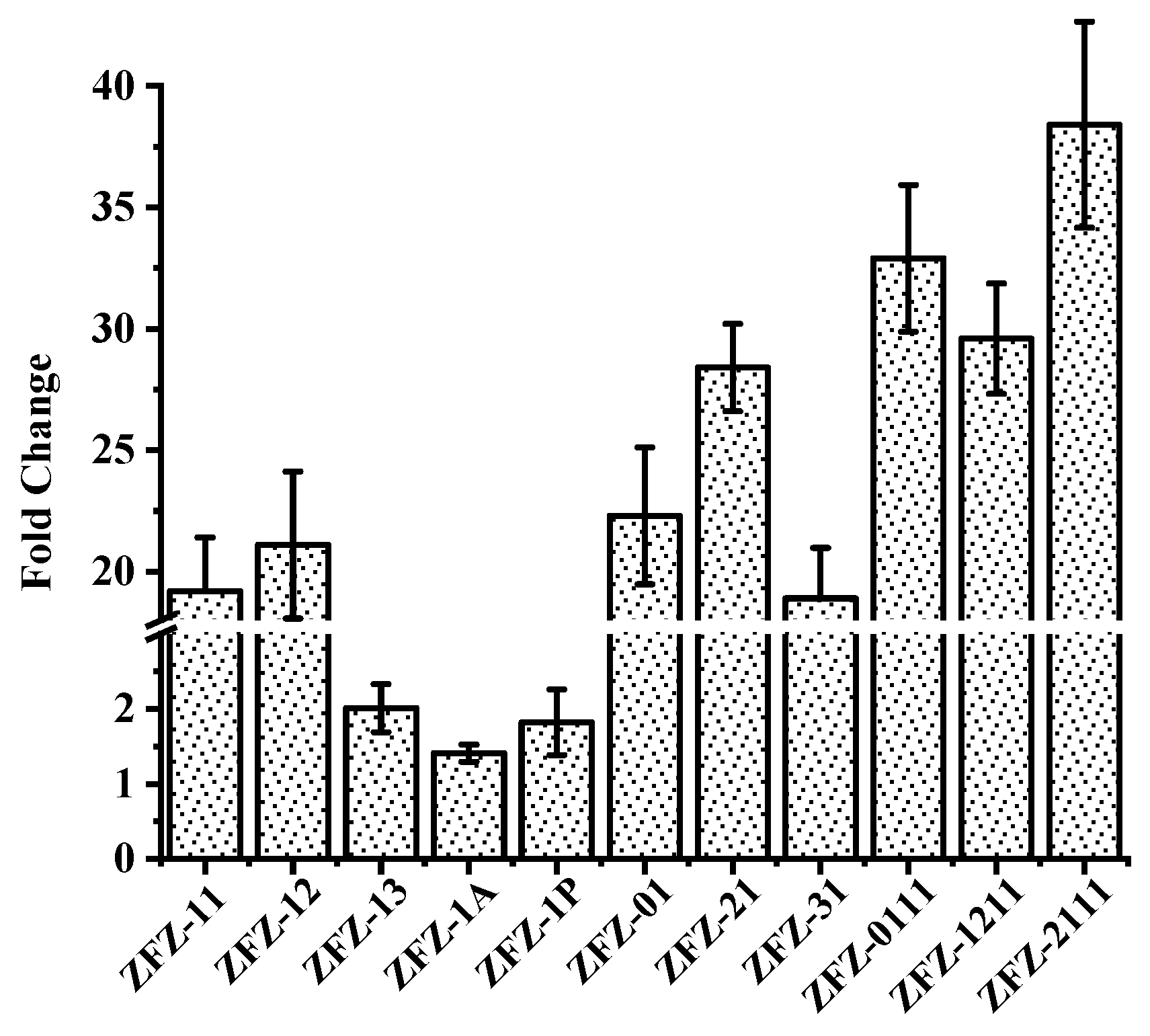Abstract
As an important hormone drug intermediate, androst-1,4-diene-3,17-dione can be bio-converted from phytosterols. However, separation and purification in the downstream process are very difficult due to the similarity in structure and physiological characteristics between ADD and androstenedione (AD). This phenomenon was correlated to the insufficient enzyme activity of 3-ketosteroid-Δ1-dehydrogenase (KSDD), which specifically catalyzes the C1,2 dehydrogenation of AD. In order to obtain a highly purified ADD from phytosterols, the dehydrogenation effect of different kinds of KSDDs and the transcription effect of four promoter sequences on ksdd were analyzed in Mycobacterium sp. ZFZ (ZFZ), the cell host that transform phytosterols to AD in the oil-aqueous system. A tandem KSDD expression cassette containing strain ZFZ-2111 yielded 2.06 ± 0.09 g L−1 ADD, with a molar ratio of ADD/AD at 41.47:1.00 in 120 h. In waste cooking oil-aqueous media, the proportion of ADD in the fermentation by ZFZ-2111 was 92%. The present study provides a reliable theoretical basis for the step-by-step transformation of phytosterols to ADD.
1. Introduction
Steroid pharmaceuticals play an irreplaceable role in the treatment of various diseases [1,2]. As the important precursor for the synthesis of hormone pharmaceuticals utilized for hormonal, anti-inflammatory and anti-tumor, androst-1,4-diene-3,17-dione (ADD) is a key derivative produced in the process of steroid degradation by microorganism [3], which can be used for the synthesis of various high-value drugs, such as estrogen, progestogen and contraceptive agents [4]. Many steroid compounds have been mainly obtained by chemical synthesis for a long time [5,6,7,8]. However, green and economical biotransformation methods have attracted widespread attention, and have been gradually applied to the pharmaceutical industry due to the depletion of raw materials and the growing concerns over the environmental damages linked to the chemical manufacturing [3,9]. At present, the ADD produced by the dehydrogenation of androstenedione (AD) catalyzed by 3-ketosteroid-Δ1-dehydrogenase has become a favorable method [10,11] due to the simple and high catalytic efficient reaction.
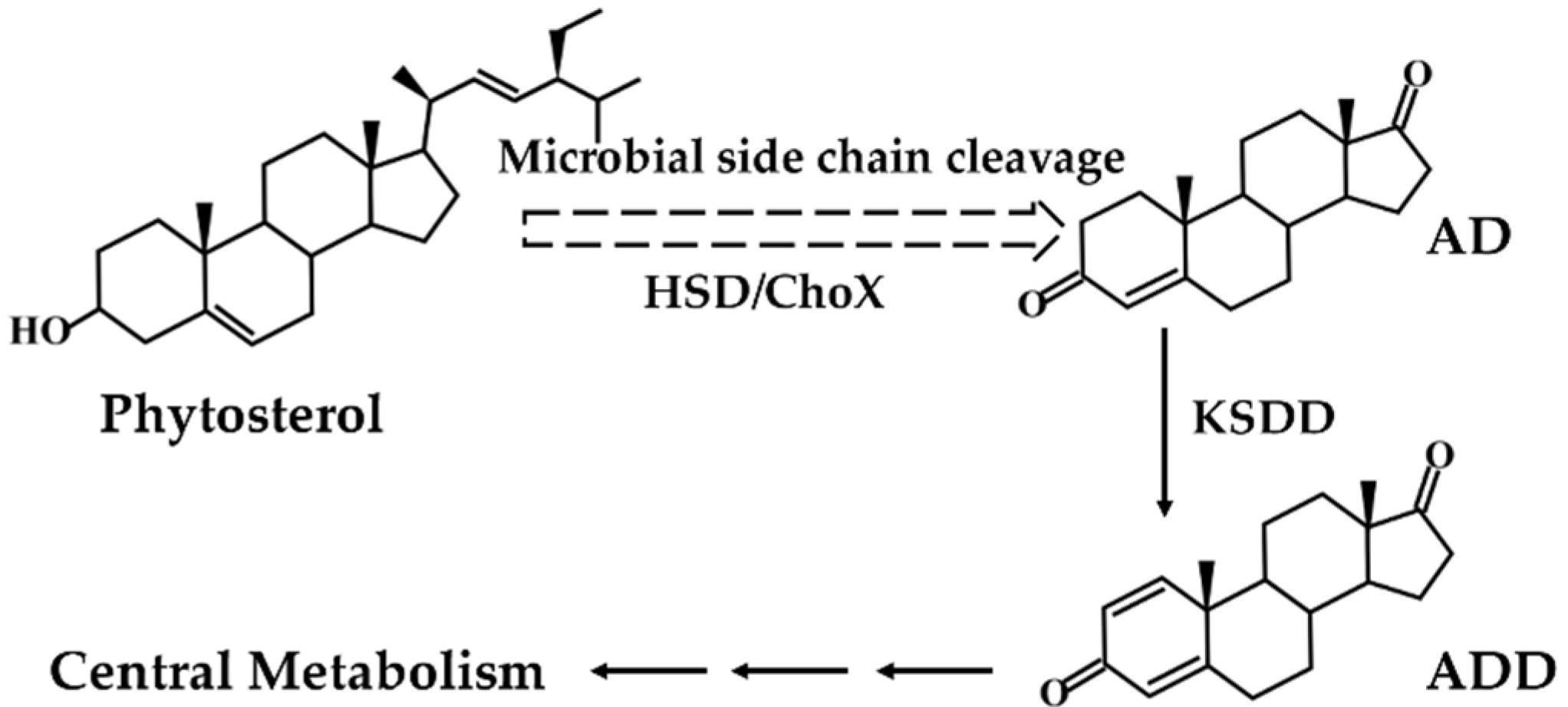
3-ketosteroid-Δ1-dehydrogenase (KSDD, EC 1.3.99.4), a flavin adenine dinucleotide (FAD) dependent enzyme, is pivotal in the process of phytosterols biodegradation and catalyzes the Δ1-dehydrogenation of AD to ADD (Figure 1) [12]. Many microorganisms that possess KSDD, such as Rhodococcus, Arthrobacter, and Nocardia, were found to be capable of converting AD to ADD [13,14]. Some researchers have focused on strain mutation or optimizing fermentation strategies to obtain ADD. For example, Shao et al. [15] reported that Mycobacterium neoaurum JC-12 converted phytosterols to ADD and 18.6 g L−1 ADD was obtained by using a three-stages strategy. Yao et al. [16] reported the transformation of phytosterols to ADD through Mycobacterium neoaurum. NwIB-01MS, and 5.57 g L−1 ADD was harvested. Besides, some laboratories have succeeded in the efficient heterologous overexpression of KSDD in E. coli, B. subtilis, etc. [17,18]. Protein engineering of the KSDD molecule to enhance dehydrogenation activity by site-directed mutagenesis has greatly increased the proportion of ADD in the product [19,20,21,22]. However, there is always residual AD in the fermentation broth of phytosterols to ADD that affects the purification of ADD. Therefore, the highly active KSDD enzyme is the key to obtaining a single ADD product.
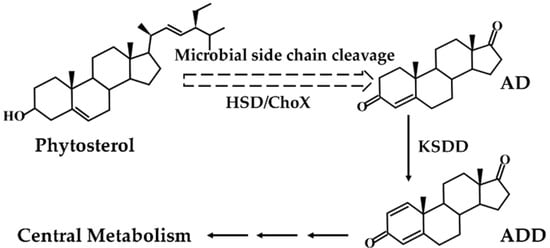
Figure 1.
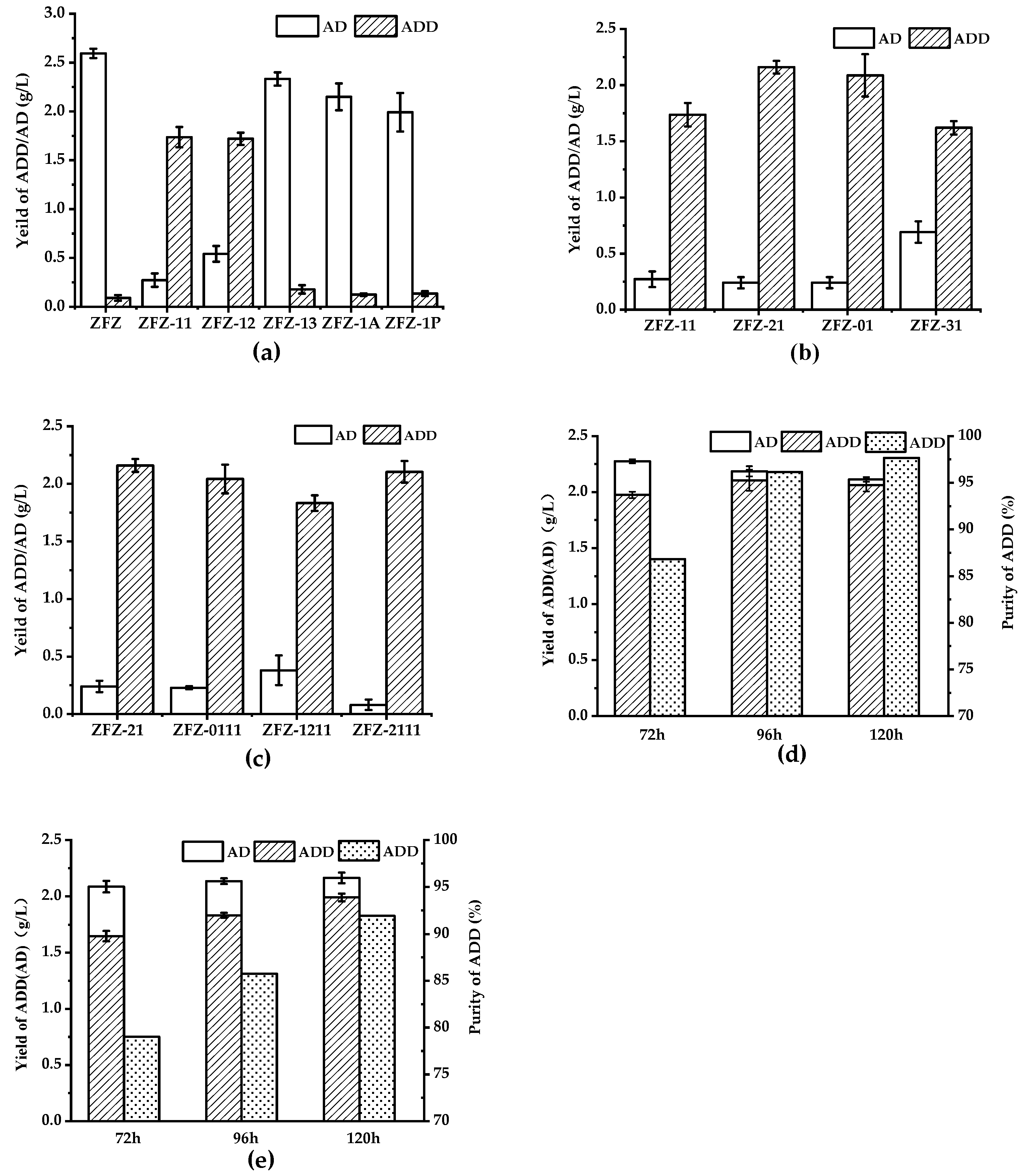
Microbial transformation pathway of phytosterols to androst-1,4-diene-3,17-dione (ADD).
The biotransformation of phytosterols to ADD was limited due to the low solubility of phytosterols [4]. To overcome the challenges of low steroid solubility, many ways such as the addition of a cosolvent, ultrasonic solubilization and water-organic biphasic fermentation system [23,24,25] were investigated. However, these methods cannot be applied to industrial production due to the high cost and complicated operation [26,27]. Another reason for the low bioconversion rate of phytosterols is that microbial cells are often exposed to toxic conditions, side effects of degradation products, and organic agents [14]. However, these two problems can be solved by the oil-aqueous two-phase system, in which the oil phase can act as a “reservoir” for the substrate and product [23,28]. On one hand, the solubility of phytosterols in soybean oil is higher (13 g L−1) than in water [29]. On the other hand, soybean oil is less aggressive toward microorganisms compared to organic solvents. It has been reported that the yield of AD carried out in the oil-aqueous biphasic system by Mycobacterium sp. MB 3683 was greatly increased to 84.8% [23]. Waste cooking oil (WCO) is easily available from restaurants, household kitchens, and agricultural manufacturers, and is rich in phytosterols, which can be used to produce AD without deep purification [3,9]. Therefore, the application of WCO as a conversion medium not only reduce the production cost of ADD but further expand the reuse of waste resources [30].
In the present study, Mycobacterium sp. ZFZ (ZFZ) was stored in our laboratory, which can efficiently transform phytosterols to AD in the oil-aqueous two-phase system. The investigators attempted to construct a strain with high activity KSDD to obtain a highly purified ADD production. Five KSDDs from M. neoaurum DSM 1381, Aspergillus fumigatus, and Pimelobacter simplex were successfully amplified, and integrated into the genome of ZFZ. The transcriptional effects of different promoters on KSDD were verified. To further increase the catalytic activity of KSDD and reduce the proportion of AD in the fermentation broth, the efficient transformation of phytosterols to ADD with a tandem promoter-ksdd cassette mutant strain in the soybean oil-aqueous two-phase system was achieved. Finally, an effective and lower-costly method that transform phytosterols to ADD through a tandem promoter-ksdd cassette mutant strain in the waste cooking oil-aqueous system was developed. This work represents a highly efficient method for ADD from phytosterols with potential industrial applications.
2. Results
2.1. Gene Cloning and Sequence Analysis
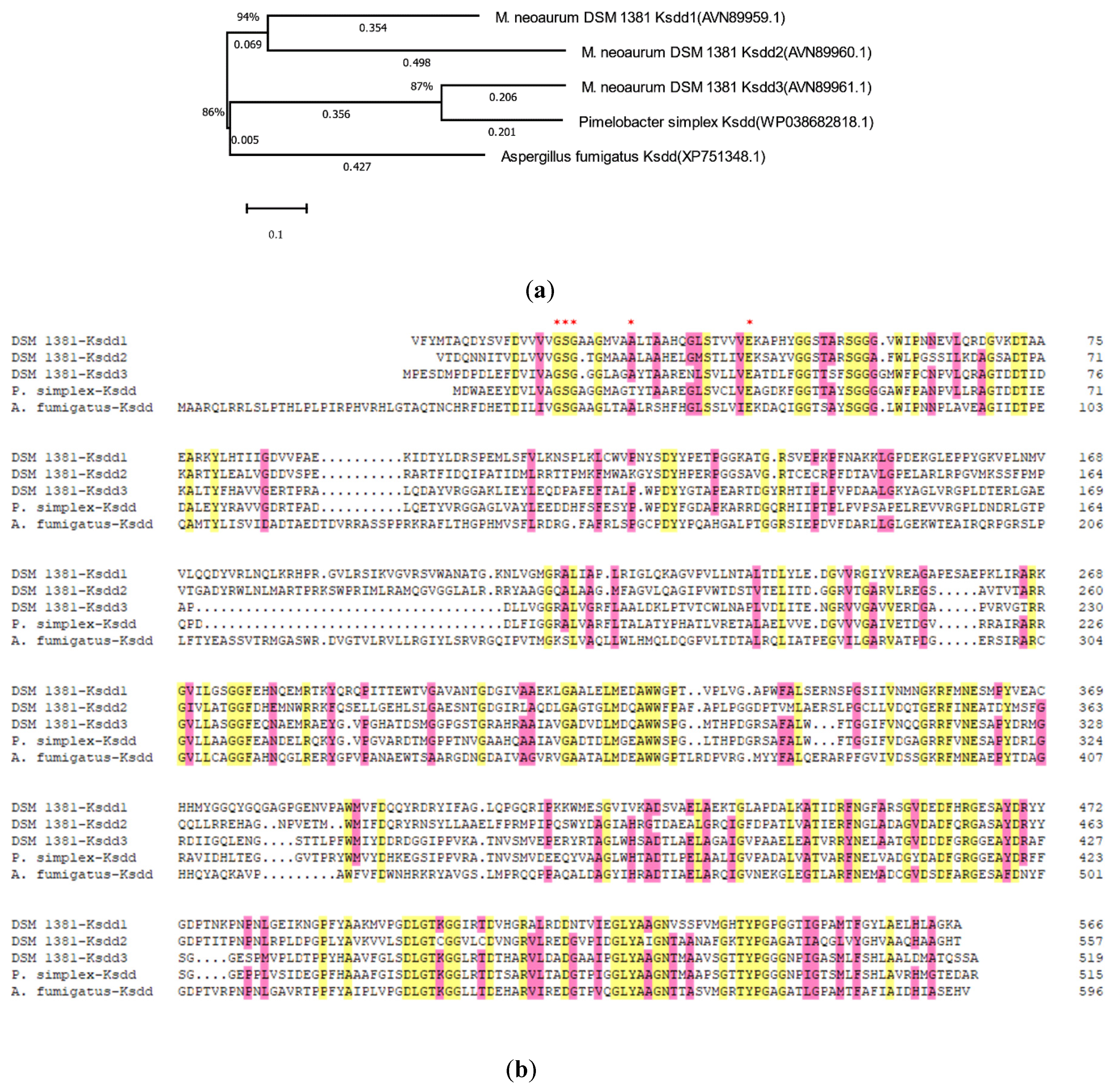
To obtain the nucleotide sequence of KSDD (ksdd1, ksdd2, and ksdd3), the genome of M. neoaurum DSM 1381 was sequenced and annotated. Three putative ksdd ORFs were identified in our previous study [31]. KSDDA from Aspergillus fumigatus and KSDDP from Pimelobacter simplex were reported to have high dehydrogenation activity on the C1,2 bond of ring A. Five KSDD were amplified, and a phylogenetic tree was subsequently constructed with MEGA X to elucidate the evolutionary relationships (Figure 2a) [32]. The BLAST result of KSDD1 from M. neoaurum DSM 1381 exhibited a high similarity at the amino acidic level (>95%) to M. neoaurum NwIBL-01, M. neoaurum ATCC 25795 and M. neoaurum VKM Ac-1816D, while the KSDD2 of M. neoaurum DSM 1381 was 85% identical at the amino acidic level to the KstD2 of M. neoaurum ATCC 25795. All five KSDDs from different genera had the putative N-terminal FAD-binding motifs G-SG-(A/G)-(A/G)-(A/G)-X17-E (Figure 2b) [33,34]. Figure S1 shows that ksdd1 and ksdd2 both have one transmembrane segment near the N-terminal, which suggest they could be membrane proteins, through analysis by TMHMM (http://www.cbs.dtu.dk/services/TMHMM/).
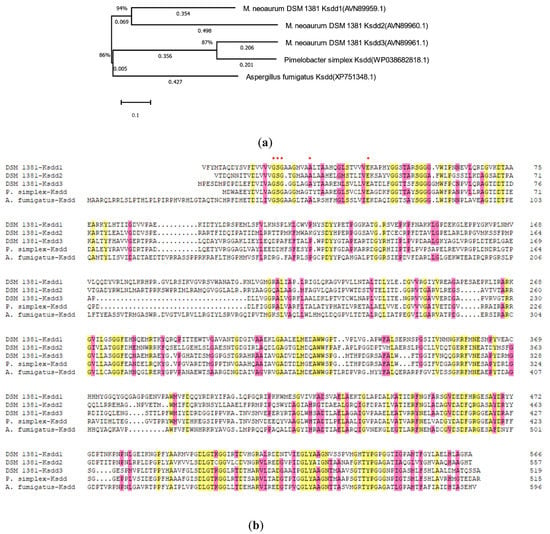
Figure 2.
Phylogenetic tree and sequence alignment analysis of 3-ketosteroid-Δ1-dehydrogenase (KSDD). (a) The phylogenetic tree of five KSDD enzymes from different species. 1,000 replicates were set to generate bootstrap values when conducting analysis. The numbers below the lines represent the evolutionary distance. (b) The sequence alignment of five KSDDs. DSM 1381-Ksdd1, DSM 1381-Ksdd2, DSM 1381-Ksdd3, Ksdd from M. neoaurum DSM 1381; A.fumigayus-Ksdd, Ksdd from Aspergillus fumigatus; P.simples-Ksdd, Ksdd from Pimelobacter simplex. A conserved sequence for FAD-binding region is indicated by asterisk.
2.2. Cell Growth of KSDD Recombinant Strains
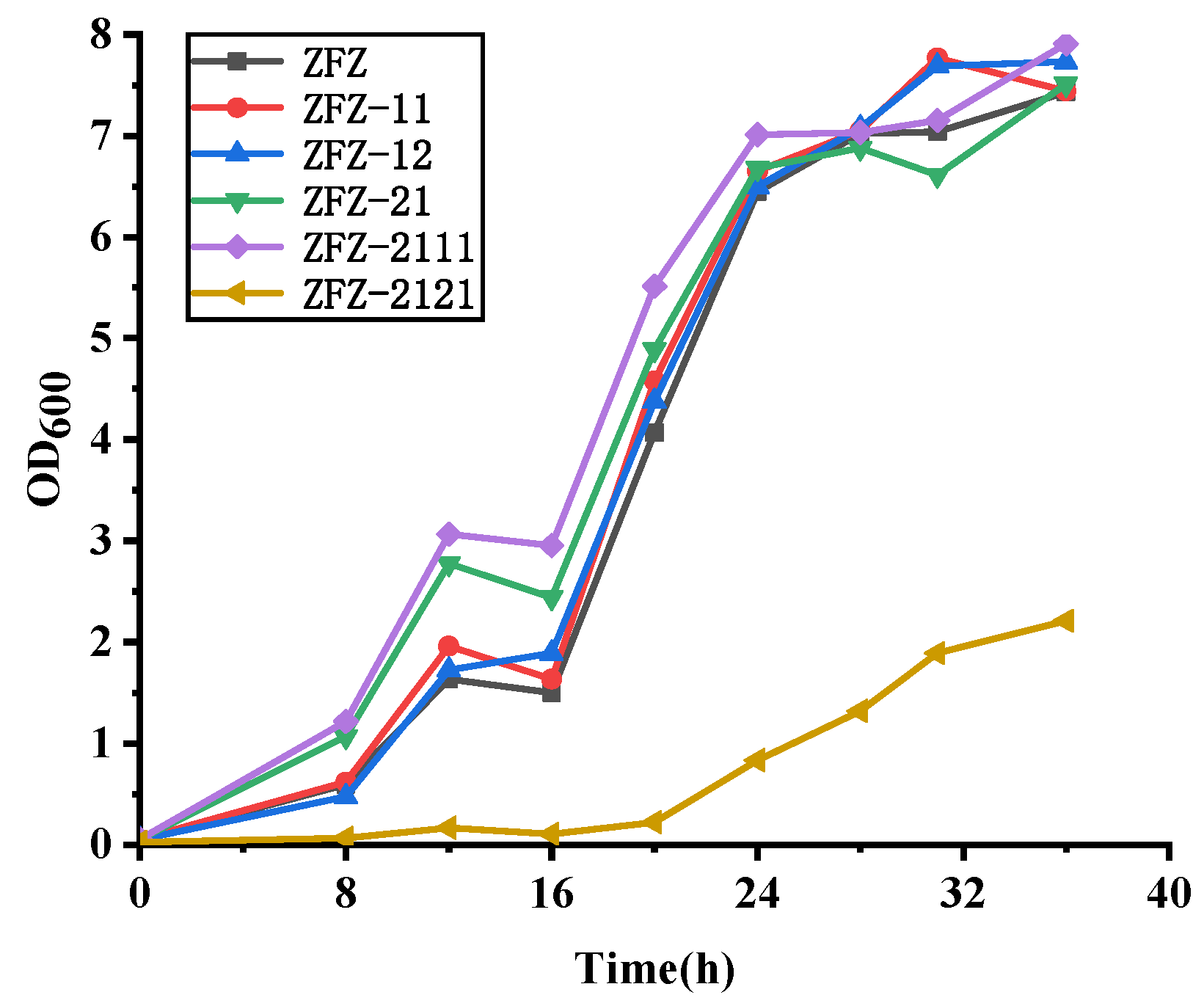
As described in Section 4.2, the recombinant KSDD integration plasmids were constructed and introduced to ZFZ by electroporation to obtain the ADD production strain. The effects of different ksdds on the growth of recombination strains were detected (Figure 3 and Figure S2). The growth curve of almost all recombinant strains was consistent with ZFZ. ZFZ-21, ZFZ-2111 and other recombinant strains have higher OD values than ZFZ, except for ZFZ-2121. This shows that the expression of KSDD has a certain promoting effect on the growth of ZFZ. It is possible that the high KSDD dehydrogenase activity would inhibit the growth of the strain, and that the OD value of ZFZ-2121 containing a tandem Psmyc-ksdd1 cassette would be much lower than ZFZ.
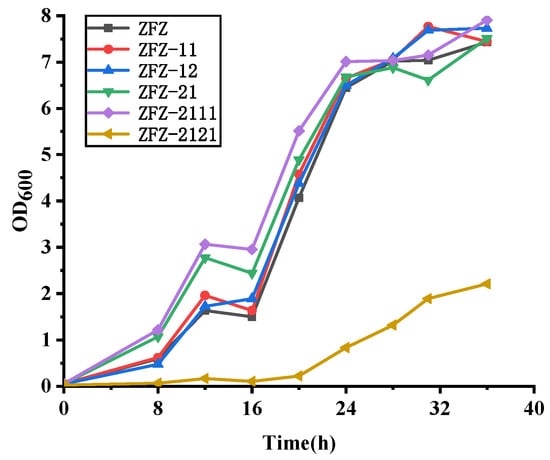
Figure 3.
Growth curve of recombinant strains.
2.3. Enzyme Activity Assay of Recombination Strains
To stabilize the expression of heterologous KSDDs in ZFZ, pMV306, which is an integrated plasmid, was used to express dehydrogenase proteins. To detect the activities of KSDDs, extracellular and intracellular KSDDs from the recombinant ZFZ were assayed using the spectrophotometrical method. As shown in Table 1, the extracellular KSDD enzyme activity from all recombinant strains on AD was not detected. This result is consistent with the notion that dehydrogenase is not a secretory enzyme, and that it is attached to the cell membrane (Figure S1) to exert a dehydrogenation effect to form a C1,2 double bond. The activities of the intracellular KSDD, which were obtained from ZFZ, were merely 0.11 ± 0.01 U mg−1. The higher intracellular activities of ZFZ-11 and ZFZ-12 were 2.03 ± 0.05 U mg−1, and 4.31 ± 0.08 U mg−1, respectively. However, the activities of ZFZ-13 that contained KSDD3 from M. neoaurum DSM 1381 was only 0.52 ± 0.05 U mg−1. This is similar to the previous research results of the investigators, in which only KSDD1 and KSDD2 from M. neoaurum DSM 1381 play an important role in the degradation of phytosterols [31]. This is probably due to the no codon optimization, in which KSDDA and KSDDP did not exhibit a high dehydrogenation activity on AD. Strong promoters and increased gene expression can better increase enzyme activity.

Table 1.
The KSDD enzyme activity of different recombinant strains.
All assays were performed in triplicate. The standard deviations of the biological replicates are shown. (1 U of KSDD enzyme activity was defined as a reduction of 1 μmol min−1 of DCPIP at pH 7.0 and 30 °C); u.d., undetectable enzyme activity.
2.4. Transcriptional Analysis of Ksdd in Recombinant Strains
Apart from the KSDD enzyme activity analysis, the change in the transcription level of all recombinant strains was investigated by RT-qPCR with the addition of phytosterols. As shown in Figure 4, phytosterols induced the transcription of KSDD in ZFZ-11 and ZFZ-12, which increased by 19.2-fold and 22.1-fold, respectively. However, the KSDD transcription only increased by 2.01-fold, 1.41-fold, and 1.82-fold in ZFZ-13, ZFZ-1A, and ZFZ-1P, respectively, relative to ZFZ, and when induced with phytosterols. In the KSDD-expressing strains, in which the transcription was controlled by different promoters, ZFZ-21 had the highest level of enzyme transcription (increased by 28.4-fold). The transcription of KSDD in ZFZ-01 increased by 22.3-fold, suggesting that the native promoter of KSDD1 did not have a great advantage in KSDD expression. The expression changes of tandem expression cassettes engineering bacteria ZFZ-0111 (increased by 32.9-fold), ZFZ-1211 (increased by 29.6-fold) and ZFZ-2111 (increased by 38.4-fold) were significantly higher than other recombinant strains.
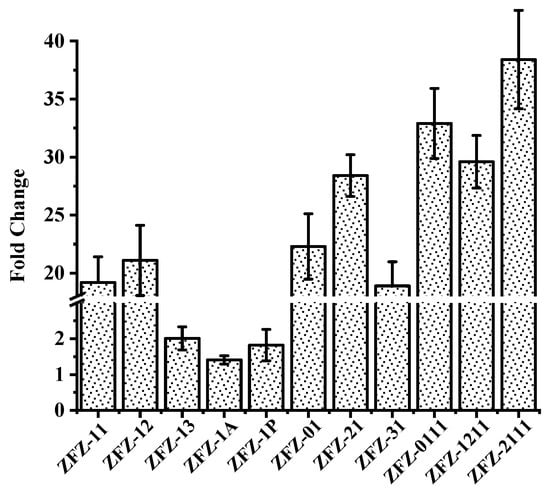
Figure 4.
Analysis of KSDD genes transcription in recombinant strains. The fold change indicates the increase in expression level of recombinant strains after removing the level in Mycobacterium sp. ZFZ (ZFZ). The data were calculated from three independent experiments.
2.5. The Phytosterols Bioconversion in an Oil-Aqueous Two-Phase System
Many studies focused on how to convert AD to ADD due to the low water solubility of phytosterols. However, these methods relatively increase the production cost of ADD. Compared to the expensive cyclodextrin, cheap soybean oil can not only increase the solubility of sterols but also shortens the fermentation time. The molar ratio of ADD and AD in the production is closely correlated to the activity of KSDD. The ADD produced by phytosterols degradation can be divided into two steps: first, the phytosterols are converted to AD, and C1,2 bond is dehydrogenated by KSDD at the A ring to form ADD. When using various KSDD recombinant Mycobacterium strains to accumulate ADD from phytosterols, the products are analyzed by HPLC and GC. This shows that both ADD and AD are present in the fermentation broth (Figure 5). As shown in Figure 5a, merely fewer ADD formations in addition to AD were detected in phytosterol fermentation by ZFZ after 96 h, and the phytosterols were completely depleted. However, the molar yields of ADD and AD in fermentation by recombinant strain ZFZ-11 are 50.20% (1.736 g L−1) and 7.92% (0.272 g L−1), and that in fermentation by recombinant strain ZFZ-12 are 49.72% (1.720 g L−1) and 15.76% (0.541 g L−1). Although the yields of ADD are similar, the molar ratios of ADD and AD were 6.83:1 and 3.17:1, respectively. However, the ZFZ-13, the ZFZ-1A, and ZFZ-1P that contained KSDD3, KSDDA, and KSDDP under the control of promoter Hsp60 did not achieve the desired effect. KSDD1 was selected as the experimental optimal enzyme in the subsequent experiments because ZFZ-11 has fewer impurities.
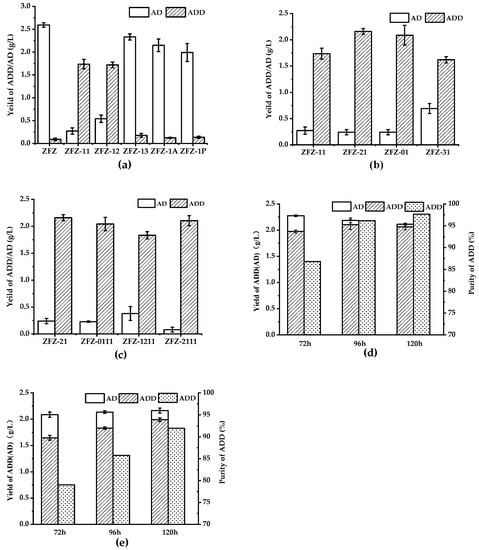
Figure 5.
The phytosterols bioconversion in the oil-aqueous two-phase system by engineered strains. The biotransformation was carried out for four days with 5 g L−1 of phytosterols as the substrate. (a) The effects of different KSDDs on the ADD yield. (b) The effects of different promoters on the ADD yield. (c) The effects of a tandem ksdd expression box on ADD yield. (d) The phytosterol biotransformation by ZFZ-2111 was carried out with soybean oil. (e) The phytosterol biotransformation by ZFZ-2111 was carried out with waste cooking oil. All assays were performed in triplicate, with three independent measurements.
The presence of AD in the fermentation broth indicates that the KSDD dehydrogenase activity of the recombinant strains is insufficient. The promoter has the function of determining the transcription level of the gene, and an appropriate promoter could elevate the activity of KSDD. In the recombinant Mycobacterium strains, in which KSDD1 is under control of a different promoter sequence (Figure 5b), the production of ADD improved to 2.16 g L−1, the AD was 0.24 g L−1, and the molar ratio of ADD/AD was 9.1:1 when the recombinant strain ZFZ-21 was used as the biocatalyst. This means that the promoter of Psmyc exhibited a higher transcriptional ability.
Surprisingly, the ADD in the fermentation broth with ZFZ-2111, which harbored the Psmyc-ksdd1-Hsp60-ksdd1 expression box, was 2.10 g L−1. Although the yield of ADD was less than that of ZFZ-21, the molar ratio of ADD/AD in the fermentation broth of the phytosterol conversion was almost 25:1 (Figure 5c) after 96 h. This also shows that the higher the activity of KSDD, the lesser the remaining byproduct AD in the fermentation broth. Figure 5d shows that the fermentation of ZFZ-2111 in the soybean oil-aqueous in 72-120 h. With the increase in fermentation time, the ratio of ADD: AD in the product increased from 6.59: 1.00 to 25:1.00. Finally, ADD reaches 97.65% at 120 h. However, the total amount of AD and ADD gradually decreased, which may be due to the presence of 3-Ketosteroid 9α-hydroxylase in ZFZ, which caused ADD to be degraded.
2.6. Phytosterol Bioconversion in the Waste Cooking Oil-Aqueous System by ZFZ-2111
WCO, which is the main waste generated by oil in food processing, is readily available from massive restaurants and household kitchens. Considering the economy and environmental protection, the effect of WCO as a co-solvent in phytosterol fermentation on ADD by ZFZ-2111 was detected (Figure 5e). The content of ADD is lower than that of soybean oil as a co-solvent in the fermentation process using WCO as a co-solvent, and some phytosterols were not completely degraded (data not shown). The concentration of ADD in the fermentation broth was 1.83 g L−1, which decreased by 13.07% when compared to that in soybean oil in 96 h. ADD increased to 92% when the fermentation time was extended to 120 h.
3. Discussion
ADD is an important steroidal intermediate in the pharmaceutical industry. Microbial degradation to produce steroidal intermediates has obtained extensive attention. Some barriers, such as low solubility, the similar properties of degradation products AD and ADD in structure and chemical properties, and the low tolerance of microbes [3,35], seriously limit the application of the biotransformation of ADD from phytosterols. Correspondingly, strain improvement through the overexpression and/or knockout of key genes is an important and effective means in industrial applications [36,37].
In the present study, the problem that the investigators wanted to solve is that the purification difficulty caused by similarities in the structure and chemical properties of the production of AD and ADD to obtain high purity ADD products. The simultaneous presence of AD and ADD in the fermentation broth was caused by the insufficient activity of KSDD. Therefore, many different KADD overexpression strains were constructed. The effect of five KSDDs and four promoters on ADD was detected in the process of phytosterol degradation by recombinant strains. Phytosterols were completely converted to AD after 72 h of inoculation with ZFZ, which were accompanied by the complete emulsification of soybean oil when utilizing the soybean oil-aqueous two-phase system, and the molar yield of AD was more than 90%. However, with the extension of the fermentation cycle, the concentration of AD gradually decreased, and a small amount of ADD was produced in the fermentation broth. This phenomenon indicates that ZFZ still contains a weak C1,2 dehydrogenation effect.
Among the five KSDD overexpression bacteria, only ZFZ-11 and ZFZ-12 had a stronger dehydrogenation effect on AD, when compared to the other three KSDDs, in which the ADD yields were 1.74 g L−1 and 1.72 g L−1, respectively. Interestingly, the enzyme activity of KSDD2 from ZFZ-12 was higher than KSDD1 from ZFZ-11 on AD. This result is consistent with the notion that the activity of KSDD2 is higher than KSDD1 when utilizing E. coli and B. subtilis as the host bacteria for AD conversion to ADD [31]. However, in the present study, the ADD yield in the fermentation of ZFZ-12 was lower than that of ZFZ-11 when utilizing the soybean oil-aqueous conversion system to produce ADD from phytosterols. This result may be correlated to the finding that a putative KstR binding site was only identified before KSDD1, and indicating that KSDD1 plays a more important role in the process of phytosterol to ADD [31]. Another possibility is that the enzyme activity of KSDD2 was reduced by oil because the concentration of ADD is also lower than that in the fermentation process without oil (data not listed in the present study).
Several promoters also exhibited significant differences in the effect of the transformation of ADD from phytosterols. ZFZ-21, which harbored the Psmyc-ksdd1 sequence and greatly reduced the AD proportion in the process of the fermentation degradation of phytosterols, had a molar ratio of ADD/AD of 9.1:1. However, the total amount of AD and ADD in the fermentation process of all single-gene overexpressing bacteria gradually decreased, revealing that there was a 3-ketosteroid-9-alpha-monooxygenase (KSH) hydroxylase activity in the mutation strains, which resulted in a decrease in the accumulation of ADD. This may be due to the lower amount of inventory and the low activity of KSH, and because 9-OHAD was not detected in the fermentation broth.
Although soybean oil increases the solubility of phytosterols, the AD conversion time was extended, because the KSDD activity was influenced by the soybean oil, when compared to the system of E. coli and B. subtilis [11,38]. Furthermore, this increased the probability of the destruction of ADD by KSH. Similarly, the degradation of ADD occurs faster in the fermentation of a tandem KSDD recombinant strains, such as ZFZ-0111, ZFZ-1211, and ZFZ-2111. There was a 95% ADD production in the fermentation broth by ZFZ-2111, and the molar ratio of ADD: AD was 25:1 in 96 h. However, in the final biotransformation with soybean oil, 2.06 g L−1 of ADD was accumulated in 120 h, and the ratio of ADD: AD increased to 41.47:1.00. Utilizing WCO as a co-solvent has a tremendous economic advantage because more than five million tons of WCO are generated per year in China [30,39]. In the process of phytosterol degradation with WCO, the highest, which is 1.98 g L−1 of ADD, was accumulated in 120 h. Although the prolonged fermentation time and proportion of ADD (92%) in the product were lower than that with soybean oil, WOC as a co-solvent in phytosterols degradation would be a new approach to utilize WOC, considering the economy and environment-friendly factors. Giving that the phenomenon on product degradation and the incomplete conversion of phytosterols, the knockout of ksh, and the amounts of WCO should be studied in the future.
4. Materials and Methods
4.1. Strains, Plasmids, and Reagents
Mycobacterium neoaurum DSM 1381 was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), and Aspergillus fumigatus, Pimelobacter Simplex, Mycobacterium sp. ZFZ and E. coli were stored in our laboratory. Supplementary Table S1 shows the plasmids, strains and primers used in this study. E. coli DH5α was cultivated with LB medium at 37 °C and 200 rpm for molecular cloning, while other strains were cultured in seed medium which including 10 g L−1 of corn steep powder, 6 g L−1 of glucose, 5.4 g L−1 of NaNO3 and 2 g L−1 of K2HPO3. Plasmids were constructed by using the ClonExpress® II One Step Cloning Kit. The Restriction endonucleases, Pfu DNA polymerase and In-Fusion Cloning Kit were purchased from Vazyme Biotech Co., Ltd. (Nanjing, China). The phytosterols, which consisted of 45% β-sitosterol, 37% campesterol, and 18% stigmasterol, were purchased from Yunnan Biological Products Co., Ltd. (Yunnan, China). The standards of AD or ADD, 2,6-dichlorophenolindophenol (DCPIP) and Phenazine methosulphate (PMS) were purchased from Sigma-Aldrich (Shanghai, China). WCO was obtained from the dining hall of Shanghai Advanced Research Institute. All other reagents were of analytical grade or higher unless noted.
4.2. Cloning and Expression
Plasmid pMV306, which is an integration plasmid without a promoter sequence, was employed to integrate the KSDD gene into the Mycobacterium genome. First, the Hsp60 promoter sequence was cloned into the XbaI/EcoRI sites of pMV306 through homologous recombination using a One Step Cloning Kit to create plasmid phsp60. For the construction of single KSDD gene expression plasmids, the ksdd1 (GB: MG251735.1), ksdd2 (GB: MG251736.1) and ksdd3 (GB: MG251737.1) from Mycobacterium neoaurum DSM 1381, ksddA (GB: XM_746255.1) from Aspergillus fumigatus Af293, and ksddP (GB: CP009896.1) from Pimelobacter simplex VKM Ac-233D were respectively amplified and cloned into the EcoRI/SalI site of phsp60. After verification by DNA sequencing, the resulting phsp60-kd1/phsp60-kd2/phsp60-kd3/phsp60-kdA/phsp60-kdp was introduced in ZFZ by electroporation methods, as described by Tauch et al. [40]. To increase the ksdd1 gene expression, different promoter sequences were cloned into the plasmid, replacing Hsp60. The promoters Psmyc (GB: MF498902.1) and pJK-F8 (GB: KR091965.1) were commercially synthesized by Genewize Co., Ltd. (Suzhou, China). The host promoter regions, together with the ORFs of ksdd1, which was named as Ymn-ksdd1, were amplified from the M. neoaurum DSM 1381 genome. The Psmyc and pJK-F8 promoters were respectively cloned into phsp-kd1 between the XbaI and EcoRI sites, and Ymn-ksdd1 was cloned into pMV360 between the XbaI and SalI sites. After being verified by DNA sequencing, the prepared ppsmyc-kd1, ppjkF8-kd1, and pYmn-kd1 were introduced in ZFZ by electroporation. For the construction of the tandem expression cassettes engineering bacteria, the different promoter-ksdd-terminator expression cassettes were amplified from the constructed plasmids, and inserted into the KpnI site of plasmid phsp60-kd1, to construct the double promoter-ksdd-terminator series expression plasmid. The recombinant plasmid was verified by digestion and DNA sequencing, and the resulting pYmn-kd1-Hsp60-kd1, pHsp60-kd2-Hsp60-kd1, pPsmyc-kd1-Hsp60-kd1, and pPsmyc-kd1-Psmyc-kd1 were introduced in ZFZ by electroporation. Kanamycin of 50 µg mL−1 was supplemented into the medium.
4.3. KSDD Enzyme Activity Assay
The activity of KSDD from different recombinant strains in cell-free extracts was measured at 600 nm (ε600 = 18.7 × 103 cm−1 M−1) by Thermo Scientific Nano Drop 2000 spectrophotometer at 30 °C. The recombinant ksdd overexpression strains were induced with 5 g L−1 of phytosterols (36–48 h), and the cells pellets collected at 7,000 rpm for 10 min from 50 mL of the culture medium were resuspended in 2 mL of 50 mM Tris–HCl buffer (pH 7.0) after being washing twice at 4 °C. Then, the cell was sonicated for 15 min under the protection of an ice-water bath and the supernatant of the cell extracts (10,000 rpm, 4 °C, 10 min) was used for the enzyme activity assay. The reaction mixture (1 mL) contained 50 mM of Tris-HCl pH 8.0, 0.12 mM of DCPIP, 1.5 mM of PMS, 500 μM of steroid substrate in methanol (2%) and appropriate concentration of cell-free extracts. Furthermore, 1 U of enzyme activity was defined as a reduction of 1 μmol min–1 of DCPIP. Three replicates were analyzed.
4.4. Total RNA Isolation, cDNA Synthesis and qPCR Conditions
500 μL of samples (Mid-log exponential phase) were harvested on seed medium with 5 g L−1 of phytosterols and immediately frozen in liquid nitrogen. Total RNA was isolated using the UNlQ-10 Column TRIzol Total RNA Isolation Kit (Sangon Biotech, China). The resulting mixture was treated with DNase I to eliminate the contaminating genomic DNA. The purity of the RNA was analyzed using NanoDrop spectrophotometer (NanoDrop 2000c, Thermo Scientific, USA) and the integrity was checked by electrophoresis on a 1.5% agarose gel. RNA samples were subjected to cDNA synthesis using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara). Thermocycling was performed using the following conditions: 5 min at 95 °C followed by 40 cycles alternating between 10 s at 95 °C and 30 s at 60 °C. Melting curve analysis (60 °C–95 °C) was routinely performed after 45 cycles to verify primer specificity. The 2−ΔΔCt method was used to calculate the relative expression level of the target genes. The 16S rRNA was used as a reference gene, and the seed medium was set as a native control. All the experiments were repeated three times.
4.5. Phytosterol Biotransformation and Analytical Methods
The recombinant cells were inoculated to 30 mL of seed medium in a 250-mL Shaker flask and cultured at 30 °C and 180 rpm. 3 mL (10% v/v) of the seed medium was transferred to the 30 mL oil-aqueous two-phase system in a 250 mL Shaker flask with baffle when the optical density (OD) reached the Mid-log exponential phase. The biotransformation medium contained the ingredient of the seed medium, 5 g L−1 of phytosterols, 1 g L−1 of Tween 80, and soybean oil or WCO. The pH of the medium was adjusted to 7.5 and was heat sterilized (20 min at 115 °C). Then, the samples were extracted three times with an equal volume of ethyl acetate after the oil phase in the fermentation broth was emulsified into small droplets. Afterward, the extracts were mixed well to analyze with HPLC and GC. The specific methodology was described by Zhang et al. [20]. All experiments were performed in triplicate.
5. Conclusions
ADD was produced by Mycobacteria using phytosterols, and the activity of KSDD affected the proportion of ADD in the production. The KSDD1 from Mycobacterium neoaurum DSM 1381 related to the ADD synthesis from phytosterols had the highest activity in five kinds of KSDDs, and promoter Psmyc had a better role in enhancing the transcription. Increasing the expression level can increase the molar ratio of ADD in the product, but cannot completely remove AD. WOC, as a co-solvent, can be applied for phytosterol transformation with a better effect. This strategy that overexpresses KSDD in ZFZ, which could convert phytosterols to AD, to achieve the one-step efficient produce ADD from low-cost phytosterols with WCO, is a convenient and economical method for the ADD production.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4344/10/6/663/s1, Figure S1: The analysis diagram of transmembrane region by TMHMM, Figure S2: Growth curve of recombinant strains, Table S1: Microorganisms, plasmids and primers used in the study.
Author Contributions
Conceptualization, X.L., J.S. (Junsong Sun) and B.Z.; data curation, X.L. and R.Z.; formal analysis, X.L. and B.Z.; funding acquisition, B.Z.; investigation, X.L., R.Z., H.C., Z.B. and C.Y.; methodology, X.L., R.Z., Z.B. and C.Y.; project administration, B.Z.; writing—original draft, X.L.; writing—review and editing, J.S. (Jiping Shi), J.S. (Junsong Sun) and B.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key R&D Program of China, grant number 2017YFE0112700.
Acknowledgments
We thank Jian Hao (Shanghai Advanced Research Institute, Chinese Academy of Sciences) for valuable suggestions on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fernandez-Cabezon, L.; Galan, B.; Garcia, J.L. New Insights on Steroid Biotechnology. Front. Microbiol. 2018, 9, 958. [Google Scholar] [CrossRef]
- Garcia, J.L.; Uhia, I.; Galan, B. Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb. Biotechnol. 2012, 5, 679–699. [Google Scholar] [CrossRef]
- Malaviya, A.; Gomes, J. Androstenedione production by biotransformation of phytosterols. Bioresour. Technol. 2008, 99, 6725–6737. [Google Scholar] [CrossRef]
- Fernandes, P.; Cruz, A.; Angelova, B.; Pinheiro, H.M.; Cabral, J.M.S. Microbial conversion of steroid compounds: Recent developments. Enzyme Microb. Techechnol. 2003, 32, 688–705. [Google Scholar] [CrossRef]
- Raymond, K.M.; David, M.S.; Alan, R.A. Guidebook to Organic Synthesis, 3rd ed.; Person Education Limited: Beijing, China, 2001; pp. 346–347. [Google Scholar]
- Jiang, B.; Shi, H.P.; Tian, W.S.; Zhou, W.S. The convergent synthesis of novel cytotoxic certonardosterol D2 from diosgenin. Tetrahedron Lett. 2008, 64, 469–476. [Google Scholar] [CrossRef]
- Minagawa, K.; Furuta, T.; Kasuya, Y.; Fujino, A.; Avery, M.A.; Tanabe, M. Total Synthesis of Cortisol—Application to Selective Deuteriation at C-1 and C-19. J. Chem. Soc. Perkin Trans. 1 1988, 587–591. [Google Scholar] [CrossRef]
- Woodward, R.B.; Sondheimer, F.; Taub, D.; Heusler, K.; McLamore, W.M. The Total Synthesis of Steroids. J. Am. Chem. Soc. 1952, 74, 4223–4251. [Google Scholar] [CrossRef]
- Fernandes, P.; Cabral, J.M.S. Phytosterols: Applications and recovery methods. Bioresour. Technol. 2007, 98, 2335–2350. [Google Scholar] [CrossRef]
- Li, Y.; Lu, F.; Sun, T.; Du, L. Expression of ksdD gene encoding 3-ketosteroid-Δ1-dehydrogenase from Arthrobacter simplex in Bacillus subtilis. Lett. Appl. Microbiol. 2007, 44, 563–568. [Google Scholar] [CrossRef]
- Zhang, W.Q.; Shao, M.L.; Rao, Z.M.; Xu, M.J.; Zhang, X.; Yang, T.W.; Li, H.; Xu, Z.H. Bioconversion of 4-androstene-3,17-dione to androst-1,4-diene-3,17-dione by recombinant Bacillus subtilis expressing ksdd gene encoding 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum JC-12. J. Steroid Biochem. Mol. Biol. 2013, 135, 36–42. [Google Scholar] [CrossRef]
- Rohman, A.; van Oosterwijk, N.; Dijkstra, B.W. Purification, crystallization and preliminary X-ray crystallographic analysis of 3-ketosteroid-Δ1-dehydrogenase from Rhodococcus erythropolis SQ1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68 Pt 5, 551–556. [Google Scholar] [CrossRef]
- De las Heras, L.F.; van der Geize, R.; Drzyzga, O.; Perera, J.; Llorens, J.M. Molecular characterization of three 3-ketosteroid-Δ1-dehydrogenase isoenzymes of Rhodococcus ruber strain Chol-4. J. Steroid Biochem. Mol. Biol. 2012, 132, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.V. Transformation of steroids by actinobacteria: A review. Appl. Biochem. Biotechnol. 2007, 43, 1–14. [Google Scholar] [CrossRef]
- Shao, M.L.; Zhang, X.; Rao, Z.M.; Xu, M.J.; Yang, T.W.; Li, H.; Xu, Z.H. Enhanced production of androst-1,4-diene-3,17-dione by Mycobacterium neoaurum JC-12 using three-stage fermentation strategy. PLoS ONE 2015, 10, e0137658. [Google Scholar] [CrossRef]
- Yao, K.; Wang, F.Q.; Zhang, H.C.; Wei, D.Z. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum. Metab. Eng. 2013, 15, 75–87. [Google Scholar] [CrossRef]
- Knol, J.; Bodewits, K.; Hessels, G.I.; Dijkhuizen, L.; van der Geize, R. 3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem. J. 2008, 410, 339–346. [Google Scholar] [CrossRef]
- Morii, S.; Fujii, C.; Miyoshi, T.; Iwami, M.; Itagaki, E. 3-Ketosteroid-Δ1-dehydrogenase of Rhodococcus rhodochrous: Sequencing of the genomic DNA and hyperexpression, purification, and characterization of the recombinant enzyme. J. Biochem. 1998, 124, 1026–1032. [Google Scholar] [CrossRef]
- Xie, R.L.; Shen, Y.B.; Qin, N.; Wang, Y.B.; Su, L.Q.; Wang, M. Genetic differences in ksdD influence on the ADD/AD ratio of Mycobacterium neoaurum. J. Ind. Microbiol. Biotechnol. 2015, 42, 507–513. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, X.; Cao, H.; Yuan, C.; Yuminaga, Y.; Zhao, S.; Shi, J.; Zhang, B. Purification, characterization, and application of a high activity 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum DSM 1381. Appl. Microbiol. Biotechnol. 2019, 103, 6605–6616. [Google Scholar] [CrossRef]
- Qin, N.; Shen, Y.B.; Yang, X.; Su, L.Q.; Tang, R.; Li, W.; Wang, M. Site-directed mutagenesis under the direction of in silico protein docking modeling reveals the active site residues of 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum. World J. Microbiol. Biotechnol. 2017, 33, 146. [Google Scholar] [CrossRef]
- Shao, M.L.; Zhang, X.; Rao, Z.M.; Xu, M.J.; Yang, T.W.; Li, H.; Xu, Z.H.; Yang, S.T. A mutant form of 3-ketosteroid-Δ1-dehydrogenase gives altered androst-1,4-diene-3, 17-dione/androst-4-ene-3,17-dione molar ratios in steroid biotransformations by Mycobacterium neoaurum ST-095. J. Ind. Microbiol. Biotechnol. 2016, 43, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.G.; Guan, Y.X.; Wang, H.Q.; Yao, S.J. Microbial side-chain cleavage of phytosterols by Mycobacteria in vegetable oil/aqueous two-phase system. Appl. Biochem. Biotechnol.. 2014, 174, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Stefanov, S.; Yankov, D.; Beschkov, V. Biotransformation of phytosterols to androstenedione in two phase water-oil systems. Chem. Biochem. Eng. Q. 2006, 20, 421–427. [Google Scholar]
- Bie, S.T.; Lu, F.P.; Du, L.X.; Qiu, Q.; Zhang, Y. Effect of phase composition on the bioconversion of methyltestosterone in a biphasic system. J. Mol. Catal. B Enzym. 2008, 55, 1–5. [Google Scholar] [CrossRef]
- Shtratnikova, V.Y.; Schelkunov, M.I.; Dovbnya, D.V.; Bragin, E.Y.; Donova, M.V. Effect of methyl-β-cyclodextrin on gene expression in microbial conversion of phytosterol. Appl. Microbiol. Biotechnol. 2017, 101, 4659–4667. [Google Scholar] [CrossRef]
- Shen, Y.; Liang, J.; Li, H.; Wang, M. Hydroxypropyl-β-cyclodextrin-mediated alterations in cell permeability, lipid and protein profiles of steroid-transforming Arthrobacter simplex. Appl. Microbiol. Biotechnol. 2015, 99, 387–397. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R.; Cruz, A.; Angelova, B.; Fernandes, P.; Pons, M.N.; Pinheiro, H.M.; Cabral, J.M.S.; da Fonseca, M.M.R. Behaviour of Mycobacterium sp NRRL B-3805 whole cells in aqueous, organic-aqueous and organic media studied by fluorescence microscopy. Appl. Microbiol. Biotechnol. 2004, 64, 695–701. [Google Scholar] [CrossRef]
- Phase, N.; Patil, S. Natural oils are better than organic-solvents for the conversion of soybean sterols to 17-ketosteroids by Mycobacterium-Fortuitum. World J. Microbiol Biotechnol. 1994, 10, 228–229. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Y.; Shen, Y.; Zhang, X.; Xu, S.; Shang, Z.; Xia, M.; Wang, M. Efficient production of androstenedione by repeated batch fermentation in waste cooking oil media through regulating NAD(+)/NADH ratio and strengthening cell vitality of Mycobacterium neoaurum. Bioresour. Technol. 2019, 279, 209–217. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, X.; Wang, Y.; Han, Y.; Sun, J.; Shi, J.; Zhang, B. Identification, function, and application of 3-ketosteroid Δ1-dehydrogenase isozymes in Mycobacterium neoaurum DSM 1381 for the production of steroidic synthons. Microb. Cell Factories. 2018, 17, 77. [Google Scholar] [CrossRef]
- Goujon, M.; McWilliam, H.; Li, W.Z.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.R.; Shah, T.; Modi, V.V.; Udupa, S.R.; Chadha, M.S. Microbial transformation of phytosterols from plant lattices. J. Ferment. Technol. 1985, 63, 279–281. [Google Scholar]
- Xu, Y.P.; Qin, J.; Sun, S.M.; Liu, T.T.; Zhang, X.L.; Qian, S.S.; Zhu, H.L. Synthesis, crystal structures, molecular docking and urease inhibitory activity of nickel(II) complexes with 3-pyridinyl-4-amino-5-mercapto-1,2,4-triazole. Inorg. Chim. Acta 2014, 423, 469–476. [Google Scholar] [CrossRef]
- Voishvillo, N.E.; Andryushina, V.A.; Savinova, T.S.; Stytsenko, T.S.; Vasil’eva, N.A.; Turova, T.P.; Kolganova, T.V.; Skryabin, K.G. Identification of a new steroid-transforming strain of mycobacteria as Mycobacterium neoaurum. Appl. Biochem. Biotechnol. 2003, 39, 152–157. [Google Scholar]
- Liu, Y.C.; Chen, G.Y.; Ge, F.L.; Li, W.; Zeng, L.H.; Cao, W.G. Efficient biotransformation of cholesterol to androsta-1,4-diene-3,17-dione by a newly isolated actinomycete Gordonia neofelifaecis. World J. Microbiol. Biotechnol. 2011, 27, 759–765. [Google Scholar] [CrossRef]
- Malaviya, A.; Gomes, J. Rapid screening and isolation of a fungus for sitosterol to androstenedione biotransformation. Appl. Biochem. Biotechnol. 2009, 158, 374–386. [Google Scholar] [CrossRef]
- Shao, M.L.; Rao, Z.M.; Zhang, X.; Xu, M.J.; Yang, T.W.; Li, H.; Xu, Z.H.; Yang, S.T. Bioconversion of cholesterol to 4-cholesten-3-one by recombinant Bacillus subtilis expressing choM gene encoding cholesterol oxidase from Mycobacterium neoaurum JC-12. J. Chem. Technol. Biotechnol. 2015, 90, 1811–1820. [Google Scholar] [CrossRef]
- Zhang, H.M.; Xu, Z.D.; Zhou, D.Q.; Cao, J. Waste cooking oil-to-energy under incomplete information: Identifying policy options through an evolutionary game. Appl. Energy 2017, 185, 547–555. [Google Scholar] [CrossRef]
- Papavinasasundaram, K.G.; Colston, M.J.; Davis, E.O. Construction and complementation of a recA deletion mutant of Mycobacterium smegmatis reveals that the intein in Mycobacterium tuberculosis recA does not affect RecA function. Mol. Microbiol. 1998, 30, 525–534. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).