and pH Profile
Abstract
1. Introduction
- (i)
- ADHs require nicotinamide cofactors—NAD(H) or NADP(H)—to perform their function. These compounds (in particular NADP+ and NADPH) are expensive, which can be a limiting factor for industrial biocatalysis. A modern solution of the problem is the application of an integrated efficient recycling system that allows process operation under economic conditions. For this purpose, two cofactor regeneration systems—enzyme-coupled and substrate-coupled—have been developed and described in detail [10,11,12]. Furthermore, the co-substrate specificity might be changed from NADP(H)- to NAD(H) dependence by site-directed mutagenesis, which then allows the utilization of the less expensive cofactor NAD(H) [13].
- (ii)
- Several ADHs are comparatively pH- and thermostable [14,15], robust against stress imposed by non-aqueous solvents [16] and can be optimized in these directions. Thermostable and solvent-resistant enzymes are particularly valuable if hydrophobic substrates with solubility problems in aqueous media are processed [17].
- (iii)
- The spectrum of substrates accepted by ADHs is rather broad ranging—for example, from small alcohols such as isopropanol [18] via sugars [12] to complex steroids [19,20]—and is often accompanied by a significant and synthetically attractive enantioselectivity. Accordingly, in case an important reaction step in asymmetric synthesis is accomplished, rather than mutating a well-known ADH, it is often more promising to screen for a novel ADH activity with the required profile [1]. With the growing number of annotated but not further characterized ADHs revealed by genome sequencing, it becomes increasingly attractive to perform such screening efforts in silico [21].
2. Results and Discussion
2.1. Identification of the Alcohol Dehydrogenase Gene
2.2. Cloning and Heterologous Overexpression of CtADH
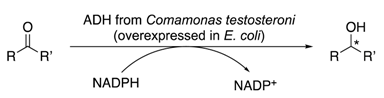
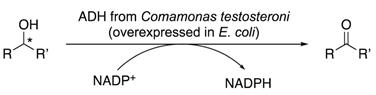
2.3. Substrate Specificity, Kinetic Parameters and Enantioselectivity of CtADH
2.4. pH-Profile of Catalytic Activity and Effect of Organic Solvents on CtADH Stability
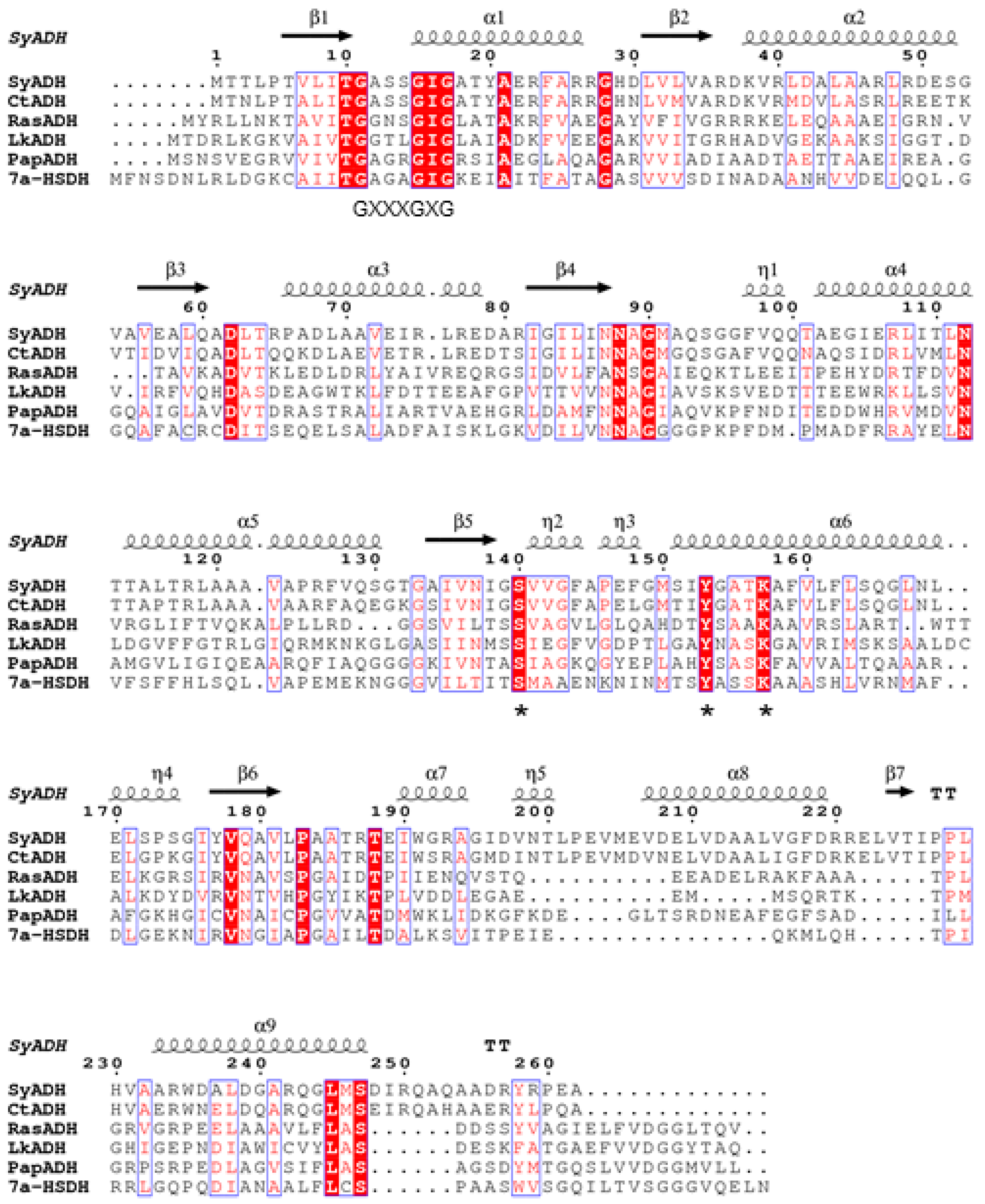
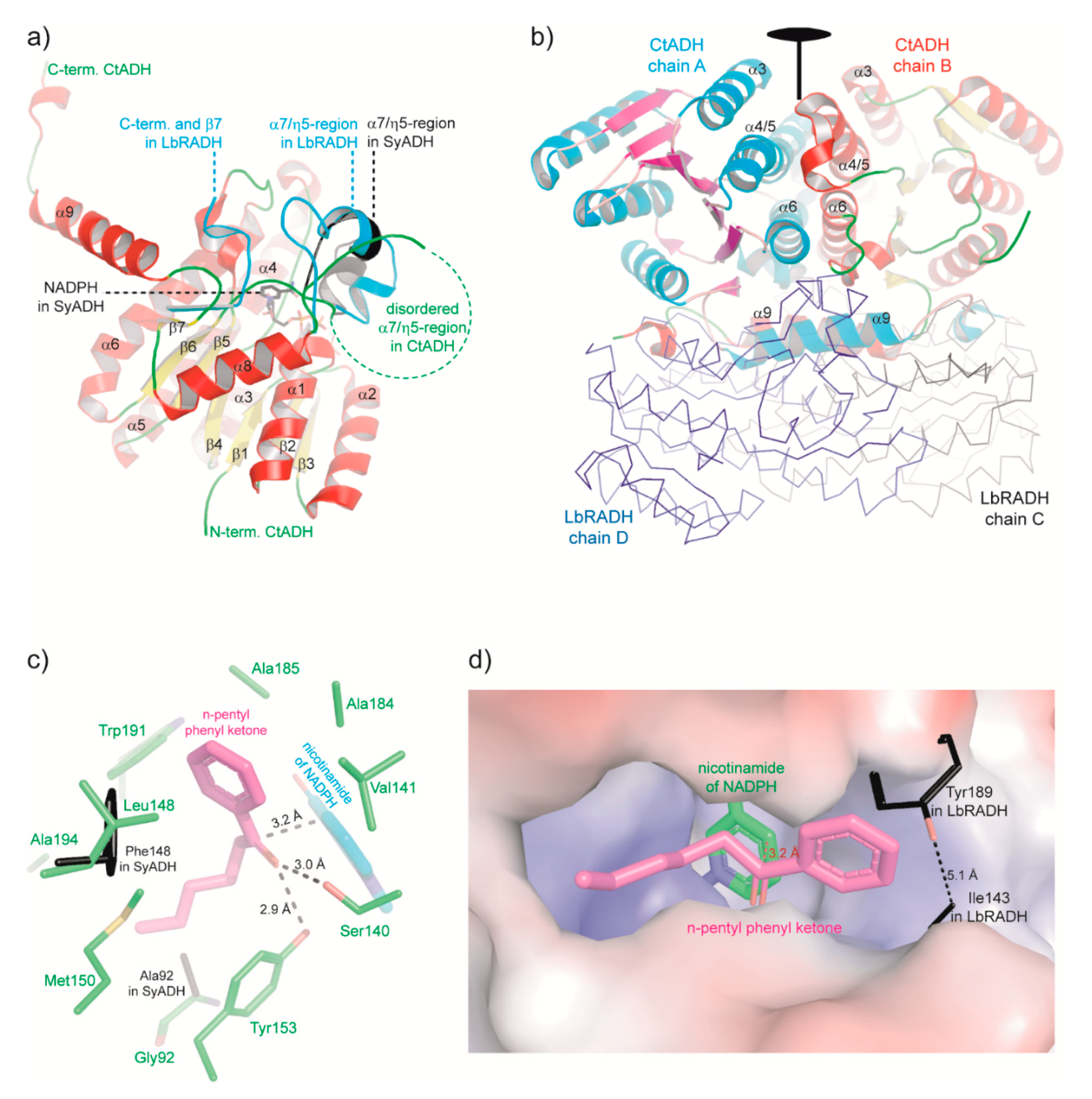
2.5. Structural Characterization of CtADH
3. Materials and Methods
3.1. Origin of Chemicals
3.2. Identification, Cloning and Heterologous Expression of CtADH and SyADH
3.3. Purification of CtADH and SyADH
3.4. Crystallization and Structure Determination of CtADH
3.5. Enzyme Activity Assays
3.6. Effect of Organic Solvents and pH on the Stability and Activity of CtADH
3.7. Determination of the Enantioselectivity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bulut, D.; Duangdee, N.; Gröger, H.; Berkessel, A.; Hummel, W.; Groeger, H. Screening, Molecular Cloning, and Biochemical Characterization of an Alcohol Dehydrogenase from Pichia pastoris Useful for the Kinetic Resolution of a Racemic β-Hydroxy-β-trifluoromethyl Ketone. ChemBioChem 2016, 17, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Gröger, H.; Chamouleau, F.; Orologas, N.; Rollmann, C.; Drauz, K.; Hummel, W.; Weckbecker, A.; May, O. Enantioselective Reduction of Ketones with “Designer Cells” at High Substrate Concentrations: Highly Efficient Access to Functionalized Optically Active Alcohols. Angew. Chem. Int. Ed. 2006, 45, 5677–5681. [Google Scholar] [CrossRef] [PubMed]
- Woodyer, R.; Wheatley, J.L.; Relyea, H.A.; Rimkus, S.; Van Der Donk, W.A. Site-Directed Mutagenesis of Active Site Residues of Phosphite Dehydrogenase. Biochemistry 2005, 44, 4765–4774. [Google Scholar] [CrossRef] [PubMed]
- Gröger, H.; Zumbrägel, N.; Wetzl, D.; Iding, H. Asymmetric Biocatalytic Reduction of Cyclic Imines: Design and Application of a Tailor-Made Whole-Cell Catalyst. Heterocycles 2017, 95, 1261. [Google Scholar] [CrossRef]
- Ni, Y.; Holtmann, D.; Hollmann, F. How green is biocatalysis? To calculate is to know. ChemCatChem 2014, 6, 930–943. [Google Scholar] [CrossRef]
- Dennig, A.; Blaschke, F.; Gandomkar, S.; Tassano, E.; Nidetzky, B. Preparative Asymmetric Synthesis of Canonical and Non-canonical α-amino Acids Through Formal Enantioselective Biocatalytic Amination of Carboxylic Acids. Adv. Synth. Catal. 2019, 361, 1348–1358. [Google Scholar] [CrossRef]
- Zhu, Y.-H.; Liu, C.-Y.; Cai, S.; Guo, L.-B.; Kim, I.-W.; Kalia, V.C.; Lee, J.-K.; Zhang, Y.-W. Cloning, Expression and Characterization of a Highly Active Alcohol Dehydrogenase for Production of Ethyl (S)-4-Chloro-3-Hydroxybutyrate. Indian J. Microbiol. 2019, 59, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Yin, S.; Wang, H.; Yang, H.; Xu, R.; Wang, Q.; Wu, Y.; Boutet, J.; Huet, R.; Wang, C. Production of Methionol from 3-Methylthiopropionaldehyde by Catalysis of the Yeast Alcohol Dehydrogenase Adh4p. J. Agric. Food Chem. 2020, 68, 4650–4656. [Google Scholar] [CrossRef]
- Kallberg, Y.; Oppermann, U.; Jörnvall, H.; Persson, B. Short-chain dehydrogenase/reductase (SDR) relationships: A large family with eight clusters common to human, animal, and plant genomes. Protein Sci. 2009, 11, 636–641. [Google Scholar] [CrossRef]
- Hummel, W.; Gröger, H. Strategies for regeneration of nicotinamide coenzymes emphasizing self-sufficient closed-loop recycling systems. J. Biotechnol. 2014, 191, 22–31. [Google Scholar] [CrossRef]
- Weckbecker, A.; Gröger, H.; Hummel, W. Regeneration of Nicotinamide Coenzymes: Principles and Applications for the Synthesis of Chiral Compounds. Biosyst. Eng. I 2010, 120, 195–242. [Google Scholar] [CrossRef]
- Weckbecker, A.; Hummel, W. Glucose Dehydrogenase for the Regeneration of NADPH and NADH; Barredo, J.L., Ed.; Humana Press: Totawa, NJ, USA, 2005; pp. 225–238. [Google Scholar]
- Schlieben, N.H.; Niefind, K.; Müller, J.; Riebel, B.; Hummel, W.; Schomburg, D. Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity. J. Mol. Biol. 2005, 349, 801–813. [Google Scholar] [CrossRef]
- Machielsen, R.; Uria, A.R.; Kengen, S.W.M.; Van Der Oost, J. Production and Characterization of a Thermostable Alcohol Dehydrogenase That Belongs to the Aldo-Keto Reductase Superfamily. Appl. Environ. Microbiol. 2006, 72, 233–238. [Google Scholar] [CrossRef]
- Petratos, K.; Gessmann, R.; Daskalakis, V.; Papadovasilaki, M.; Papanikolau, Y.; Tsigos, I.; Bouriotis, V. Structure and Dynamics of a Thermostable Alcohol Dehydrogenase from the Antarctic Psychrophile Moraxella sp. TAE123. ACS Omega 2020, 5, 14523–14534. [Google Scholar] [CrossRef] [PubMed]
- Lavandera, I.; Kern, A.; Schaffenberger, M.; Gross, J.; Glieder, A.; De Wildeman, S.; Kroutil, W. An Exceptionally DMSO-Tolerant Alcohol Dehydrogenase for the Stereoselective Reduction of Ketones. ChemSusChem 2008, 1, 431–436. [Google Scholar] [CrossRef]
- Lavandera, I.; Kern, A.; Resch, V.; Ferreira-Silva, B.; Glieder, A.; Fabian, W.M.F.; De Wildeman, S.; Kroutil, W. One-Way Biohydrogen Transfer for Oxidation of sec-Alcohols. Org. Lett. 2008, 10, 2155–2158. [Google Scholar] [CrossRef]
- Hu, J.; Li, G.; Liang, C.; Shams, S.; Zhu, S.; Zheng, G. Stereoselective synthesis of the key intermediate of ticagrelor and its diverse analogs using a new alcohol dehydrogenase from Rhodococcus kyotonensis. Process Biochem. 2020, 92, 232–243. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Kudo, T. Steroid degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 2012, 129, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Kurita, T.; Hayashi, T.; Kudo, T. Steroid degradation genes in Comamonas testosteroni TA441: Isolation of genes encoding a Δ4(5)-isomerase and 3α- and 3β-dehydrogenases and evidence for a 100kb steroid degradation gene hot spot. J. Steroid Biochem. Mol. Biol. 2010, 122, 253–263. [Google Scholar] [CrossRef]
- Wang, L.-J.; Li, C.-X.; Ni, Y.; Zhang, J.; Liu, X.; Xu, J.-H. Highly efficient synthesis of chiral alcohols with a novel NADH-dependent reductase from Streptomyces coelicolor. Bioresour. Technol. 2011, 102, 7023–7028. [Google Scholar] [CrossRef] [PubMed]
- Niefind, K.; Müller, J.; Riebel, B.; Hummel, W.; Schomburg, I. The crystal structure of R-specific alcohol dehydrogenase from Lactobacillus brevis suggests the structural basis of its metal dependency. J. Mol. Biol. 2003, 327, 317–328. [Google Scholar] [CrossRef]
- Lavandera, I.; Kern, A.; Ferreira-Silva, B.; Glieder, A.; De Wildeman, S.; Kroutil, W. Stereoselective Bioreduction of Bulky-Bulky Ketones by a Novel ADH from Ralstonia sp. J. Org. Chem. 2008, 73, 6003–6005. [Google Scholar] [CrossRef] [PubMed]
- Kulig, J.; Frese, A.; Kroutil, W.; Pohl, M.; Rother, D. Biochemical characterization of an alcohol dehydrogenase from Ralstonia sp. Biotechnol. Bioeng. 2013, 110, 1838–1848. [Google Scholar] [CrossRef]
- Lavandera, I.; Oberdorfer, G.; Gross, J.; De Wildeman, S.; Kroutil, W. Stereocomplementary Asymmetric Reduction of Bulky–Bulky Ketones by Biocatalytic Hydrogen Transfer. Eur. J. Org. Chem. 2008, 2008, 2539–2543. [Google Scholar] [CrossRef]
- Man, H.; Kedziora, K.W.; Kulig, J.K.; Frank, A.; Lavandera, I.; Gotor-Fernández, V.; Rother, D.; Hart, S.; Turkenburg, J.P.; Grogan, G. Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium spp. Reveal the Molecular Basis for Their Recognition of ‘Bulky–Bulky’ Ketones. Top. Catal. 2013, 57, 356–365. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Wang, C.-H.; Yang, F.-C.; Ismail, W.; Wang, P.-H.; Shih, C.-J.; Wu, Y.-C.; Chiang, Y.-R. Identification of Comamonas testosteroni as an androgen degrader in sewage. Sci. Rep. 2016, 6, 35386. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Hosoyama, A.; Tsuchikane, K.; Ohji, S.; Yamazoe, A.; Fujita, N.; Shintani, M.; Kimbara, K. Complete Genome Sequence of Polychlorinated Biphenyl Degrader Comamonas testosteroni TK102 (NBRC 109938). Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-F.; Zhang, Y.; Zhang, J.-Y.; Chen, D.; Zhu, Y.; Zheng, H.; Wang, S.-Y.; Jiang, C.-Y.; Zhao, G.-P.; Liu, S.-J. The Complete Genome of Comamonas testosteroni Reveals Its Genetic Adaptations to Changing Environments. Appl. Environ. Microbiol. 2009, 75, 6812–6819. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, W.; Cao, Z.; Xu, B.; Wang, G.; Wang, G. High correlation between genotypes and phenotypes of environmental bacteria Comamonas testosteroni strains. BMC Genom. 2015, 16, 110. [Google Scholar] [CrossRef]
- Liu, C.; Liu, K.; Zhao, C.; Gong, P.; Yu, Y. The characterization of a short chain dehydrogenase/reductase (SDRx) in Comamonas testosteroni. Toxicol. Rep. 2020, 7, 460–467. [Google Scholar] [CrossRef]
- Liu, L.; Jiang, C.; Liu, X.-Y.; Wu, J.-F.; Han, J.-G.; Liu, S.-J. Plant-microbe association for rhizoremediation of chloronitroaromatic pollutants with Comamonas sp. strain CNB-1. Environ. Microbiol. 2007, 9, 465–473. [Google Scholar] [CrossRef]
- Loffhagen, N.; Babel, W. Energization of Comamonas testosteroni ATCC 17454 for Indicating Toxic Effects of Chlorophenoxy Herbicides. Arch. Environ. Contam. Toxicol. 2003, 45, 317–323. [Google Scholar] [CrossRef]
- Benson, D.A.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2014, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, A.S.; Simon, R.C.; Grischek, B.; De Paula, G.C.; Horta, B.A.C.; De Miranda, L.S.M.; Kroutil, W.; Kappe, C.O.; De Souza, R.O.M.A. Chiral Chlorohydrins from the Biocatalyzed Reduction of Chloroketones: Chiral Building Blocks for Antiretroviral Drugs. ChemCatChem 2015, 7, 984–992. [Google Scholar] [CrossRef]
- Yang, Z.-Y.; Ye, W.-J.; Xie, Y.-Y.; Liu, Q.-H.; Chen, R.; Wang, H.-L.; Wei, D.-Z. Efficient Asymmetric Synthesis of Ethyl (S)-4-Chloro-3-hydroxybutyrate Using Alcohol Dehydrogenase SmADH31 with High Tolerance of Substrate and Product in a Monophasic Aqueous System. Org. Process Res. Dev. 2020, 24, 1068–1076. [Google Scholar] [CrossRef]
- Liu, Z.-Q.; Dong, S.-C.; Yin, H.-H.; Xue, Y.; Tang, X.-L.; Zhang, X.-J.; He, J.-Y.; Zheng, Y.-G. Enzymatic synthesis of an ezetimibe intermediate using carbonyl reductase coupled with glucose dehydrogenase in an aqueous-organic solvent system. Bioresour. Technol. 2017, 229, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. Biocatalysis in Organic Chemistry and Biotechnology: Past, Present, and Future. J. Am. Chem. Soc. 2013, 135, 12480–12496. [Google Scholar] [CrossRef]
- Riebel, B. Biochemische und molekularbiologische Charakterisierung neuer mikrobieller NAD(P)-abhängiger Alkoholdehydrogenasen. Ph.D. Thesis, Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany, 1996. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 293–302. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P.R. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 62, 72–82. [Google Scholar] [CrossRef]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of theCCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Tickle, I.J.; Flensburg, C.; Keller, P.; Paciorek, W.; Sharff, A.; Vonrhein, C.; Bricogne, G. STARANISO; Global Phasing Ltd.: Cambridge, UK, 2018. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, version 1.7.0.3; Schrödinger, LLC: New York, NY, USA, 2012.
- Vázquez-Figueroa, E.; Chaparro-Riggers, J.; Bommarius, A.S. Development of a Thermostable Glucose Dehydrogenase by a Structure-Guided Consensus Concept. ChemBioChem 2007, 8, 2295–2301. [Google Scholar] [CrossRef]

| # | Substrate | Concentration [mM] | Spec. Activity [U mg−1] |
|---|---|---|---|
| 1 | 2-aminoacetophenone hydrochloride | 50 | 0.1 |
| 2 | 4-phenyl-2-butanone | 10 | 8.1 |
| 3 | ethyl 2-oxo-4-phenylbutyrate | 10 | 0.4 |
| 4 | 4-chlorobutyrophenone | <10 | 3.7 |
| 5 | acetophenone | 10 | 0.8 |
| 6 | propiophenone | 10 | 1.4 |
| 7 | (R)-carvone | <10 | 0.5 |
| 8 | ethyl 4-chloroacetoacetate | 10 | 3.4 |
| 9 | cyclohexanone | 10 | 1.2 |
| 10 | ethyl benzoylformate | 1 | 82.9 |
| 11 | ethyl acetoacetate | 50 | 18.5 |
| 12 | 2,2,2-trifluoroacetophenone | 10 | 127.7 |
| 13 | 4-hydroxy-2-butanone | 50 | 0.3 |
| 14 | 2-heptanone | 10 | 6.1 |
| 15 | 2,3-pentandione | 50 | 85.9 |
| 16 | 4-nonanone | <10 | 2.8 |
| 17 | 2-octanone | <10 | 5.6 |
| 18 | ethyl 4,4,4-trifluoroacetoacetate | 50 | 4.6 |
| 19 | 2-chlorocyclopentanone | 25 | 12.1 |
| 20 | 2-chlorocyclohexanone | 25 | 64.2 |
| # | Substrate | Spec. Activity [U mg−1] |
|---|---|---|
| 21 | 4-phenyl-2-butanol | 3.4 |
| 22 | rac-1-phenylethanol | 1.0 |
| 23 | (R)-1-phenylethanol | 0.1 |
| 24 | (S)-1-phenylethanol | 0.7 |
| 25 | rac-1-phenylpropanol | 1.0 |

| Substrate | CtADH | SyADH | ||
|---|---|---|---|---|
| Conversion [%] | Selectivity [ee] | Conversion [%] | Selectivity [ee] | |
| acetophenone | >99 | (S) 97% | >99 | (S) 98% |
| ethyl benzoylformate | >99 | (S) 65% | >99 | (S) 57% |
| ethyl 4-chloroacetoacetate | >99 | (S) 96% | >99 | (R) 41% |
| 4-chlorobutyrophenone | >99 | (S) 99% | >99 | (S) 99% |
| 2,2,2-trifluoroacetophenone | >99 | (S) 77% | >99 | (S) 58% |
| ethyl 4,4,4-trifluoroacetoacetate | >99 | (S) 99% | >99 | (S) 83% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakonyi, D.; Toelzer, C.; Stricker, M.; Hummel, W.; Niefind, K.; Gröger, H. Expanding the Application Range of Microbial Oxidoreductases by an Alcohol Dehydrogenase from Comamonas testosteroni with a Broad Substrate Spectrum and pH Profile. Catalysts 2020, 10, 1281. https://doi.org/10.3390/catal10111281
Bakonyi D, Toelzer C, Stricker M, Hummel W, Niefind K, Gröger H. Expanding the Application Range of Microbial Oxidoreductases by an Alcohol Dehydrogenase from Comamonas testosteroni with a Broad Substrate Spectrum and pH Profile. Catalysts. 2020; 10(11):1281. https://doi.org/10.3390/catal10111281
Chicago/Turabian StyleBakonyi, Daniel, Christine Toelzer, Michael Stricker, Werner Hummel, Karsten Niefind, and Harald Gröger. 2020. "Expanding the Application Range of Microbial Oxidoreductases by an Alcohol Dehydrogenase from Comamonas testosteroni with a Broad Substrate Spectrum and pH Profile" Catalysts 10, no. 11: 1281. https://doi.org/10.3390/catal10111281
APA StyleBakonyi, D., Toelzer, C., Stricker, M., Hummel, W., Niefind, K., & Gröger, H. (2020). Expanding the Application Range of Microbial Oxidoreductases by an Alcohol Dehydrogenase from Comamonas testosteroni with a Broad Substrate Spectrum and pH Profile. Catalysts, 10(11), 1281. https://doi.org/10.3390/catal10111281