Enzymatic Hydrolysis of Softwood Derived Paper Sludge by an In Vitro Recombinant Cellulase Cocktail for the Production of Fermentable Sugars
Abstract
1. Introduction
2. Results and Discussion
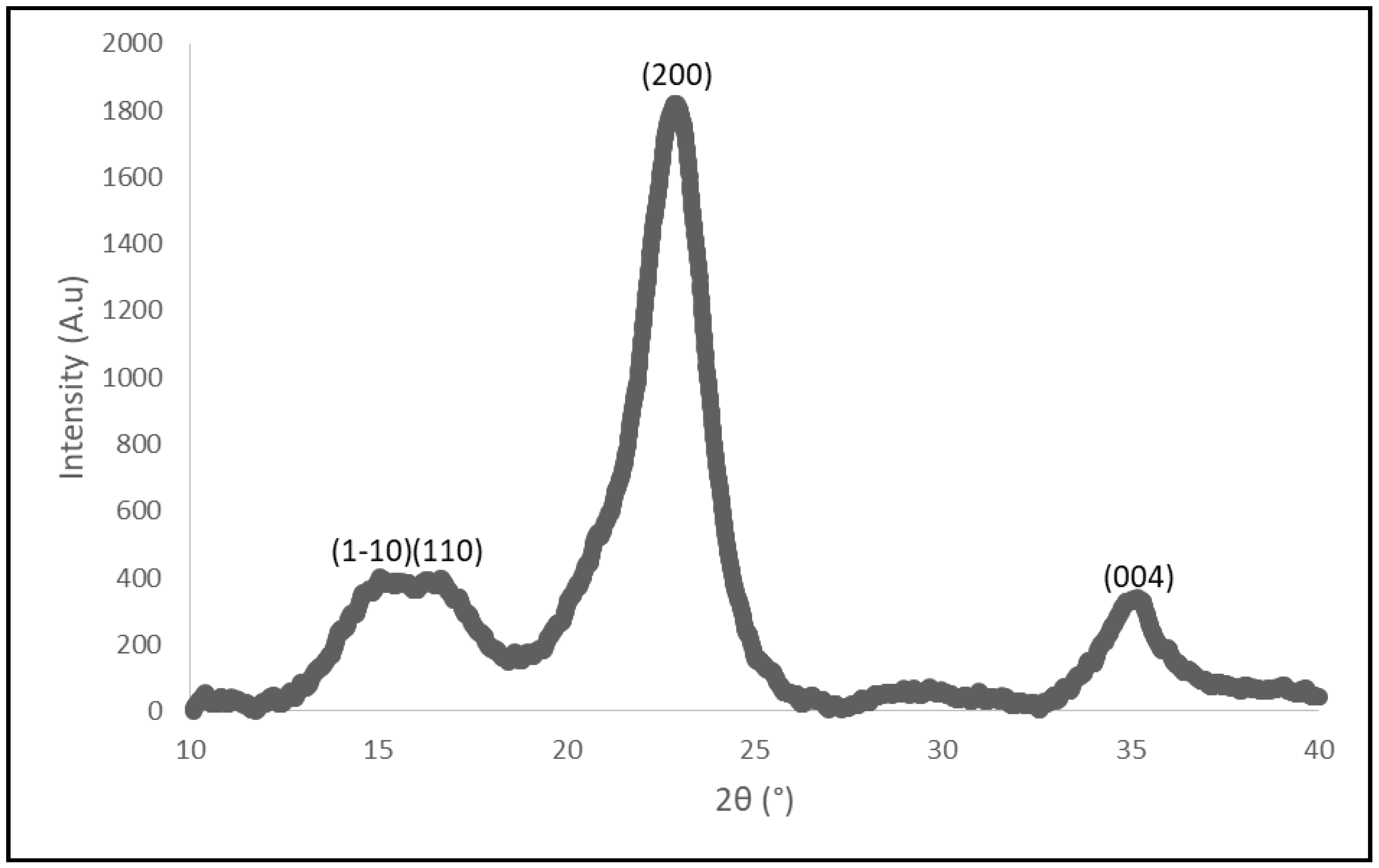
2.1. Composition and Structural Analysis of Paper Sludge
2.2. Enzyme Production and Substrate Specificities Using “Model” Substrates
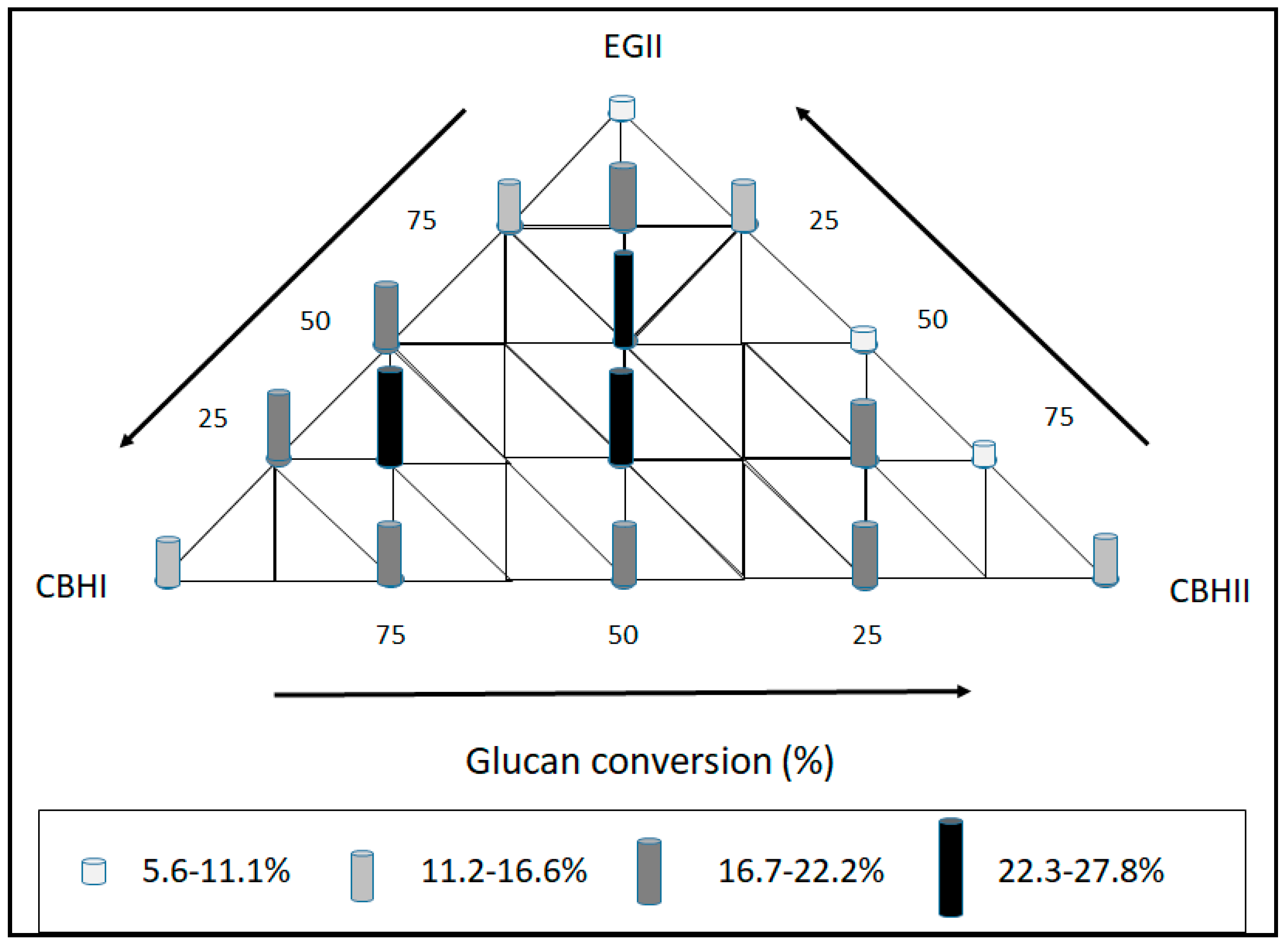
2.3. Enzyme Cocktail Formulation
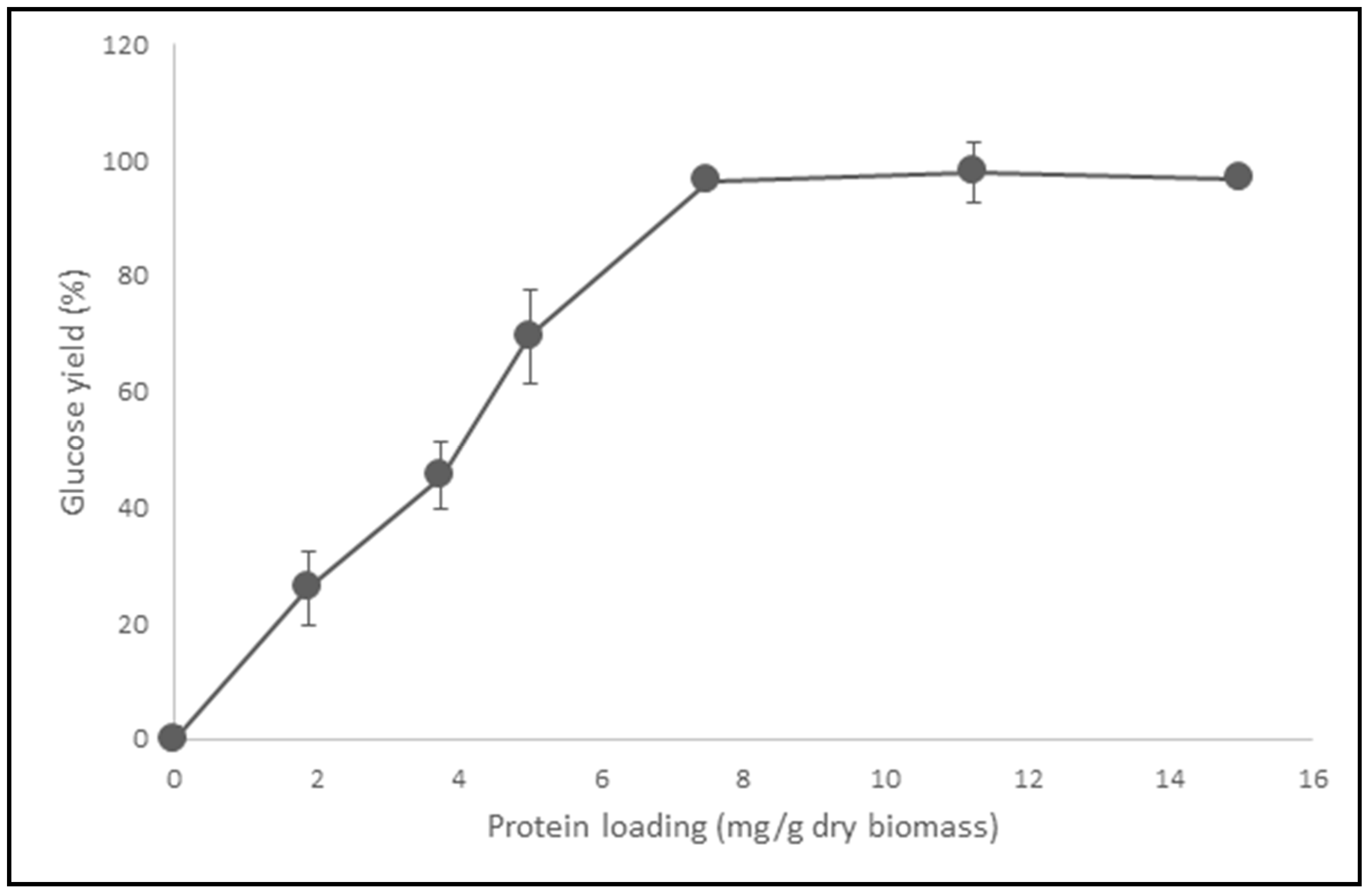
2.4. Effect of Opt CelMix Loading on the Hydrolysis Yields of Paper Sludge
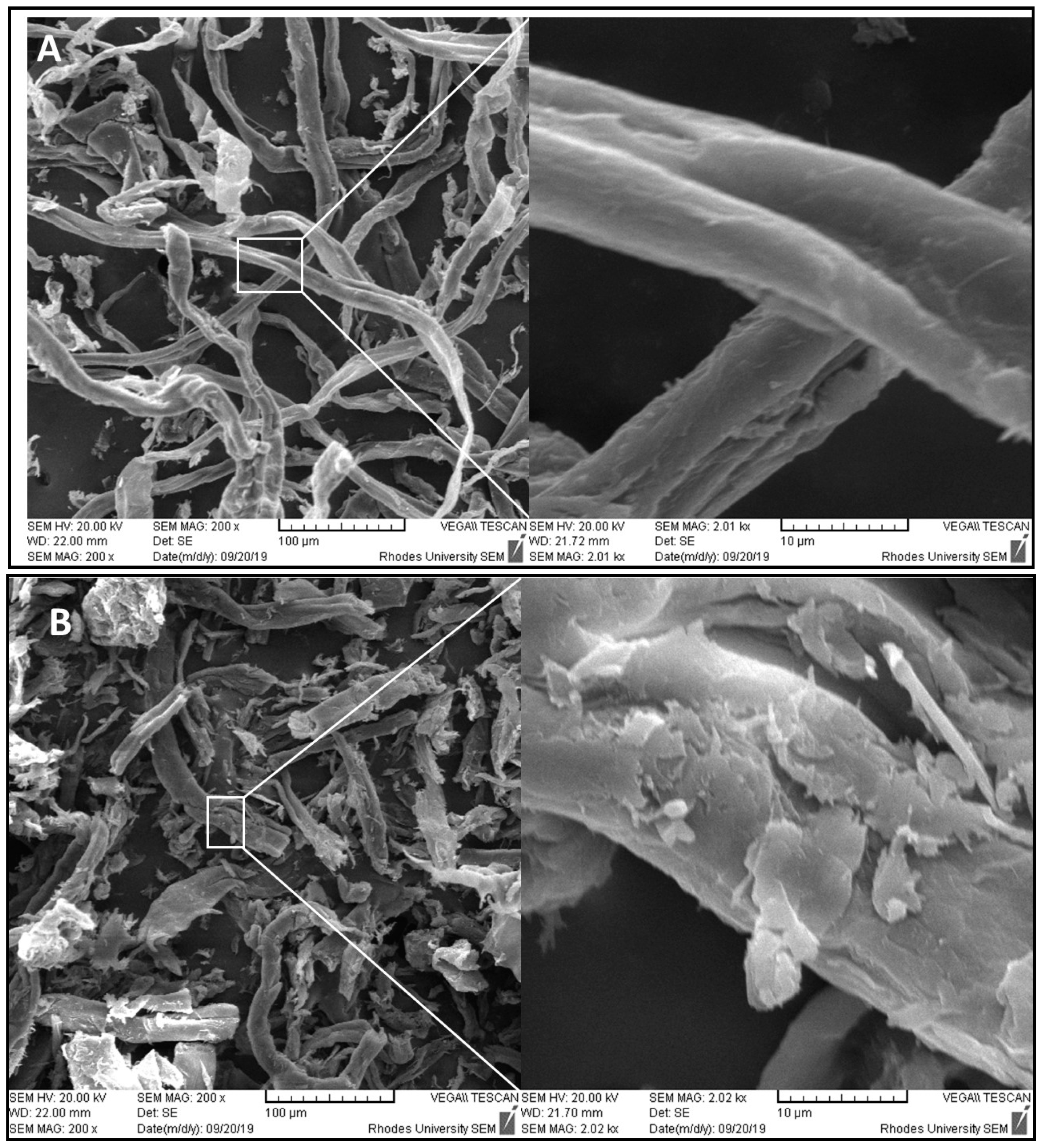
2.5. Scanning Electron Microscopy (SEM)
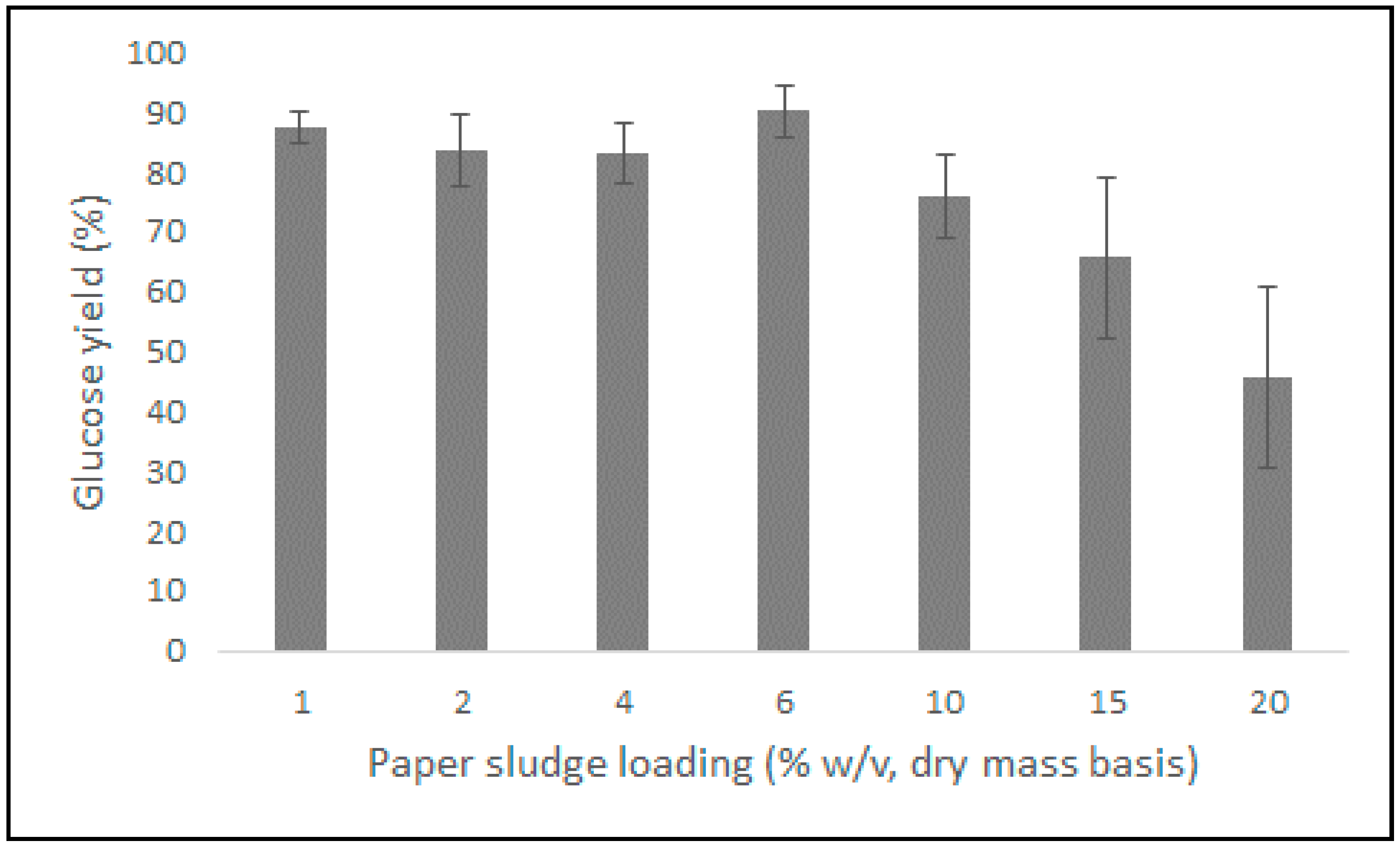
2.6. Evaluation of the Performance of the Opt CelMix at Varying Paper Sludge Loadings
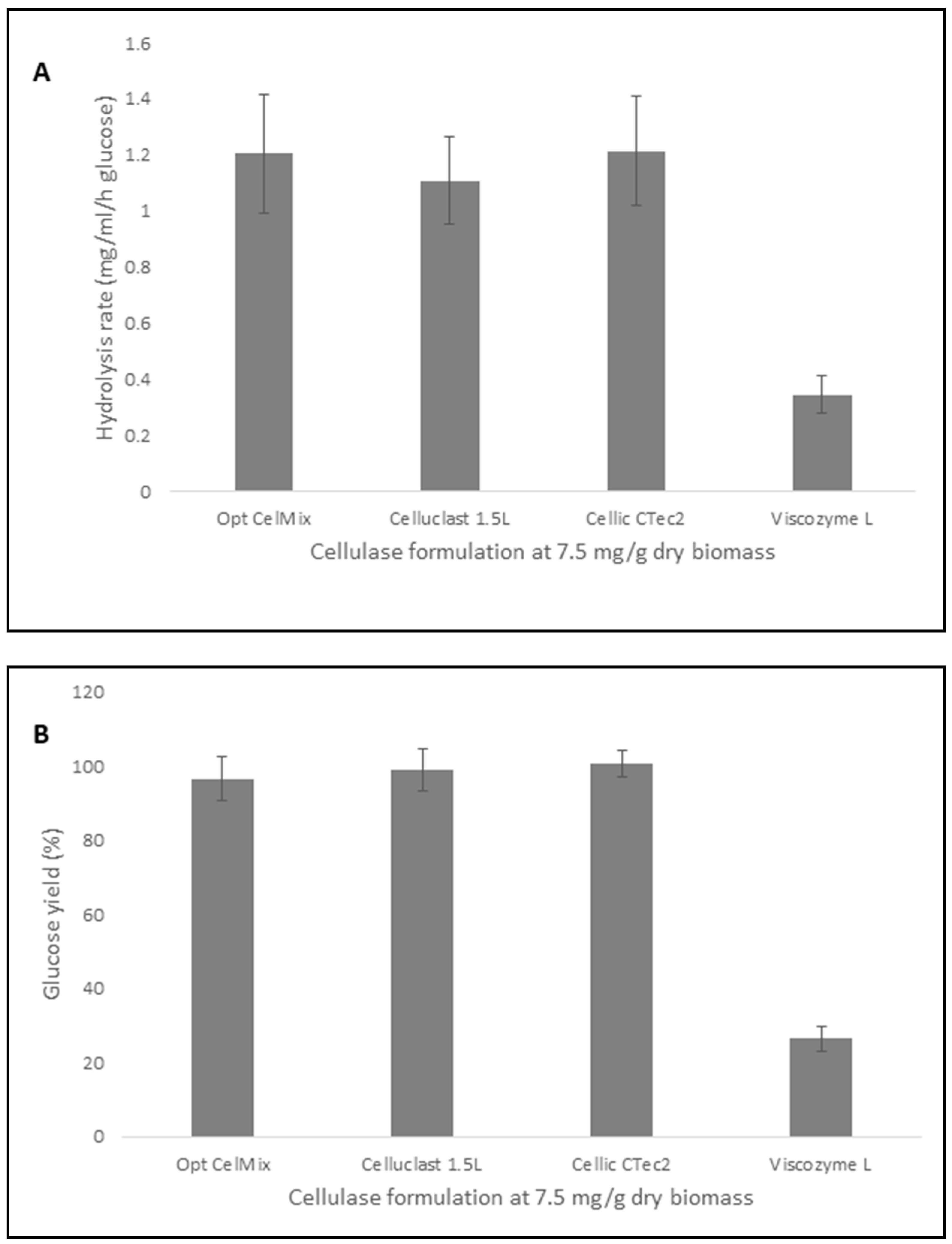
2.7. Comparison of the Hydrolytic Efficiency of the Opt CelMix to Commercial Cellulase Preparations
3. Materials and Methods
3.1. Materials
3.2. Paper Sludge Moisture Content Determination
3.3. Paper Sludge Chemical Composition
3.4. Paper Sludge Crystallinity Index (CrI)
3.5. Simon’s Staining (SS) of Paper Sludge
3.6. Media, Yeast Strains and Culture Conditions
3.7. Preparation of Partially Purified Enzymes
3.8. Protein Content and Purity Determination
3.9. Enzyme Assays
3.9.1. Substrate Specificity Determination
3.9.2. Enzyme Cocktail Formulation
3.9.3. Effect of Cellulase Cocktail Loading on the Hydrolysis Yields of Paper Sludge
3.9.4. Effect of Paper Sludge Loading on Cellulase Cocktail Hydrolytic Efficiency
3.9.5. Comparison of the Formulated Cellulase Cocktail to Commercial Enzyme Preparations
3.10. Analytical Methods
3.11. Scanning Electron Microscopy (SEM)
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Prasetyo, J.; Naruse, K.; Kato, T.; Boonchird, C.; Harashima, S.; Park, E.Y. Bioconversion of paper sludge to biofuel by simultaneous saccharification and fermentation using a cellulase of paper sludge origin and thermotolerant Saccharomyces cerevisiae TJ14. Biotechnol. Biofuels 2011, 4, 35. [Google Scholar] [CrossRef]
- Gottumukkala, L.D.; Haigh, K.; Collard, F.X.; van Rensburg, E.; Görgens, J. Opportunities and prospects of biorefinery-based valorisation of pulp and paper sludge. Bioresour. Technol. 2016, 215, 37–49. [Google Scholar] [CrossRef]
- Mendes, C.V.T.; Cruz, C.H.G.; Reis, D.F.N.; Carvalho, M.G.V.S.; Rocha, J.M.S. Integrated bioconversion of pulp and paper primary sludge to second generation bioethanol using Saccharomyces cerevisiae ATCC 26602. Bioresour. Technol. 2016, 220, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Wang, W.; Lee, Y.Y. Bioconversion of kraft paper mill sludges to ethanol by SSF and SSCF. Appl. Biochem. Biotechnol. 2010, 161, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Gomes, D.G.; Serna-Loaiza, S.; Cardona, C.A.; Gama, M.; Domingues, L. Insights into the economic viability of cellulases recycling on bioethanol production from recycled paper sludge. Bioresour. Technol. 2018, 267, 347–355. [Google Scholar] [CrossRef]
- Ganner, T.; Bubner, P.; Eibinger, M.; Mayrhofers, C.; Planks, H.; Nidetzky, B. Dissecting and reconstructing synergism: In situ visualization of cooperativity among cellulases. J. Biol. Chem. 2012, 287, 43215–43222. [Google Scholar] [CrossRef] [PubMed]
- Kostylev, M.; Wilson, D. A distinct model of synergism between a processive endocellulase (TfCel9A) and an exocellulase (TfCel48A) from Thermobifida fusca. Appl. Environ. Microbiol. 2014, 80, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Malgas, S.; Thoresen, M.; van Dyk, S.J.; Pletschke, B.I. Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates. Enzyme Microb. Technol. 2017, 103, 1–11. [Google Scholar] [CrossRef]
- Thoresen, M.; Malgas, S.; Pletschke, B.I. Enzyme adsorption-desorption and evaluation of various cellulase recycling strategies for steam pre-treated Eucalyptus enzymatic degradation. Biomass Convers. Biorefinery 2020. [Google Scholar] [CrossRef]
- Rizk, M.; Antranikian, G.; Elleuche, S. End-to-end gene fusions and their impact on the production of multifunctional biomass degrading enzymes. Biochem. Biophys. Res. Commun. 2012, 428, 1–5. [Google Scholar] [CrossRef]
- Ilmén, M.; Den Haan, R.; Brevnova, E.; McBride, J.; Wiswall, E.; Froehlich, A.; Koivula, A.; Voutilainen, S.P.; Siika-Aho, M.; La Grange, D.C.; et al. High level secretion of cellobiohydrolases by Saccharomyces cerevisiae. Biotechnol. Biofuels 2011, 4, 30. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, S.I.; den Haan, R.; Viljoen-Bloom, M.; van Zyl, W.H. Lignocellulosic hydrolysate inhibitors selectively inhibit/deactivate cellulase performance. Enzyme Microb. Technol. 2015, 81, 16–22. [Google Scholar] [CrossRef] [PubMed]
- den Haan, R.; van Rensburg, E.; Rose, S.H.; Görgens, J.F.; van Zyl, W.H. Progress and challenges in the engineering of non-cellulolytic microorganisms for consolidated bioprocessing. Curr. Opin. Biotechnol. 2015, 33, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Davison, S.A.; den Haan, R.; van Zyl, W.H. Heterologous expression of cellulase genes in natural Saccharomyces cerevisiae strains. Appl. Microbiol. Biotechnol. 2016, 100, 8241–8254. [Google Scholar] [CrossRef]
- Voutilainen, S.P.; Murray, P.G.; Tuohy, M.G.; Koivula, A. Expression of Talaromyces emersonii cellobiohydrolase Cel7A in Saccharomyces cerevisiae and rational mutagenesis to improve its thermostability and activity. Protein Eng. Des. Sel. 2010, 23, 69–79. [Google Scholar] [CrossRef]
- Kroukamp, H.; den Haan, R.; van Zyl, J.-H.; van Zyl, W.H. Rational strain engineering interventions to enhance cellulase secretion by Saccharomyces cerevisiae. Biofuels Bioprod. Biorefining 2018, 12, 108–124. [Google Scholar] [CrossRef]
- Davison, S.A.; den Haan, R.; van Zyl, W.H. Exploiting strain diversity and rational engineering strategies to enhance recombinant cellulase secretion by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Park, S.; Baker, J.O.; Himmel, M.E.; Parilla, P.A.; Johnson, D.K. Cellulose crystallinity index: Measurement techniques and their impact on interpreting cellulase performance. Biotechnol. Biofuels 2010, 3, 10. [Google Scholar] [CrossRef]
- Segal, L.; Creely, J.; Martin, A.; Conrad, C. An empirical method for estimating the degree of crystallinity of native cellulose using the x-ray diffractometer. Text. Res. J. 1959, 29. [Google Scholar] [CrossRef]
- Malgas, S.; Kwanya Minghe, V.M.; Pletschke, B.I. The effect of hemicellulose on the binding and activity of cellobiohydrolase I, Cel7A, from Trichoderma reesei to cellulose. Cellulose 2020, 27, 781–797. [Google Scholar] [CrossRef]
- Thygesen, A.; Oddershede, J.; Lilholt, H.; Thomsen, A.B.; Ståhl, K. On the determination of crystallinity and cellulose content in plant fibres. Cellulose 2005, 12, 563–576. [Google Scholar] [CrossRef]
- den Haan, R.; van Zyl, J.M.; Harms, T.M.; van Zyl, W.H. Modeling the minimum enzymatic requirements for optimal cellulose conversion. Environ. Res. Lett. 2013, 8, 025013. [Google Scholar] [CrossRef]
- Tsukada, T.; Igarashi, K.; Yoshida, M.; Samejima, M. Molecular cloning and characterization of two intracellular β-glucosidases belonging to glycoside hydrolase family 1 from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 2006, 73, 807–814. [Google Scholar] [CrossRef]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef] [PubMed]
- Njokweni, A.P.; Rose, S.H.; Van Zyl, W.H. Fungal β-glucosidase expression in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2012, 39, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Malgas, S.; Chandra, R.; Van Dyk, J.S.; Saddler, J.N.; Pletschke, B.I. Formulation of an optimized synergistic enzyme cocktail, HoloMix, for effective degradation of various pre-treated hardwoods. Bioresour. Technol. 2017, 245, 52–65. [Google Scholar] [CrossRef]
- Nakamura, A.; Tsukada, T.; Auer, S.; Furuta, T.; Wada, M.; Koivula, A.; Igarashi, K.; Samejima, M. The tryptophan residue at the active site tunnel entrance of Trichoderma reesei cellobiohydrolase Cel7A is Important for initiation of degradation of crystalline cellulose. J. Biol. Chem. 2013, 288, 13503–13510. [Google Scholar] [CrossRef]
- Zhong, L.; Matthews, J.F.; Hansen, P.I.; Crowley, M.F.; Cleary, J.M.; Walker, R.C.; Nimlos, M.R.; Brooks, C.L.; Adney, W.S.; Himmel, M.E.; et al. Computational simulations of the Trichoderma reesei cellobiohydrolase I acting on microcrystalline cellulose Iβ: The enzyme-substrate complex. Carbohydr. Res. 2009, 344, 1984–1992. [Google Scholar] [CrossRef]
- Igarashi, K.; Koivula, A.; Wada, M.; Kimura, S.; Penttila, M.; Samejima, M. High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 2009, 284, 36186–36190. [Google Scholar] [CrossRef]
- Hoshino, E.; Shiroishi, M.; Amano, Y.; Nomura, M.; Kanda, T. Synergistic actions of exo-type cellulases in the hydrolysis of cellulose with different crystallinities. J. Ferment. Bioeng. 1997, 84, 300–306. [Google Scholar] [CrossRef]
- Väljamäe, P.; Sild, V.; Pettersson, G.; Johansson, G. The initial kinetics of hydrolysis by cellobiohydrolases I and II is consistent with a cellulose surface-erosion model. Eur. J. Biochem. 1998, 253, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.M.; McCrae, S.I. Synergism Between Enzymes Involved in the Solubilization of Native Cellulose. In Hydrolysis of Cellulose: Mechanisms of Enzymatic and Acid Catalysis; Advances in Chemistry; American Chemical Society: Washington, DC, USA, 1979; Volume 181, pp. 181–209. ISBN 0-8412-0460-8. [Google Scholar]
- Jalak, J.; Kurašin, M.; Teugjas, H.; Väljamä, P. Endo-exo synergism in cellulose hydrolysis revisited. J. Biol. Chem. 2012, 287, 28802–28815. [Google Scholar] [CrossRef] [PubMed]
- Boisset, C.; Fraschini, C.; Schülein, M.; Henrissat, B.; Chanzy, H. Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 2000, 66, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Boisset, C.; Pétrequin, C.; Chanzy, H.; Henrissat, B.; Schlein, M. Optimized mixtures of recombinant Humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 2001, 72, 339–345. [Google Scholar] [CrossRef]
- Woodward, J.; Lima, M.; Lee, N.E. The role of cellulase concentration in determining the degree of synergism in the hydrolysis of microcrystalline cellulose. Biochem. J. 1988, 255, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Kallioinen, A.; Puranen, T.; Siika-Aho, M. Mixtures of thermostable enzymes show high performance in biomass saccharification. Appl. Biochem. Biotechnol. 2014, 173, 1038–1056. [Google Scholar] [CrossRef]
- Väljamäe, P.; Sild, V.; Nutt, A.; Pettersson, G.; Johansson, G. Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur. J. Biochem. 1999, 266, 327–334. [Google Scholar] [CrossRef]
- Zhang, Y.H.P.; Lynd, L.R. A functionally based model for hydrolysis of cellulose by fungal cellulase. Biotechnol. Bioeng. 2006, 94, 888–898. [Google Scholar] [CrossRef]
- Zhang, Y.H.P.; Lynd, L.R. Determination of the number-average degree of polymerization of cellodextrins and cellulose with application to enzymatic hydrolysis. Biomacromolecules 2005, 6, 1510–1515. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Y.H.; Chu, J.; Luo, L.Z.; Zhuang, Y.P.; Zhang, S.L. Optimization of cellulase mixture for efficient hydrolysis of steam-exploded corn stover by statistically designed experiments. Bioresour. Technol. 2009, 100, 819–825. [Google Scholar] [CrossRef]
- Przybysz Buzała, K.; Kalinowska, H.; Przybysz, P.; Małachowska, E. Conversion of various types of lignocellulosic biomass to fermentable sugars using kraft pulping and enzymatic hydrolysis. Wood Sci. Technol. 2017, 51, 873–885. [Google Scholar] [CrossRef]
- Hu, J.; Arantes, V.; Saddler, J.N. The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: Is it an additive or synergistic effect? Biotechnol. Biofuels 2011, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.D.; Percival Zhang, Y.H. Analysis of biofuels production from sugar based on three criteria: Thermodynamics, bioenergetics, and product separation. Energy Environ. Sci. 2011, 4, 784–792. [Google Scholar] [CrossRef]
- Mukasekuru, M.R.; Hu, J.; Zhao, X.; Sun, F.F.; Pascal, K.; Ren, H.; Zhang, J. Enhanced high-solids fed-batch enzymatic hydrolysis of sugar cane bagasse with accessory enzymes and additives at low cellulase loading. ACS Sustain. Chem. Eng. 2018, 6, 12787–12796. [Google Scholar] [CrossRef]
- Gomes, D.; Gama, M.; Domingues, L. Determinants on an efficient cellulase recycling process for the production of bioethanol from recycled paper sludge under high solid loadings. Biotechnol. Biofuels 2018, 11, 111. [Google Scholar] [CrossRef]
- Steffen, F.; Janzon, R.; Saake, B. Enzymatic treatment of deinking sludge–effect on fibre and drainage properties. Environ. Technol. 2018, 39, 2810–2821. [Google Scholar] [CrossRef]
- Sluiter, J.B.; Ruiz, R.O.; Scarlata, C.J.; Sluiter, A.D.; Templeton, D.W. Compositional analysis of lignocellulosic feedstocks. 1. Review and description of methods. J. Agric. Food Chem. 2010, 58, 9043–9053. [Google Scholar] [CrossRef]
- Chandra, R.; Ewanick, S.; Hsieh, C.; Saddler, J.N. The characterization of pretreated lignocellulosic substrates prior to enzymatic hydrolysis. Biotechnol. Prog. 2008, 24, 1178–1185. [Google Scholar] [CrossRef]
- Chandra, R.P.; Saddler, J.N. Use of the Simons’ staining technique to assess cellulose accessibility in pretreated substrates. Ind. Biotechnol. 2012, 8, 230–237. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structura l Proteins during the Assembly of the Head of Bacteriop hage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]

| Component/Property | Content/Accessibility |
|---|---|
| Glucan * | 89.7% |
| Mannan * | 2.73% |
| Xylan * | 1.65% |
| Galactan * | 0.08% |
| Arabinan * | Nd |
| Lignin * | 0.8% |
| Ash * | 1.7% |
| Moisture | 74.3% |
| Crystallinity index | 91.5% |
| Substrate accessibility (mg/g) | 87 mg/g |
| Enzyme | CMC-Na | Cellopentaitol | pNPC | pNPG | Reference |
|---|---|---|---|---|---|
| BGL | 0.67 | - | 5.74 | 56 | [10,14,25] |
| CBHI | 0.24 | Nd | 0.20 | 0.35 | [12,26] |
| CBHII | 1.69 | 0.38 | Nd | 0.11 | [26] |
| EGII | 44 | - | Nd | 0.11 | [10,12,14,26] |
| No | CBHI% | CBHII% | EGII% |
|---|---|---|---|
| 1. | 100 | 0 | 0 |
| 2. | 75 | 25 | 0 |
| 3. | 50 | 50 | 0 |
| 4. | 25 | 75 | 0 |
| 5. | 0 | 100 | 0 |
| 6. | 0 | 75 | 25 |
| 7. | 0 | 50 | 50 |
| 8. | 0 | 25 | 75 |
| 9. | 0 | 0 | 100 |
| 10. | 25 | 0 | 75 |
| 11. | 50 | 0 | 50 |
| 12. | 75 | 0 | 25 |
| 13. | 33.3 | 33.3 | 33.3 |
| 14. | 33.5 | 33.5 | 25 |
| 15. | 12.5 | 12.5 | 75 |
| 16. | 18.75 | 56.25 | 25 |
| 17. | 56.25 | 18.75 | 25 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malgas, S.; Rose, S.H.; van Zyl, W.H.; Pletschke, B.I. Enzymatic Hydrolysis of Softwood Derived Paper Sludge by an In Vitro Recombinant Cellulase Cocktail for the Production of Fermentable Sugars. Catalysts 2020, 10, 775. https://doi.org/10.3390/catal10070775
Malgas S, Rose SH, van Zyl WH, Pletschke BI. Enzymatic Hydrolysis of Softwood Derived Paper Sludge by an In Vitro Recombinant Cellulase Cocktail for the Production of Fermentable Sugars. Catalysts. 2020; 10(7):775. https://doi.org/10.3390/catal10070775
Chicago/Turabian StyleMalgas, Samkelo, Shaunita H. Rose, Willem H. van Zyl, and Brett I. Pletschke. 2020. "Enzymatic Hydrolysis of Softwood Derived Paper Sludge by an In Vitro Recombinant Cellulase Cocktail for the Production of Fermentable Sugars" Catalysts 10, no. 7: 775. https://doi.org/10.3390/catal10070775
APA StyleMalgas, S., Rose, S. H., van Zyl, W. H., & Pletschke, B. I. (2020). Enzymatic Hydrolysis of Softwood Derived Paper Sludge by an In Vitro Recombinant Cellulase Cocktail for the Production of Fermentable Sugars. Catalysts, 10(7), 775. https://doi.org/10.3390/catal10070775






