Membrane Protein Modified Electrodes in Bioelectrocatalysis
Abstract
:1. Introduction
2. Membrane Proteins in Biofuel Conversion and Photosynthesis
2.1. Membrane Enzymes in Biofuel Conversion
2.2. Membrane Proteins in Photosynthesis
3. Membrane Protein Electrode Design
3.1. Unmodified Electrode
3.2. SAM Modified Electrode
3.3. Nanoparticle Modified Electrode
3.4. Redox Polymers
3.5. Membrane Modified Electrode
3.6. Multi-Layer Assembly, Multilayered Lipid Membrane Stacks, 3D Structure Electrode
4. Methods to Characterise Membrane Protein Modified Electrode
4.1. Electrochemical Methods
4.2. Spectroscopic Methods
4.3. Spectroelectrochemistry
4.4. Microscopy
4.4.1. Electron Microscopy
4.4.2. Atomic Force Microscopy
4.5. Quartz Crystal Microbalance
5. Membrane Protein Modified Electrodes for Bioelectrocatalysis
5.1. Enzymatic Biofuel Cells (EBCs)

5.2. Biophotoelectrocatalysis (PEC)
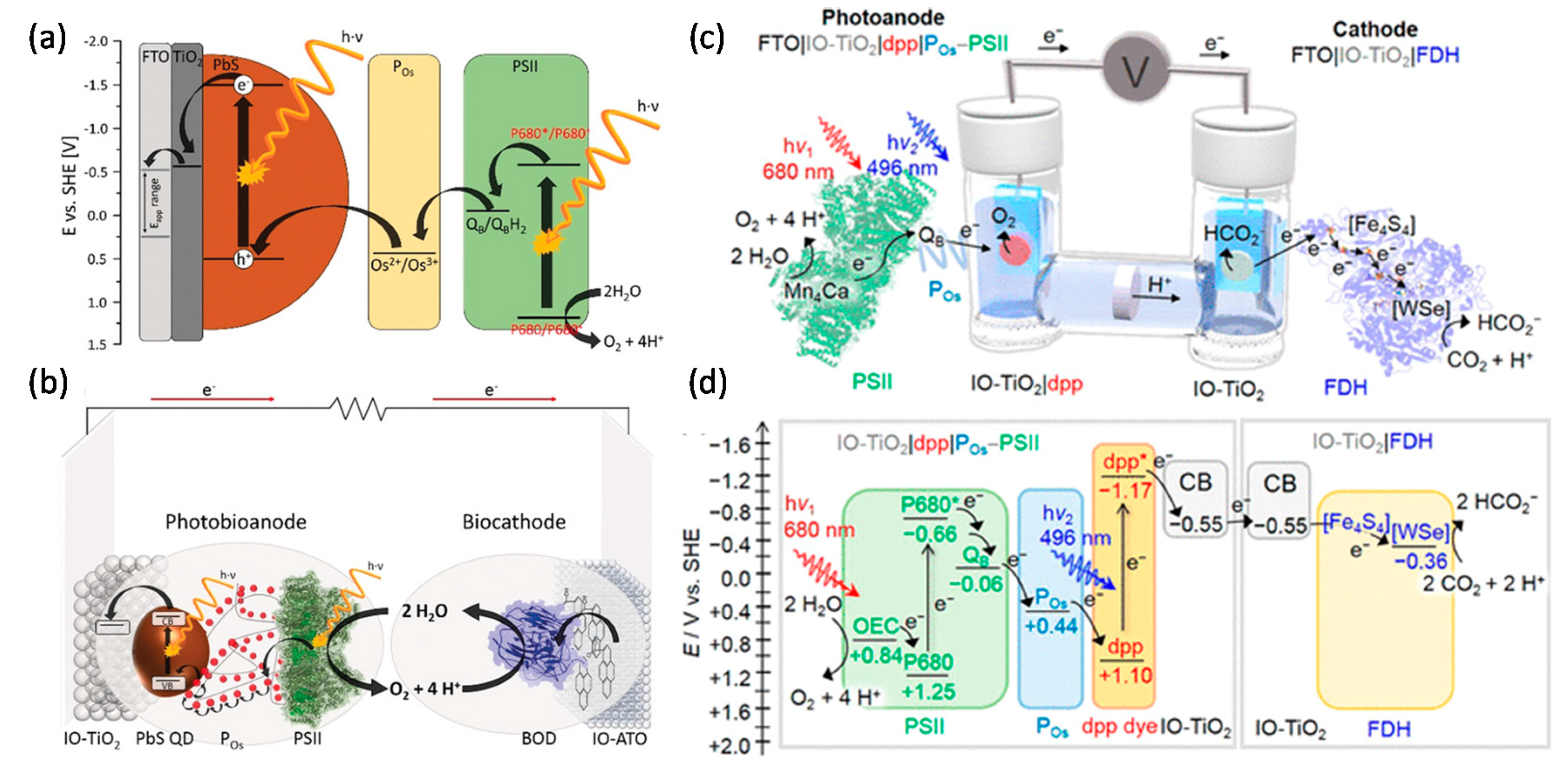
6. Emerging Methods and Future Considerations
6.1. Extending the Lifetime of Membrane Enzymes
6.2. Microbial Electrosynthesis and Whole-Cell Semi-Artificial Photosynthesis
6.2.1. Microbial Electrosynthesis
6.2.2. Whole-Cell Based Semi-Artificial Photosynthesis
7. Conclusions
Funding
Conflicts of Interest
References
- von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006, 7, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Hedin, L.E.; Illergård, K.; Elofsson, A. An introduction to membrane proteins. J. Proteome Res. 2011, 10, 3324–3331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.; Tan, H.T.; Chung, M.C.M. Membrane proteins and membrane proteomics. Proteomics 2008, 8, 3924–3932. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Kim, S.-H.; Lay, C.-H.; Ponnusamy, V.K. Recent developments on alternative fuels, energy and environment for sustainability. Bioresour. Technol. 2020, 317, 124010. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, N.; Zhang, J.Z.; Sakimoto, K.K.; Yang, P.; Reisner, E. Interfacing nature’s catalytic machinery with synthetic materials for semi-artificial photosynthesis. Nat. Nanotechnol. 2018, 13, 890–899. [Google Scholar] [CrossRef]
- Vignais, P.M.; Billoud, B.; Meyer, J. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 2001, 25, 455–501. [Google Scholar] [CrossRef]
- Shima, S.; Pilak, O.; Vogt, S.; Schick, M.; Stagni, M.S.; Meyer-Klaucke, W.; Warkentin, E.; Thauer, R.K.; Ermler, U. The crystal structure of Fe-hydrogenase reveals the geometry of the active site. Science 2008, 321, 572–575. [Google Scholar] [CrossRef] [Green Version]
- Fritsch, J.; Lenz, O.; Friedrich, B. Structure, function and biosynthesis of O2-tolerant hydrogenases. Nat. Rev. Microbiol. 2013, 11, 106–114. [Google Scholar] [CrossRef]
- Sousa, F.L.; Alves, R.J.; Ribeiro, M.A.; Pereira-Leal, J.B.; Teixeira, M.; Pereira, M.M. The superfamily of heme-copper oxygen reductases: Types and evolutionary considerations. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Borisov, V.B.; Gennis, R.B.; Hemp, J.; Verkhovsky, M.I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 1398–1413. [Google Scholar] [CrossRef] [Green Version]
- Vanlerberghe, G.C. Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 2013, 14, 6805–6847. [Google Scholar] [CrossRef] [PubMed]
- García-Horsman, J.A.; Barquera, B.; Rumbley, J.; Ma, J.; Gennis, R.B. The superfamily of heme-copper respiratory oxidases. J. Bacteriol. 1994, 176, 5587–5600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safarian, S.; Hahn, A.; Mills, D.J.; Radloff, M.; Eisinger, M.L.; Nikolaev, A.; Meier-Credo, J.; Melin, F.; Miyoshi, H.; Gennis, R.B.; et al. Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science 2019, 366, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Vivián, C.; Cabello, P.; Martínez-Luque, M.; Blasco, R.; Castillo, F. Prokaryotic nitrate reduction: Molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 1999, 181, 6573–6584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berks, B.C.; Ferguson, S.J.; Moir, J.W.B.; Richardson, D.J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta Bioenerg. 1995, 1232, 97–173. [Google Scholar] [CrossRef] [Green Version]
- Bertero, M.G.; Rothery, R.A.; Palak, M.; Hou, C.; Lim, D.; Blasco, F.; Weiner, J.H.; Strynadka, N.C.J. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 2003, 10, 681–687. [Google Scholar] [CrossRef]
- Nelson, N.; Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 2004, 5, 971–982. [Google Scholar] [CrossRef]
- Gao, J.; Wang, H.; Yuan, Q.; Feng, Y. Structure and Function of the Photosystem Supercomplexes. Front. Plant. Sci. 2018, 9, 357. [Google Scholar] [CrossRef]
- Hu, X.; Damjanović, A.; Ritz, T.; Schulten, K. Architecture and mechanism of the light-harvesting apparatus of purple bacteria. Proc. Natl. Acad. Sci. USA 1998, 95, 5935–5941. [Google Scholar] [CrossRef] [Green Version]
- Bahatyrova, S.; Frese, R.N.; Siebert, C.A.; Olsen, J.D.; van der Werf, K.O.; van Grondelle, R.; Niederman, R.A.; Bullough, P.A.; Otto, C.; Hunter, C.N. The native architecture of a photosynthetic membrane. Nature 2004, 430, 1058–1062. [Google Scholar] [CrossRef]
- Mirkovic, T.; Ostroumov, E.E.; Anna, J.M.; van Grondelle, R.; Govindjee; Scholes, G.D. Light Absorption and Energy Transfer in the Antenna Complexes of Photosynthetic Organisms. Chem. Rev. 2017, 117, 249–293. [Google Scholar] [CrossRef] [PubMed]
- Plumeré, N.; Nowaczyk, M.M. Biophotoelectrochemistry of Photosynthetic Proteins. In Biophotoelectrochemistry: From Bioelectrochemistry to Biophotovoltaics; Jeuken, L., Ed.; Springer: Cham, Switzerland, 2016; Volume 158, pp. 111–136. [Google Scholar]
- Yehezkeli, O.; Tel-Vered, R.; Michaeli, D.; Willner, I.; Nechushtai, R. Photosynthetic reaction center-functionalized electrodes for photo-bioelectrochemical cells. Photosynth. Res. 2014, 120, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Simoska, O.; Lim, K.; Grattieri, M.; Yuan, M.; Dong, F.; Lee, Y.S.; Beaver, K.; Weliwatte, S.; Gaffney, E.M.; et al. Fundamentals, Applications, and Future Directions of Bioelectrocatalysis. Chem. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, E.P.; Beis, K.; Cameron, A.D.; Iwata, S. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 2008, 18, 581–586. [Google Scholar] [CrossRef] [Green Version]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta 2004, 1666, 105–117. [Google Scholar] [CrossRef] [Green Version]
- Palazzo, G. Colloidal aspects of photosynthetic membrane proteins. Curr. Opin. Colloid Interface Sci. 2006, 11, 65–73. [Google Scholar] [CrossRef]
- Rawlings, A.E. Membrane proteins: Always an insoluble problem? Biochem. Soc. Trans. 2016, 44, 790–795. [Google Scholar] [CrossRef]
- Gillam, E.M.J. Engineering cytochrome P450 enzymes. Chem. Res. Toxicol. 2008, 21, 220–231. [Google Scholar] [CrossRef]
- Hunte, C.; Richers, S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 2008, 18, 406–411. [Google Scholar] [CrossRef]
- Mazurenko, I.; Hitaishi, V.P.; Lojou, E. Recent advances in surface chemistry of electrodes to promote direct enzymatic bioelectrocatalysis. Curr. Opin. Electrochem. 2020, 19, 113–121. [Google Scholar] [CrossRef]
- Yates, N.D.J.; Fascione, M.A.; Parkin, A. Methodologies for “Wiring” Redox Proteins/Enzymes to Electrode Surfaces. Chem. Eur. J. 2018, 24, 12164–12182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sucheta, A.; Ackrell, B.A.; Cochran, B.; Armstrong, F.A. Diode-like behaviour of a mitochondrial electron-transport enzyme. Nature 1992, 356, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.J.; Richardson, D.J.; Butt, J.N. Catalytic protein film voltammetry from a respiratory nitrate reductase provides evidence for complex electrochemical modulation of enzyme activity. Biochemistry 2001, 40, 11294–11307. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.J.; Hoke, K.R.; Heffron, K.; Palak, M.; Rothery, R.A.; Weiner, J.H.; Armstrong, F.A. Voltammetric studies of the catalytic mechanism of the respiratory nitrate reductase from Escherichia coli: How nitrate reduction and inhibition depend on the oxidation state of the active site. Biochemistry 2004, 43, 799–807. [Google Scholar] [CrossRef]
- den Hollander, M.-J.; Magis, J.G.; Fuchsenberger, P.; Aartsma, T.J.; Jones, M.R.; Frese, R.N. Enhanced photocurrent generation by photosynthetic bacterial reaction centers through molecular relays, light-harvesting complexes, and direct protein-gold interactions. Langmuir 2011, 27, 10282–10294. [Google Scholar] [CrossRef]
- Kamran, M.; Delgado, J.D.; Friebe, V.; Aartsma, T.J.; Frese, R.N. Photosynthetic protein complexes as bio-photovoltaic building blocks retaining a high internal quantum efficiency. Biomacromolecules 2014, 15, 2833–2838. [Google Scholar] [CrossRef]
- Yasuda, Y.; Sugino, H.; Toyotama, H.; Hirata, Y.; Hara, M.; Miyake, J. Control of protein orientation in molecular photoelectric devices using Langmuir—Blodgett films of photosynthetic reaction centers from Rhodopseudomonas viridis. Bioelectrochem. Bioenerg. 1994, 34, 135–139. [Google Scholar] [CrossRef]
- Uphaus, R.A.; Fang, J.Y.; Picorel, R.; Chumanov, G.; Wang, J.Y.; Cotton, T.M.; Seibert, M. Langmuir-Blodgett and X-ray diffraction studies of isolated photosystem II reaction centers in monolayers and multilayers: Physical dimensions of the complex. Photochem. Photobiol. 1997, 65, 673–679. [Google Scholar] [CrossRef] [Green Version]
- Kernen, P.; Gruszecki, W.I.; Matuła, M.; Wagner, P.; Ziegler, U.; Krupa, Z. Light-harvesting complex II in monocomponent and mixed lipid-protein monolayers. Biochim. Biophys. Acta Biomembr. 1998, 1373, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Pepe, I.M.; Nicolini, C. Langmuir-Blodgett films of photosensitive proteins. J. Photochem. Photobiol. B 1996, 33, 191–200. [Google Scholar] [CrossRef]
- Ko, B.S.; Babcock, B.; Jennings, G.K.; Tilden, S.G.; Peterson, R.R.; Cliffel, D.; Greenbaum, E. Effect of surface composition on the adsorption of photosystem I onto alkanethiolate self-assembled monolayers on gold. Langmuir 2004, 20, 4033–4038. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Lee, J.W.; Greenbaum, E. Biomolecular Electronics: Vectorial Arrays of Photosynthetic Reaction Centers. Phys. Rev. Lett. 1997, 79, 3294–3297. [Google Scholar] [CrossRef]
- Trammell, S.A.; Wang, L.; Zullo, J.M.; Shashidhar, R.; Lebedev, N. Orientated binding of photosynthetic reaction centers on gold using Ni-NTA self-assembled monolayers. Biosens. Bioelectron. 2004, 19, 1649–1655. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, C.; Hasegawa, M.; Yasuda, Y.; Miyake, J. Self-Assembling Photosynthetic Reaction Centers on Electrodes for Current Generation. In Twenty-First Symposium on Biotechnology for Fuels and Chemicals; Finkelstein, M., Davison, B.H., Eds.; Humana Press: Totowa, NJ, USA, 2000; Volume 84–86, pp. 401–408. [Google Scholar]
- Maly, J.; Krejci, J.; Ilie, M.; Jakubka, L.; Masojídek, J.; Pilloton, R.; Sameh, K.; Steffan, P.; Stryhal, Z.; Sugiura, M. Monolayers of photosystem II on gold electrodes with enhanced sensor response-effect of porosity and protein layer arrangement. Anal. Bioanal. Chem. 2005, 381, 1558–1567. [Google Scholar] [CrossRef]
- Faulkner, C.J.; Lees, S.; Ciesielski, P.N.; Cliffel, D.E.; Jennings, G.K. Rapid assembly of photosystem I monolayers on gold electrodes. Langmuir 2008, 24, 8409–8412. [Google Scholar] [CrossRef]
- Friebe, V.M.; Millo, D.; Swainsbury, D.J.K.; Jones, M.R.; Frese, R.N. Cytochrome c Provides an Electron-Funneling Antenna for Efficient Photocurrent Generation in a Reaction Center Biophotocathode. ACS Appl. Mater. Interfaces 2017, 9, 23379–23388. [Google Scholar] [CrossRef] [Green Version]
- Badura, A.; Kothe, T.; Schuhmann, W.; Rögner, M. Wiring photosynthetic enzymes to electrodes. Energy Environ. Sci. 2011, 4, 3263. [Google Scholar] [CrossRef]
- Xiao, Y.; Patolsky, F.; Katz, E.; Hainfeld, J.F.; Willner, I. “Plugging into Enzymes”: Nanowiring of redox enzymes by a gold nanoparticle. Science 2003, 299, 1877–1881. [Google Scholar] [CrossRef]
- Fournier, E.; Nikolaev, A.; Nasiri, H.R.; Hoeser, J.; Friedrich, T.; Hellwig, P.; Melin, F. Creation of a gold nanoparticle based electrochemical assay for the detection of inhibitors of bacterial cytochrome bd oxidases. Bioelectrochemistry 2016, 111, 109–114. [Google Scholar] [CrossRef]
- Meyer, T.; Melin, F.; Richter, O.-M.H.; Ludwig, B.; Kannt, A.; Müller, H.; Michel, H.; Hellwig, P. Electrochemistry suggests proton access from the exit site to the binuclear center in Paracoccus denitrificans cytochrome c oxidase pathway variants. FEBS Lett. 2015, 589, 565–568. [Google Scholar] [CrossRef]
- Meyer, T.; Melin, F.; Xie, H.; von der Hocht, I.; Choi, S.K.; Noor, M.R.; Michel, H.; Gennis, R.B.; Soulimane, T.; Hellwig, P. Evidence for distinct electron transfer processes in terminal oxidases from different origin by means of protein film voltammetry. J. Am. Chem. Soc. 2014, 136, 10854–10857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melin, F.; Meyer, T.; Lankiang, S.; Choi, S.K.; Gennis, R.B.; Blanck, C.; Schmutz, M.; Hellwig, P. Direct electrochemistry of cytochrome bo3 oxidase at a series of gold nanoparticles-modified electrodes. Electrochem. Commun. 2013, 26, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolaev, A.; Makarchuk, I.; Thesseling, A.; Hoeser, J.; Friedrich, T.; Melin, F.; Hellwig, P. Stabilization of the Highly Hydrophobic Membrane Protein, Cytochrome bd Oxidase, on Metallic Surfaces for Direct Electrochemical Studies. Molecules 2020, 25, 3240. [Google Scholar] [CrossRef]
- Asadian, E.; Ghalkhani, M.; Shahrokhian, S. Electrochemical sensing based on carbon nanoparticles: A review. Sens. Actuators B 2019, 293, 183–209. [Google Scholar] [CrossRef]
- Vincent, K.A.; Li, X.; Blanford, C.F.; Belsey, N.A.; Weiner, J.H.; Armstrong, F.A. Enzymatic catalysis on conducting graphite particles. Nat. Chem. Biol. 2007, 3, 761–762. [Google Scholar] [CrossRef]
- Duca, M.; Weeks, J.R.; Fedor, J.G.; Weiner, J.H.; Vincent, K.A. Combining Noble Metals and Enzymes for Relay Cascade Electrocatalysis of Nitrate Reduction to Ammonia at Neutral pH. ChemElectroChem 2015, 2, 1086–1089. [Google Scholar] [CrossRef]
- Ruff, A. Redox polymers in bioelectrochemistry: Common playgrounds and novel concepts. Curr. Opin. Electrochem. 2017, 5, 66–73. [Google Scholar] [CrossRef]
- Badura, A.; Guschin, D.; Kothe, T.; Kopczak, M.J.; Schuhmann, W.; Rögner, M. Photocurrent generation by photosystem 1 integrated in crosslinked redox hydrogels. Energy Environ. Sci. 2011, 4, 2435. [Google Scholar] [CrossRef]
- Badura, A.; Guschin, D.; Esper, B.; Kothe, T.; Neugebauer, S.; Schuhmann, W.; Rögner, M. Photo-Induced Electron Transfer Between Photosystem 2 via Cross-linked Redox Hydrogels. Electroanalysis 2008, 20, 1043–1047. [Google Scholar] [CrossRef]
- Kothe, T.; Plumeré, N.; Badura, A.; Nowaczyk, M.M.; Guschin, D.A.; Rögner, M.; Schuhmann, W. Combination of a photosystem 1-based photocathode and a photosystem 2-based photoanode to a Z-scheme mimic for biophotovoltaic applications. Angew. Chem. Int. Ed. 2013, 52, 14233–14236. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, V.; Kothe, T.; Pöller, S.; El-Mohsnawy, E.; Nowaczyk, M.M.; Plumeré, N.; Schuhmann, W.; Rögner, M. Redox hydrogels with adjusted redox potential for improved efficiency in Z-scheme inspired biophotovoltaic cells. Phys. Chem. Chem. Phys. 2014, 16, 11936–11941. [Google Scholar] [CrossRef] [PubMed]
- Kothe, T.; Pöller, S.; Zhao, F.; Fortgang, P.; Rögner, M.; Schuhmann, W.; Plumeré, N. Engineered electron-transfer chain in photosystem 1 based photocathodes outperforms electron-transfer rates in natural photosynthesis. Chem. Eur. J. 2014, 20, 11029–11034. [Google Scholar] [CrossRef] [PubMed]
- Laftsoglou, T.; Jeuken, L.J.C. Supramolecular electrode assemblies for bioelectrochemistry. Chem. Commun. 2017, 53, 3801–3809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeuken, L.J.C. Electrodes for integral membrane enzymes. Nat. Prod. Rep. 2009, 26, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Malmagro, J.; García-Molina, G.; López De Lacey, A. Electrochemical Biosensors Based on Membrane-Bound Enzymes in Biomimetic Configurations. Sensors 2020, 20, 3393. [Google Scholar] [CrossRef] [PubMed]
- Burgess, J.D.; Rhoten, M.C.; Hawkridge, F.M. Cytochrome c Oxidase Immobilized in Stable Supported Lipid Bilayer Membranes. Langmuir 1998, 14, 2467–2475. [Google Scholar] [CrossRef]
- Rhoten, M.C.; Hawkridge, F.M.; Wilczek, J. The reaction of cytochrome c with bovine and Bacillus stearothermophilus cytochrome c oxidase immobilized in electrode-supported lipid bilayer membranes. J. Electroanal. Chem. 2002, 535, 97–106. [Google Scholar] [CrossRef]
- Kong, J.; Lu, Z.; Lvov, Y.M.; Desamero, R.Z.B.; Frank, H.A.; Rusling, J.F. Direct Electrochemistry of Cofactor Redox Sites in a Bacterial Photosynthetic Reaction Center Protein. J. Am. Chem. Soc. 1998, 120, 7371–7372. [Google Scholar] [CrossRef]
- Munge, B.; Pendon, Z.; Frank, H.A.; Rusling, J.F. Electrochemical reactions of redox cofactors in Rhodobacter sphaeroides reaction center proteins in lipid films. Bioelectrochemistry 2001, 54, 145–150. [Google Scholar] [CrossRef]
- Munge, B.; Das, S.K.; Ilagan, R.; Pendon, Z.; Yang, J.; Frank, H.A.; Rusling, J.F. Electron transfer reactions of redox cofactors in spinach photosystem I reaction center protein in lipid films on electrodes. J. Am. Chem. Soc. 2003, 125, 12457–12463. [Google Scholar] [CrossRef]
- Alcantara, K.; Munge, B.; Pendon, Z.; Frank, H.A.; Rusling, J.F. Thin film voltammetry of spinach photosystem II. Proton-gated electron transfer involving the Mn4 cluster. J. Am. Chem. Soc. 2006, 128, 14930–14937. [Google Scholar] [CrossRef] [PubMed]
- Noji, T.; Matsuo, M.; Takeda, N.; Sumino, A.; Kondo, M.; Nango, M.; Itoh, S.; Dewa, T. Lipid-Controlled Stabilization of Charge-Separated States (P+QB−) and Photocurrent Generation Activity of a Light-Harvesting-Reaction Center Core Complex (LH1-RC) from Rhodopseudomonas palustris. J. Phys. Chem. B 2018, 122, 1066–1080. [Google Scholar] [CrossRef] [PubMed]
- Jeuken, L.J.C.; Connell, S.D.; Henderson, P.J.F.; Gennis, R.B.; Evans, S.D.; Bushby, R.J. Redox enzymes in tethered membranes. J. Am. Chem. Soc. 2006, 128, 1711–1716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radu, V.; Frielingsdorf, S.; Evans, S.D.; Lenz, O.; Jeuken, L.J.C. Enhanced oxygen-tolerance of the full heterotrimeric membrane-bound NiFe-hydrogenase of Ralstonia eutropha. J. Am. Chem. Soc. 2014, 136, 8512–8515. [Google Scholar] [CrossRef]
- Pelster, L.N.; Minteer, S.D. Mitochondrial Inner Membrane Biomimic for the Investigation of Electron Transport Chain Supercomplex Bioelectrocatalysis. ACS Catal. 2016, 6, 4995–4999. [Google Scholar] [CrossRef]
- Ataka, K.; Giess, F.; Knoll, W.; Naumann, R.; Haber-Pohlmeier, S.; Richter, B.; Heberle, J. Oriented attachment and membrane reconstitution of His-tagged cytochrome c oxidase to a gold electrode: In situ monitoring by surface-enhanced infrared absorption spectroscopy. J. Am. Chem. Soc. 2004, 126, 16199–16206. [Google Scholar] [CrossRef] [Green Version]
- Ataka, K.; Richter, B.; Heberle, J. Orientational control of the physiological reaction of cytochrome c oxidase tethered to a gold electrode. J. Phys. Chem. B 2006, 110, 9339–9347. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, M.G.; Giebeta, F.; Naumann, R.; Knoll, W.; Ataka, K.; Heberle, J.; Hrabakova, J.; Murgida, D.H.; Hildebrandt, P. Active site structure and redox processes of cytochrome c oxidase immobilised in a novel biomimetic lipid membrane on an electrode. Chem. Commun. 2004, 2376–2377. [Google Scholar] [CrossRef]
- Lisdat, F. Trends in the layer-by-layer assembly of redox proteins and enzymes in bioelectrochemistry. Curr. Opin. Electrochem. 2017, 5, 165–172. [Google Scholar] [CrossRef]
- Stieger, K.R.; Ciornii, D.; Kölsch, A.; Hejazi, M.; Lokstein, H.; Feifel, S.C.; Zouni, A.; Lisdat, F. Engineering of supramolecular photoactive protein architectures: The defined co-assembly of photosystem I and cytochrome c using a nanoscaled DNA-matrix. Nanoscale 2016, 8, 10695–10705. [Google Scholar] [CrossRef]
- Zhang, Y.; LaFountain, A.M.; Magdaong, N.; Fuciman, M.; Allen, J.P.; Frank, H.A.; Rusling, J.F. Thin film voltammetry of wild type and mutant reaction center proteins from photosynthetic bacteria. J. Phys. Chem. B 2011, 115, 3226–3232. [Google Scholar] [CrossRef] [PubMed]
- Mallardi, A.; Giustini, M.; Lopez, F.; Dezi, M.; Venturoli, G.; Palazzo, G. Functionality of photosynthetic reaction centers in polyelectrolyte multilayers: Toward an herbicide biosensor. J. Phys. Chem. B 2007, 111, 3304–3314. [Google Scholar] [CrossRef] [PubMed]
- Heath, G.R.; Li, M.; Polignano, I.L.; Richens, J.L.; Catucci, G.; O’Shea, P.; Sadeghi, S.J.; Gilardi, G.; Butt, J.N.; Jeuken, L.J.C. Layer-by-Layer Assembly of Supported Lipid Bilayer Poly-L-Lysine Multilayers. Biomacromolecules 2016, 17, 324–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, G.R.; Li, M.; Rong, H.; Radu, V.; Frielingsdorf, S.; Lenz, O.; Butt, J.N.; Jeuken, L.J.C. Multilayered Lipid Membrane Stacks for Biocatalysis Using Membrane Enzymes. Adv. Funct. Mater. 2017, 27, 1606265. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Zhu, J.; Baumann, D.; Peng, L.; Xu, Y.; Shakir, I.; Huang, Y.; Duan, X. Hierarchical 3D electrodes for electrochemical energy storage. Nat. Rev. Mater. 2019, 4, 45–60. [Google Scholar] [CrossRef]
- Friebe, V.M.; Delgado, J.D.; Swainsbury, D.J.K.; Gruber, J.M.; Chanaewa, A.; van Grondelle, R.; von Hauff, E.; Millo, D.; Jones, M.R.; Frese, R.N. Plasmon-Enhanced Photocurrent of Photosynthetic Pigment Proteins on Nanoporous Silver. Adv. Funct. Mater. 2016, 26, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Ciesielski, P.N.; Scott, A.M.; Faulkner, C.J.; Berron, B.J.; Cliffel, D.E.; Jennings, G.K. Functionalized nanoporous gold leaf electrode films for the immobilization of photosystem I. ACS Nano 2008, 2, 2465–2472. [Google Scholar] [CrossRef]
- Lu, Y.; Yuan, M.; Liu, Y.; Tu, B.; Xu, C.; Liu, B.; Zhao, D.; Kong, J. Photoelectric performance of bacteria photosynthetic proteins entrapped on tailored mesoporous WO3-TiO2 films. Langmuir 2005, 21, 4071–4076. [Google Scholar] [CrossRef]
- Kato, M.; Cardona, T.; Rutherford, A.W.; Reisner, E. Photoelectrochemical water oxidation with photosystem II integrated in a mesoporous indium-tin oxide electrode. J. Am. Chem. Soc. 2012, 134, 8332–8335. [Google Scholar] [CrossRef]
- Kato, M.; Cardona, T.; Rutherford, A.W.; Reisner, E. Covalent immobilization of oriented photosystem II on a nanostructured electrode for solar water oxidation. J. Am. Chem. Soc. 2013, 135, 10610–10613. [Google Scholar] [CrossRef]
- Mersch, D.; Lee, C.-Y.; Zhang, J.Z.; Brinkert, K.; Fontecilla-Camps, J.C.; Rutherford, A.W.; Reisner, E. Wiring of Photosystem II to Hydrogenase for Photoelectrochemical Water Splitting. J. Am. Chem. Soc. 2015, 137, 8541–8549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, K.P.; Mersch, D.; Hartmann, V.; Zhang, J.Z.; Nowaczyk, M.M.; Rögner, M.; Ruff, A.; Schuhmann, W.; Plumeré, N.; Reisner, E. Rational wiring of photosystem II to hierarchical indium tin oxide electrodes using redox polymers. Energy Environ. Sci. 2016, 9, 3698–3709. [Google Scholar] [CrossRef] [Green Version]
- Stieger, K.R.; Feifel, S.C.; Lokstein, H.; Hejazi, M.; Zouni, A.; Lisdat, F. Biohybrid architectures for efficient light-to-current conversion based on photosystem I within scalable 3D mesoporous electrodes. J. Mater. Chem. A 2016, 4, 17009–17017. [Google Scholar] [CrossRef]
- Wolfe, K.D.; Dervishogullari, D.; Stachurski, C.D.; Passantino, J.M.; Kane Jennings, G.; Cliffel, D.E. Photosystem I Multilayers within Porous Indium Tin Oxide Cathodes Enhance Mediated Electron Transfer. ChemElectroChem 2020, 7, 596–603. [Google Scholar] [CrossRef]
- Armstrong, F.A.; Butt, J.N.; Sucheta, A. [18] Voltammetric studies of redox-active centers in metalloproteins adsorbed on electrodes. Methods Enzymol. 1993, 227, 479–500. [Google Scholar] [PubMed]
- Léger, C.; Elliott, S.J.; Hoke, K.R.; Jeuken, L.J.C.; Jones, A.K.; Armstrong, F.A. Enzyme electrokinetics: Using protein film voltammetry to investigate redox enzymes and their mechanisms. Biochemistry 2003, 42, 8653–8662. [Google Scholar] [CrossRef]
- Armstrong, F.A. Recent developments in dynamic electrochemical studies of adsorbed enzymes and their active sites. Curr. Opin. Chem. Biol. 2005, 9, 110–117. [Google Scholar] [CrossRef]
- Melin, F.; Hellwig, P. Recent advances in the electrochemistry and spectroelectrochemistry of membrane proteins. Biol. Chem. 2013, 394, 593–609. [Google Scholar] [CrossRef] [Green Version]
- Vacek, J.; Zatloukalova, M.; Novak, D. Electrochemistry of membrane proteins and protein–lipid assemblies. Curr. Opin. Electrochem. 2018, 12, 73–80. [Google Scholar] [CrossRef]
- Armstrong, F.A.; Heering, H.A.; Hirst, J. Reaction of complex metalloproteins studied by protein-film voltammetry. Chem. Soc. Rev. 1997, 26, 169–179. [Google Scholar] [CrossRef]
- Léger, C.; Bertrand, P. Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem. Rev. 2008, 108, 2379–2438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, J.N.; Armstrong, F.A. Voltammetry of Adsorbed Redox Enzymes: Mechanisms in the Potential Dimension. In Bioinorganic Electrochemistry; Hammerich, O., Ulstrup, J., Eds.; Springer: Dordrecht, The Netherlands, 2008; Volume 101, pp. 91–128. [Google Scholar]
- Fourmond, V.; Léger, C. Protein Electrochemistry: Questions and Answers. In Biophotoelectrochemistry: From Bioelectrochemistry to Biophotovoltaics; Jeuken, L., Ed.; Springer: Cham, Switzerland, 2016; Volume 158, pp. 1–41. [Google Scholar]
- Fourmond, V.; Léger, C. Modelling the voltammetry of adsorbed enzymes and molecular catalysts. Curr. Opin. Electrochem. 2017, 1, 110–120. [Google Scholar] [CrossRef]
- Lang, H.; Duschl, C.; Vogel, H. A new class of thiolipids for the attachment of lipid bilayers on gold surfaces. Langmuir 1994, 10, 197–210. [Google Scholar] [CrossRef]
- Millo, D.; Bonifacio, A.; Moncelli, M.R.; Sergo, V.; Gooijer, C.; van der Zwan, G. Characterization of hybrid bilayer membranes on silver electrodes as biocompatible SERS substrates to study membrane-protein interactions. Colloids Surf. B 2010, 81, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Zhang, J.Z.; Paul, N.; Reisner, E. Protein film photoelectrochemistry of the water oxidation enzyme photosystem II. Chem. Soc. Rev. 2014, 43, 6485–6497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornienko, N.; Ly, K.H.; Robinson, W.E.; Heidary, N.; Zhang, J.Z.; Reisner, E. Advancing Techniques for Investigating the Enzyme-Electrode Interface. Acc. Chem. Res. 2019, 52, 1439–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.Z.; Reisner, E. Advancing photosystem II photoelectrochemistry for semi-artificial photosynthesis. Nat. Rev. Chem. 2020, 4, 6–21. [Google Scholar] [CrossRef]
- Kornienko, N.; Zhang, J.Z.; Sokol, K.P.; Lamaison, S.; Fantuzzi, A.; van Grondelle, R.; Rutherford, A.W.; Reisner, E. Oxygenic Photoreactivity in Photosystem II Studied by Rotating Ring Disk Electrochemistry. J. Am. Chem. Soc. 2018, 140, 17923–17931. [Google Scholar] [CrossRef]
- Zhao, F.; Conzuelo, F.; Hartmann, V.; Li, H.; Nowaczyk, M.M.; Plumeré, N.; Rögner, M.; Schuhmann, W. Light Induced H2 Evolution from a Biophotocathode Based on Photosystem 1--Pt Nanoparticles Complexes Integrated in Solvated Redox Polymers Films. J. Phys. Chem. B 2015, 119, 13726–13731. [Google Scholar] [CrossRef]
- Zhao, F.; Plumeré, N.; Nowaczyk, M.M.; Ruff, A.; Schuhmann, W.; Conzuelo, F. Interrogation of a PS1-Based Photocathode by Means of Scanning Photoelectrochemical Microscopy. Small 2017, 13, 1604093. [Google Scholar] [CrossRef]
- Zhao, F.; Hardt, S.; Hartmann, V.; Zhang, H.; Nowaczyk, M.M.; Rögner, M.; Plumeré, N.; Schuhmann, W.; Conzuelo, F. Light-induced formation of partially reduced oxygen species limits the lifetime of photosystem 1-based biocathodes. Nat. Commun. 2018, 9, 1973. [Google Scholar] [CrossRef] [PubMed]
- Demirel, G.; Usta, H.; Yilmaz, M.; Celik, M.; Alidagi, H.A.; Buyukserin, F. Surface-enhanced Raman spectroscopy (SERS): An adventure from plasmonic metals to organic semiconductors as SERS platforms. J. Mater. Chem. C 2018, 6, 5314–5335. [Google Scholar] [CrossRef]
- Grytsyk, N.; Boubegtiten-Fezoua, Z.; Javahiraly, N.; Omeis, F.; Devaux, E.; Hellwig, P. Surface-enhanced resonance Raman spectroscopy of heme proteins on a gold grid electrode. Spectrochim. Acta Part A 2020, 230, 118081. [Google Scholar] [CrossRef] [PubMed]
- Hrabakova, J.; Ataka, K.; Heberle, J.; Hildebrandt, P.; Murgida, D.H. Long distance electron transfer in cytochrome c oxidase immobilised on electrodes. A surface enhanced resonance Raman spectroscopic study. Phys. Chem. Chem. Phys. 2006, 8, 759–766. [Google Scholar] [CrossRef] [Green Version]
- Sezer, M.; Frielingsdorf, S.; Millo, D.; Heidary, N.; Utesch, T.; Mroginski, M.-A.; Friedrich, B.; Hildebrandt, P.; Zebger, I.; Weidinger, I.M. Role of the HoxZ subunit in the electron transfer pathway of the membrane-bound NiFe-hydrogenase from Ralstonia eutropha immobilized on electrodes. J. Phys. Chem. B 2011, 115, 10368–10374. [Google Scholar] [CrossRef]
- Ataka, K.; Stripp, S.T.; Heberle, J. Surface-enhanced infrared absorption spectroscopy (SEIRAS) to probe monolayers of membrane proteins. Biochim. Biophys. Acta 2013, 1828, 2283–2293. [Google Scholar] [CrossRef] [Green Version]
- Wiebalck, S.; Kozuch, J.; Forbrig, E.; Tzschucke, C.C.; Jeuken, L.J.C.; Hildebrandt, P. Monitoring the Transmembrane Proton Gradient Generated by Cytochrome bo3 in Tethered Bilayer Lipid Membranes Using SEIRA Spectroscopy. J. Phys. Chem. B 2016, 120, 2249–2256. [Google Scholar] [CrossRef]
- López-Lorente, Á.I.; Kranz, C. Recent advances in biomolecular vibrational spectroelectrochemistry. Curr. Opin. Electrochem. 2017, 5, 106–113. [Google Scholar] [CrossRef]
- Zhai, Y.; Zhu, Z.; Zhou, S.; Zhu, C.; Dong, S. Recent advances in spectroelectrochemistry. Nanoscale 2018, 10, 3089–3111. [Google Scholar] [CrossRef]
- Melin, F.; Hellwig, P. Redox Properties of the Membrane Proteins from the Respiratory Chain. Chem. Rev. 2020, 120, 10244–10297. [Google Scholar] [CrossRef]
- Białek, R.; Friebe, V.; Ruff, A.; Jones, M.R.; Frese, R.; Gibasiewicz, K. In situ spectroelectrochemical investigation of a biophotoelectrode based on photoreaction centers embedded in a redox hydrogel. Electrochim. Acta 2020, 330, 135190. [Google Scholar] [CrossRef]
- Ash, P.A.; Vincent, K.A. Spectroscopic analysis of immobilised redox enzymes under direct electrochemical control. Chem. Commun. 2012, 48, 1400–1409. [Google Scholar] [CrossRef] [PubMed]
- Ritter, M.; Anderka, O.; Ludwig, B.; Mäntele, W.; Hellwig, P. Electrochemical and FTIR spectroscopic characterization of the cytochrome bc1 complex from Paracoccus denitrificans: Evidence for protonation reactions coupled to quinone binding. Biochemistry 2003, 42, 12391–12399. [Google Scholar] [CrossRef] [PubMed]
- Baymann, F.; Robertson, D.E.; Dutton, P.L.; Mäntele, W. Electrochemical and spectroscopic investigations of the cytochrome bc1 complex from Rhodobacter capsulatus. Biochemistry 1999, 38, 13188–13199. [Google Scholar] [CrossRef]
- Kato, Y.; Sugiura, M.; Oda, A.; Watanabe, T. Spectroelectrochemical determination of the redox potential of pheophytin a, the primary electron acceptor in photosystem II. Proc. Natl. Acad. Sci. USA 2009, 106, 17365–17370. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, A.; Suzawa, T.; Kato, Y.; Watanabe, T. Species dependence of the redox potential of the primary electron donor P700 in photosystem I of oxygenic photosynthetic organisms revealed by spectroelectrochemistry. Plant. Cell Physiol. 2011, 52, 815–823. [Google Scholar] [CrossRef] [Green Version]
- Haas, A.S.; Pilloud, D.L.; Reddy, K.S.; Babcock, G.T.; Moser, C.C.; Blasie, J.K.; Dutton, P.L. Cytochrome c and Cytochrome c Oxidase: Monolayer Assemblies and Catalysis. J. Phys. Chem. B 2001, 105, 11351–11362. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Reuillard, B.; Sokol, K.P.; Laftsoglou, T.; Lockwood, C.W.J.; Rowe, S.F.; Hwang, E.T.; Fontecilla-Camps, J.C.; Jeuken, L.J.C.; Butt, J.N.; et al. A decahaem cytochrome as an electron conduit in protein-enzyme redox processes. Chem. Commun. 2016, 52, 7390–7393. [Google Scholar] [CrossRef] [Green Version]
- Nowak, C.; Laredo, T.; Gebert, J.; Lipkowski, J.; Gennis, R.B.; Ferguson-Miller, S.; Knoll, W.; Naumann, R.L.C. 2D-SEIRA spectroscopy to highlight conformational changes of the cytochrome c oxidase induced by direct electron transfer. Metallomics 2011, 3, 619–627. [Google Scholar] [CrossRef] [Green Version]
- Steininger, C.; Reiner-Rozman, C.; Schwaighofer, A.; Knoll, W.; Naumann, R.L.C. Kinetics of cytochrome c oxidase from R. sphaeroides initiated by direct electron transfer followed by tr-SEIRAS. Bioelectrochemistry 2016, 112, 1–8. [Google Scholar] [CrossRef]
- Bogner, A.; Jouneau, P.-H.; Thollet, G.; Basset, D.; Gauthier, C. A history of scanning electron microscopy developments: Towards “wet-STEM” imaging. Micron 2007, 38, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Monsalve, K.; Roger, M.; Gutierrez-Sanchez, C.; Ilbert, M.; Nitsche, S.; Byrne-Kodjabachian, D.; Marchi, V.; Lojou, E. Hydrogen bioelectrooxidation on gold nanoparticle-based electrodes modified by Aquifex aeolicus hydrogenase: Application to hydrogen/oxygen enzymatic biofuel cells. Bioelectrochemistry 2015, 106, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Gaub, H.E.; Newton, R.; Pfreundschuh, M.; Gerber, C.; Müller, D.J. Atomic force microscopy-based characterization and design of biointerfaces. Nat. Rev. Mater. 2017, 2, 17008. [Google Scholar] [CrossRef]
- Connell, S.D.; Smith, D.A. The atomic force microscope as a tool for studying phase separation in lipid membranes. Mol. Membr. Biol. 2006, 23, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, J.A.; Walsh, D.L.; Connell, S.D. Nanoscale Substrate Roughness Hinders Domain Formation in Supported Lipid Bilayers. Langmuir 2019, 35, 15352–15363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aufderhorst-Roberts, A.; Chandra, U.; Connell, S.D. Three-Phase Coexistence in Lipid Membranes. Biophys. J. 2017, 112, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frederix, P.L.T.M.; Bosshart, P.D.; Engel, A. Atomic force microscopy of biological membranes. Biophys. J. 2009, 96, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, F.A.; Connell, S.; Miltenberger-Miltenyi, G.; Pereira, S.V.; Tavares, A.; Ariëns, R.A.S.; Santos, N.C. Atomic force microscopy-based molecular recognition of a fibrinogen receptor on human erythrocytes. ACS Nano 2010, 4, 4609–4620. [Google Scholar] [CrossRef]
- Heath, G.R.; Roth, J.; Connell, S.D.; Evans, S.D. Diffusion in low-dimensional lipid membranes. Nano Lett. 2014, 14, 5984–5988. [Google Scholar] [CrossRef]
- Heath, G.R.; Scheuring, S. Advances in high-speed atomic force microscopy (HS-AFM) reveal dynamics of transmembrane channels and transporters. Curr. Opin. Struct. Biol. 2019, 57, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Hao, X.; Zhang, J.; Christensen, H.E.M.; Wang, H.; Ulstrup, J. Electrochemical single-molecule AFM of the redox metalloenzyme copper nitrite reductase in action. ChemPhysChem 2012, 13, 2919–2924. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yan, X.; Huang, W.; Engelbrekt, C.; Duus, J.Ø.; Ulstrup, J.; Xiao, X.; Zhang, J. Bilirubin oxidase oriented on novel type three-dimensional biocathodes with reduced graphene aggregation for biocathode. Biosens. Bioelectron. 2020, 167, 112500. [Google Scholar] [CrossRef] [PubMed]
- González Arzola, K.; Gimeno, Y.; Arévalo, M.C.; Falcón, M.A.; Hernández Creus, A. Electrochemical and AFM characterization on gold and carbon electrodes of a high redox potential laccase from Fusarium proliferatum. Bioelectrochemistry 2010, 79, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Ciaccafava, A.; Infossi, P.; Ilbert, M.; Guiral, M.; Lecomte, S.; Giudici-Orticoni, M.T.; Lojou, E. Electrochemistry, AFM, and PM-IRRA spectroscopy of immobilized hydrogenase: Role of a hydrophobic helix in enzyme orientation for efficient H2 oxidation. Angew. Chem. Int. Ed. 2012, 51, 953–956. [Google Scholar] [CrossRef]
- Gutiérrez-Sanz, O.; Olea, D.; Pita, M.; Batista, A.P.; Alonso, A.; Pereira, M.M.; Vélez, M.; de Lacey, A.L. Reconstitution of respiratory complex I on a biomimetic membrane supported on gold electrodes. Langmuir 2014, 30, 9007–9015. [Google Scholar] [CrossRef] [Green Version]
- Neupane, S.; de Smet, Y.; Renner, F.U.; Losada-Pérez, P. Quartz Crystal Microbalance with Dissipation Monitoring: A Versatile Tool to Monitor Phase Transitions in Biomimetic Membranes. Front. Mater. 2018, 5, 46. [Google Scholar] [CrossRef] [Green Version]
- Giess, F.; Friedrich, M.G.; Heberle, J.; Naumann, R.L.; Knoll, W. The protein-tethered lipid bilayer: A novel mimic of the biological membrane. Biophys. J. 2004, 87, 3213–3220. [Google Scholar] [CrossRef] [Green Version]
- Lam, K.B.; Irwin, E.F.; Healy, K.E.; Lin, L. Bioelectrocatalytic self-assembled thylakoids for micro-power and sensing applications. Sens. Actuators B 2006, 117, 480–487. [Google Scholar] [CrossRef]
- Chen, H.; Dong, F.; Minteer, S.D. The progress and outlook of bioelectrocatalysis for the production of chemicals, fuels and materials. Nat. Catal. 2020, 3, 225–244. [Google Scholar] [CrossRef]
- Ruff, A.; Conzuelo, F.; Schuhmann, W. Bioelectrocatalysis as the basis for the design of enzyme-based biofuel cells and semi-artificial biophotoelectrodes. Nat. Catal. 2020, 3, 214–224. [Google Scholar] [CrossRef]
- Grattieri, M.; Beaver, K.; Gaffney, E.M.; Dong, F.; Minteer, S.D. Advancing the fundamental understanding and practical applications of photo-bioelectrocatalysis. Chem. Commun. 2020, 56, 8553–8568. [Google Scholar] [CrossRef] [PubMed]
- Milton, R.D.; Minteer, S.D. Direct enzymatic bioelectrocatalysis: Differentiating between myth and reality. J. R. Soc. Interface 2017, 14, 20170253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Xia, H.-Q.; Wu, R.; Bai, L.; Yan, L.; Magner, E.; Cosnier, S.; Lojou, E.; Zhu, Z.; Liu, A. Tackling the Challenges of Enzymatic (Bio)Fuel Cells. Chem. Rev. 2019, 119, 9509–9558. [Google Scholar] [CrossRef] [PubMed]
- Cooney, M.J.; Svoboda, V.; Lau, C.; Martin, G.; Minteer, S.D. Enzyme catalysed biofuel cells. Energy Environ. Sci. 2008, 1, 320–337. [Google Scholar] [CrossRef]
- Minteer, S.D.; Liaw, B.Y.; Cooney, M.J. Enzyme-based biofuel cells. Curr. Opin. Biotechnol. 2007, 18, 228–234. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Woodley, J.M. Role of Biocatalysis in Sustainable Chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef]
- Mazurenko, I.; Wang, X.; de Poulpiquet, A.; Lojou, E. H2/O2 enzymatic fuel cells: From proof-of-concept to powerful devices. Sustain. Energy Fuels 2017, 1, 1475–1501. [Google Scholar] [CrossRef]
- Vincent, K.A.; Parkin, A.; Armstrong, F.A. Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem. Rev. 2007, 107, 4366–4413. [Google Scholar] [CrossRef]
- Plumeré, N.; Rüdiger, O.; Oughli, A.A.; Williams, R.; Vivekananthan, J.; Pöller, S.; Schuhmann, W.; Lubitz, W. A redox hydrogel protects hydrogenase from high-potential deactivation and oxygen damage. Nat. Chem. 2014, 6, 822–827. [Google Scholar] [CrossRef]
- Ruff, A.; Szczesny, J.; Marković, N.; Conzuelo, F.; Zacarias, S.; Pereira, I.A.C.; Lubitz, W.; Schuhmann, W. A fully protected hydrogenase/polymer-based bioanode for high-performance hydrogen/glucose biofuel cells. Nat. Commun. 2018, 9, 3675. [Google Scholar] [CrossRef]
- Vincent, K.A.; Cracknell, J.A.; Lenz, O.; Zebger, I.; Friedrich, B.; Armstrong, F.A. Electrocatalytic hydrogen oxidation by an enzyme at high carbon monoxide or oxygen levels. Proc. Natl. Acad. Sci. USA 2005, 102, 16951–16954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincent, K.A.; Cracknell, J.A.; Clark, J.R.; Ludwig, M.; Lenz, O.; Friedrich, B.; Armstrong, F.A. Electricity from low-level H2 in still air-an ultimate test for an oxygen tolerant hydrogenase. Chem. Commun. 2006, 5033–5035. [Google Scholar] [CrossRef] [PubMed]
- So, K.; Kitazumi, Y.; Shirai, O.; Nishikawa, K.; Higuchi, Y.; Kano, K. Direct electron transfer-type dual gas diffusion H2/O2 biofuel cells. J. Mater. Chem. A 2016, 4, 8742–8749. [Google Scholar] [CrossRef]
- Xia, H.-Q.; So, K.; Kitazumi, Y.; Shirai, O.; Nishikawa, K.; Higuchi, Y.; Kano, K. Dual gas-diffusion membrane- and mediatorless dihydrogen/air-breathing biofuel cell operating at room temperature. J. Power Sources 2016, 335, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Mano, N.; de Poulpiquet, A. O2 Reduction in Enzymatic Biofuel Cells. Chem. Rev. 2018, 118, 2392–2468. [Google Scholar] [CrossRef] [Green Version]
- Katz, E.; Willner, I.; Kotlyar, A.B. A non-compartmentalized glucose∣O2 biofuel cell by bioengineered electrode surfaces. J. Electroanal. Chem. 1999, 479, 64–68. [Google Scholar] [CrossRef]
- Katz, E.; Willner, I. A biofuel cell with electrochemically switchable and tunable power output. J. Am. Chem. Soc. 2003, 125, 6803–6813. [Google Scholar] [CrossRef]
- Wang, X.; Clément, R.; Roger, M.; Bauzan, M.; Mazurenko, I.; de Poulpiquet, A.; Ilbert, M.; Lojou, E. Bacterial Respiratory Chain Diversity Reveals a Cytochrome c Oxidase Reducing O2 at Low Overpotentials. J. Am. Chem. Soc. 2019, 141, 11093–11102. [Google Scholar] [CrossRef] [Green Version]
- Sakai, K.; Kitazumi, Y.; Shirai, O.; Takagi, K.; Kano, K. High-Power Formate/Dioxygen Biofuel Cell Based on Mediated Electron Transfer Type Bioelectrocatalysis. ACS Catal. 2017, 7, 5668–5673. [Google Scholar] [CrossRef]
- Reda, T.; Plugge, C.M.; Abram, N.J.; Hirst, J. Reversible interconversion of carbon dioxide and formate by an electroactive enzyme. Proc. Natl. Acad. Sci. USA 2008, 105, 10654–10658. [Google Scholar] [CrossRef] [Green Version]
- Milton, R.D.; Minteer, S.D. Enzymatic Bioelectrosynthetic Ammonia Production: Recent Electrochemistry of Nitrogenase, Nitrate Reductase, and Nitrite Reductase. ChemPlusChem 2017, 82, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Teodor, A.H.; Bruce, B.D. Putting Photosystem I to Work: Truly Green Energy. Trends Biotechnol. 2020, 38, 1329–1342. [Google Scholar] [CrossRef] [PubMed]
- Friebe, V.M.; Frese, R.N. Photosynthetic reaction center-based biophotovoltaics. Curr. Opin. Electrochem. 2017, 5, 126–134. [Google Scholar] [CrossRef]
- Koblizek, M.; Masojidek, J.; Komenda, J.; Kucera, T.; Pilloton, R.; Mattoo, A.K.; Giardi, M.T. A sensitive photosystem II-based biosensor for detection of a class of herbicides. Biotechnol. Bioeng. 1998, 60, 664–669. [Google Scholar] [CrossRef]
- Swainsbury, D.J.K.; Friebe, V.M.; Frese, R.N.; Jones, M.R. Evaluation of a biohybrid photoelectrochemical cell employing the purple bacterial reaction centre as a biosensor for herbicides. Biosens. Bioelectron. 2014, 58, 172–178. [Google Scholar] [CrossRef] [Green Version]
- Yehezkeli, O.; Tel-Vered, R.; Wasserman, J.; Trifonov, A.; Michaeli, D.; Nechushtai, R.; Willner, I. Integrated photosystem II-based photo-bioelectrochemical cells. Nat. Commun. 2012, 3, 781. [Google Scholar] [CrossRef] [Green Version]
- Riedel, M.; Wersig, J.; Ruff, A.; Schuhmann, W.; Zouni, A.; Lisdat, F. A Z-Scheme-Inspired Photobioelectrochemical H2O/O2 Cell with a 1 V Open-Circuit Voltage Combining Photosystem II and PbS Quantum Dots. Angew. Chem. Int. Ed. 2019, 58, 801–805. [Google Scholar] [CrossRef]
- Nam, D.H.; Zhang, J.Z.; Andrei, V.; Kornienko, N.; Heidary, N.; Wagner, A.; Nakanishi, K.; Sokol, K.P.; Slater, B.; Zebger, I.; et al. Solar Water Splitting with a Hydrogenase Integrated in Photoelectrochemical Tandem Cells. Angew. Chem. Int. Ed. 2018, 57, 10595–10599. [Google Scholar] [CrossRef]
- Sokol, K.P.; Robinson, W.E.; Warnan, J.; Kornienko, N.; Nowaczyk, M.M.; Ruff, A.; Zhang, J.Z.; Reisner, E. Bias-free photoelectrochemical water splitting with photosystem II on a dye-sensitized photoanode wired to hydrogenase. Nat. Energy 2018, 3, 944–951. [Google Scholar] [CrossRef]
- Sokol, K.P.; Robinson, W.E.; Oliveira, A.R.; Warnan, J.; Nowaczyk, M.M.; Ruff, A.; Pereira, I.A.C.; Reisner, E. Photoreduction of CO2 with a Formate Dehydrogenase Driven by Photosystem II Using a Semi-artificial Z-Scheme Architecture. J. Am. Chem. Soc. 2018, 140, 16418–16422. [Google Scholar] [CrossRef] [Green Version]
- Lubner, C.E.; Grimme, R.; Bryant, D.A.; Golbeck, J.H. Wiring photosystem I for direct solar hydrogen production. Biochemistry 2010, 49, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Tapia, C.; Milton, R.D.; Pankratova, G.; Minteer, S.D.; Åkerlund, H.-E.; Leech, D.; De Lacey, A.L.; Pita, M.; Gorton, L. Wiring of Photosystem I and Hydrogenase on an Electrode for Photoelectrochemical H2 Production by using Redox Polymers for Relatively Positive Onset Potential. ChemElectroChem 2017, 4, 90–95. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, P.; Ruff, A.; Hartmann, V.; Zacarias, S.; Pereira, I.A.C.; Nowaczyk, M.M.; Rögner, M.; Conzuelo, F.; Schuhmann, W. A photosystem I monolayer with anisotropic electron flow enables Z-scheme like photosynthetic water splitting. Energy Environ. Sci. 2019, 12, 3133–3143. [Google Scholar] [CrossRef]
- Hancock, A.M.; Meredith, S.A.; Connell, S.D.; Jeuken, L.J.C.; Adams, P.G. Proteoliposomes as energy transferring nanomaterials: Enhancing the spectral range of light-harvesting proteins using lipid-linked chromophores. Nanoscale 2019, 11, 16284–16292. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Friebe, V.M.; Frese, R.N.; Jones, M.R. Polychromatic solar energy conversion in pigment-protein chimeras that unite the two kingdoms of (bacterio)chlorophyll-based photosynthesis. Nat. Commun. 2020, 11, 1542. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Mantell, J.; Jones, M.R. Minding the Gap between Plant and Bacterial Photosynthesis within a Self-Assembling Biohybrid Photosystem. ACS Nano 2020, 14, 4536–4549. [Google Scholar] [CrossRef]
- Moehlenbrock, M.J.; Minteer, S.D. Extended lifetime biofuel cells. Chem. Soc. Rev. 2008, 37, 1188–1196. [Google Scholar] [CrossRef]
- Rideau, E.; Dimova, R.; Schwille, P.; Wurm, F.R.; Landfester, K. Liposomes and polymersomes: A comparative review towards cell mimicking. Chem. Soc. Rev. 2018, 47, 8572–8610. [Google Scholar] [CrossRef] [Green Version]
- Discher, D.E.; Eisenberg, A. Polymer vesicles. Science 2002, 297, 967–973. [Google Scholar] [CrossRef] [Green Version]
- Poschenrieder, S.T.; Schiebel, S.K.; Castiglione, K. Stability of polymersomes with focus on their use as nanoreactors. Eng. Life Sci. 2018, 18, 101–113. [Google Scholar] [CrossRef]
- Beales, P.A.; Khan, S.; Muench, S.P.; Jeuken, L.J.C. Durable vesicles for reconstitution of membrane proteins in biotechnology. Biochem. Soc. Trans. 2017, 45, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Li, M.; Muench, S.P.; Jeuken, L.J.C.; Beales, P.A. Durable proteo-hybrid vesicles for the extended functional lifetime of membrane proteins in bionanotechnology. Chem. Commun. 2016, 52, 11020–11023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seneviratne, R.; Khan, S.; Moscrop, E.; Rappolt, M.; Muench, S.P.; Jeuken, L.J.C.; Beales, P.A. A reconstitution method for integral membrane proteins in hybrid lipid-polymer vesicles for enhanced functional durability. Methods 2018, 147, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Marušič, N.; Otrin, L.; Zhao, Z.; Lira, R.B.; Kyrilis, F.L.; Hamdi, F.; Kastritis, P.L.; Vidaković-Koch, T.; Ivanov, I.; Sundmacher, K.; et al. Constructing artificial respiratory chain in polymer compartments: Insights into the interplay between bo3 oxidase and the membrane. Proc. Natl. Acad. Sci. USA 2020, 117, 15006–15017. [Google Scholar] [CrossRef] [PubMed]
- Otrin, L.; Marušič, N.; Bednarz, C.; Vidaković-Koch, T.; Lieberwirth, I.; Landfester, K.; Sundmacher, K. Toward Artificial Mitochondrion: Mimicking Oxidative Phosphorylation in Polymer and Hybrid Membranes. Nano Lett. 2017, 17, 6816–6821. [Google Scholar] [CrossRef] [Green Version]
- Smirnova, I.A.; Ädelroth, P.; Brzezinski, P. Extraction and liposome reconstitution of membrane proteins with their native lipids without the use of detergents. Sci. Rep. 2018, 8, 14950. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Knowles, T.J.; Postis, V.L.G.; Jamshad, M.; Parslow, R.A.; Lin, Y.-P.; Goldman, A.; Sridhar, P.; Overduin, M.; Muench, S.P.; et al. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016, 11, 1149–1162. [Google Scholar] [CrossRef]
- Kumar, A.; Hsu, L.H.-H.; Kavanagh, P.; Barrière, F.; Lens, P.N.L.; Lapinsonnière, L.; Lienhard V, J.H.; Schröder, U.; Jiang, X.; Leech, D. The ins and outs of microorganism–electrode electron transfer reactions. Nat. Rev. Chem. 2017, 1, 24. [Google Scholar] [CrossRef]
- Rabaey, K.; Rozendal, R.A. Microbial electrosynthesis—Revisiting the electrical route for microbial production. Nat. Rev. Microbiol. 2010, 8, 706–716. [Google Scholar] [CrossRef]
- Logan, B.E.; Rossi, R.; Ragab, A.; Saikaly, P.E. Electroactive microorganisms in bioelectrochemical systems. Nat. Rev. Microbiol. 2019, 17, 307–319. [Google Scholar] [CrossRef]
- Jiang, Y.; Zeng, R.J. Bidirectional extracellular electron transfers of electrode-biofilm: Mechanism and application. Bioresour. Technol. 2019, 271, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Jiang, Y.; Shi, L. Electromicrobiology and biotechnological applications of the exoelectrogens Geobacter and Shewanella spp. Sci. China Technol. Sci. 2019, 62, 1670–1678. [Google Scholar] [CrossRef]
- Gregory, K.B.; Bond, D.R.; Lovley, D.R. Graphite electrodes as electron donors for anaerobic respiration. Environ. Microbiol. 2004, 6, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Geelhoed, J.S.; Stams, A.J.M. Electricity-assisted biological hydrogen production from acetate by Geobacter sulfurreducens. Environ. Sci. Technol. 2011, 45, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.E.; Flynn, J.M.; Baron, D.B.; Gralnick, J.A.; Bond, D.R. Towards electrosynthesis in shewanella: Energetics of reversing the Mtr pathway for reductive metabolism. PLoS ONE 2011, 6, e16649. [Google Scholar] [CrossRef]
- Le, Q.A.T.; Kim, H.G.; Kim, Y.H. Electrochemical synthesis of formic acid from CO2 catalyzed by Shewanella oneidensis MR-1 whole-cell biocatalyst. Enzyme Microb. Technol. 2018, 116, 1–5. [Google Scholar] [CrossRef]
- La Cava, E.; Guionet, A.; Saito, J.; Okamoto, A. Involvement of Proton Transfer for Carbon Dioxide Reduction Coupled with Extracellular Electron Uptake in Shewanella oneidensis MR-1. Electroanalysis 2020, 32, 1659–1663. [Google Scholar] [CrossRef]
- Shin, H.J.; Jung, K.A.; Nam, C.W.; Park, J.M. A genetic approach for microbial electrosynthesis system as biocommodities production platform. Bioresour. Technol. 2017, 245, 1421–1429. [Google Scholar] [CrossRef]
- Min, D.; Cheng, L.; Zhang, F.; Huang, X.-N.; Li, D.-B.; Liu, D.-F.; Lau, T.-C.; Mu, Y.; Yu, H.-Q. Enhancing Extracellular Electron Transfer of Shewanella oneidensis MR-1 through Coupling Improved Flavin Synthesis and Metal-Reducing Conduit for Pollutant Degradation. Environ. Sci. Technol. 2017, 51, 5082–5089. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, Y.; Hu, Y.; Cao, B.; Rice, S.A.; Kjelleberg, S.; Song, H. Enhancing Bidirectional Electron Transfer of Shewanella oneidensis by a Synthetic Flavin Pathway. ACS Synth. Biol. 2015, 4, 815–823. [Google Scholar] [CrossRef]
- Glaven, S.M. Bioelectrochemical systems and synthetic biology: More power, more products. Microb. Biotechnol. 2019, 12, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Ueki, T.; Nevin, K.P.; Woodard, T.L.; Aklujkar, M.A.; Holmes, D.E.; Lovley, D.R. Construction of a Geobacter Strain With Exceptional Growth on Cathodes. Front. Microbiol. 2018, 9, 1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La, J.A.; Jeon, J.-M.; Sang, B.-I.; Yang, Y.-H.; Cho, E.C. A Hierarchically Modified Graphite Cathode with Au Nanoislands, Cysteamine, and Au Nanocolloids for Increased Electricity-Assisted Production of Isobutanol by Engineered Shewanella oneidensis MR-1. ACS Appl. Mater. Interfaces 2017, 9, 43563–43574. [Google Scholar] [CrossRef] [PubMed]
- Rowe, A.R.; Rajeev, P.; Jain, A.; Pirbadian, S.; Okamoto, A.; Gralnick, J.A.; El-Naggar, M.Y.; Nealson, K.H. Tracking Electron Uptake from a Cathode into Shewanella Cells: Implications for Energy Acquisition from Solid-Substrate Electron Donors. mBio 2018, 9, e02203-17. [Google Scholar] [CrossRef] [Green Version]
- Tefft, N.M.; TerAvest, M.A. Reversing an Extracellular Electron Transfer Pathway for Electrode-Driven Acetoin Reduction. ACS Synth. Biol. 2019, 8, 1590–1600. [Google Scholar] [CrossRef]
- Xie, Q.; Lu, Y.; Tang, L.; Zeng, G.; Yang, Z.; Fan, C.; Wang, J.; Atashgahi, S. The mechanism and application of bidirectional extracellular electron transport in the field of energy and environment. Crit. Rev. Environ. Sci. Technol. 2020, 6, 1–46. [Google Scholar]
- Hasan, K.; Çevik, E.; Sperling, E.; Packer, M.A.; Leech, D.; Gorton, L. Photoelectrochemical Wiring of Paulschulzia pseudovolvox (Algae) to Osmium Polymer Modified Electrodes for Harnessing Solar Energy. Adv. Energy Mater. 2015, 5, 1501100. [Google Scholar] [CrossRef]
- Bombelli, P.; Müller, T.; Herling, T.W.; Howe, C.J.; Knowles, T.P.J. A High Power-Density, Mediator-Free, Microfluidic Biophotovoltaic Device for Cyanobacterial Cells. Adv. Energy Mater. 2015, 5, 1–6. [Google Scholar] [CrossRef] [Green Version]
- McCormick, A.J.; Bombelli, P.; Scott, A.M.; Philips, A.J.; Smith, A.G.; Fisher, A.C.; Howe, C.J. Photosynthetic biofilms in pure culture harness solar energy in a mediatorless bio-photovoltaic cell (BPV) system. Energy Environ. Sci. 2011, 4, 4699. [Google Scholar] [CrossRef]
- Sekar, N.; Umasankar, Y.; Ramasamy, R.P. Photocurrent generation by immobilized cyanobacteria via direct electron transport in photo-bioelectrochemical cells. Phys. Chem. Chem. Phys. 2014, 16, 7862–7871. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.Z.; Bombelli, P.; Sokol, K.P.; Fantuzzi, A.; Rutherford, A.W.; Howe, C.J.; Reisner, E. Photoelectrochemistry of Photosystem II in Vitro vs in Vivo. J. Am. Chem. Soc. 2018, 140, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Grattieri, M.; Patterson, S.; Copeland, J.; Klunder, K.; Minteer, S.D. Purple Bacteria and 3D Redox Hydrogels for Bioinspired Photo-bioelectrocatalysis. ChemSusChem 2020, 13, 230–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankratov, D.; Pankratova, G.; Gorton, L. Thylakoid membrane–based photobioelectrochemical systems: Achievements, limitations, and perspectives. Curr. Opin. Electrochem. 2020, 19, 49–54. [Google Scholar] [CrossRef]
- Wenzel, T.; Härtter, D.; Bombelli, P.; Howe, C.J.; Steiner, U. Porous translucent electrodes enhance current generation from photosynthetic biofilms. Nat. Commun. 2018, 9, 1299. [Google Scholar] [CrossRef]
- Sakimoto, K.K.; Wong, A.B.; Yang, P. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production. Science 2016, 351, 74–77. [Google Scholar] [CrossRef] [Green Version]
- Cestellos-Blanco, S.; Zhang, H.; Kim, J.M.; Shen, Y.-X.; Yang, P. Photosynthetic semiconductor biohybrids for solar-driven biocatalysis. Nat. Catal. 2020, 3, 245–255. [Google Scholar] [CrossRef]
- Hartshorne, R.S.; Reardon, C.L.; Ross, D.; Nuester, J.; Clarke, T.A.; Gates, A.J.; Mills, P.C.; Fredrickson, J.K.; Zachara, J.M.; Shi, L.; et al. Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. USA 2009, 106, 22169–22174. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Deng, S.; Marshall, M.J.; Wang, Z.; Kennedy, D.W.; Dohnalkova, A.C.; Mottaz, H.M.; Hill, E.A.; Gorby, Y.A.; Beliaev, A.S.; et al. Direct involvement of type II secretion system in extracellular translocation of Shewanella oneidensis outer membrane cytochromes MtrC and OmcA. J. Bacteriol. 2008, 190, 5512–5516. [Google Scholar] [CrossRef] [Green Version]
- Ainsworth, E.V.; Lockwood, C.W.J.; White, G.F.; Hwang, E.T.; Sakai, T.; Gross, M.A.; Richardson, D.J.; Clarke, T.A.; Jeuken, L.J.C.; Reisner, E.; et al. Photoreduction of Shewanella oneidensis Extracellular Cytochromes by Organic Chromophores and Dye-Sensitized TiO2. Chembiochem 2016, 17, 2324–2333. [Google Scholar] [CrossRef] [Green Version]
- Hwang, E.T.; Sheikh, K.; Orchard, K.L.; Hojo, D.; Radu, V.; Lee, C.-Y.; Ainsworth, E.; Lockwood, C.; Gross, M.A.; Adschiri, T.; et al. A Decaheme Cytochrome as a Molecular Electron Conduit in Dye-Sensitized Photoanodes. Adv. Funct. Mater. 2015, 25, 2308–2315. [Google Scholar] [CrossRef] [Green Version]
- Hwang, E.T.; Orchard, K.L.; Hojo, D.; Beton, J.; Lockwood, C.W.J.; Adschiri, T.; Butt, J.N.; Reisner, E.; Jeuken, L.J.C. Exploring Step-by-Step Assembly of Nanoparticle:Cytochrome Biohybrid Photoanodes. ChemElectroChem 2017, 4, 1959–1968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowe, S.F.; Le Gall, G.; Ainsworth, E.V.; Davies, J.A.; Lockwood, C.W.J.; Shi, L.; Elliston, A.; Roberts, I.N.; Waldron, K.W.; Richardson, D.J.; et al. Light-Driven H2 Evolution and C═C or C═O Bond Hydrogenation by Shewanella oneidensis: A Versatile Strategy for Photocatalysis by Nonphotosynthetic Microorganisms. ACS Catal. 2017, 7, 7558–7566. [Google Scholar] [CrossRef] [Green Version]
- Stikane, A.; Hwang, E.T.; Ainsworth, E.V.; Piper, S.E.H.; Critchley, K.; Butt, J.N.; Reisner, E.; Jeuken, L.J.C. Towards compartmentalized photocatalysis: Multihaem proteins as transmembrane molecular electron conduits. Faraday Discuss. 2019, 215, 26–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Catania, R.; Jeuken, L.J.C. Membrane Protein Modified Electrodes in Bioelectrocatalysis. Catalysts 2020, 10, 1427. https://doi.org/10.3390/catal10121427
Zhang H, Catania R, Jeuken LJC. Membrane Protein Modified Electrodes in Bioelectrocatalysis. Catalysts. 2020; 10(12):1427. https://doi.org/10.3390/catal10121427
Chicago/Turabian StyleZhang, Huijie, Rosa Catania, and Lars J. C. Jeuken. 2020. "Membrane Protein Modified Electrodes in Bioelectrocatalysis" Catalysts 10, no. 12: 1427. https://doi.org/10.3390/catal10121427
APA StyleZhang, H., Catania, R., & Jeuken, L. J. C. (2020). Membrane Protein Modified Electrodes in Bioelectrocatalysis. Catalysts, 10(12), 1427. https://doi.org/10.3390/catal10121427