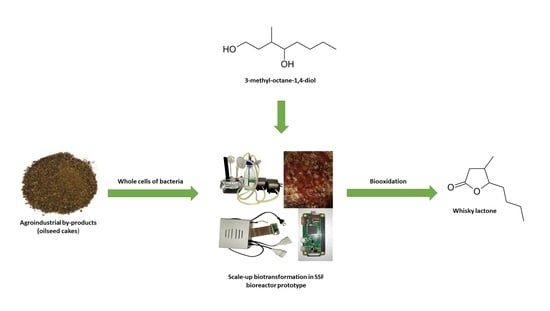
Bacterial Whole Cells Synthesis of Whisky Lactones in a Solid-State Fermentation Bioreactor Prototype
Abstract
1. Introduction
2. Results and Discussion
2.1. Preliminary Screening Scale Biotransformations with Anti-3-methyl-octane-1,4-diol (1a) on Different Oil Cakes
2.2. Preliminary Screening Scale Biotransformations with Syn-3-methyl-octane-1,4-diol (1b) on Different Oil Cakes
2.3. Preliminary Screening Scale Biotransformations with a Diastereoisomeric Mixture of Anti- and Syn-3-methyl-octane-1,4-diols (1a–b)
2.4. Screening Scale Biotransformations with Anti-3-methyl-octane-1,4-diol (1a) on Linseed Cake
2.5. Screening Scale Biotransformations with Syn-3-methyl-octane-1,4-diol (1b) on Linseed Cake
2.6. Selection of the Extraction Method
2.7. Calibration of Moisture Sensor
2.8. Preparative Biotransformations with anti- and Syn-3-methyl-octane-1,4-diols (1a–b)
3. Materials and Methods
3.1. Microorganisms
3.2. Materials
3.3. Measurement of Oilseed Cake Moisture
3.4. Separation of the cis/trans-Whisky Lactones
3.5. Chemical Reduction of Whisky Lactones
3.6. Screening Scale Biotransformations
3.7. Statystical Analysis
3.8. Preparative Biotransformations
3.9. Design of the Bioreactor
3.10. Extraction Methods
3.10.1. Simple Extraction
3.10.2. Steam Distillation
3.10.3. Steam Distillation from the Extract
3.10.4. Extraction with a Dryng Apparatus
3.11. Analysis Procedure
3.12. Moisiture Analysis
- Ww—Water weight [g]
- Ow—Oilcake weight [g]
- W—Moisture content that we need to obtain [%]
- Mw—Moisture contet in oilcakes [%]
3.13. Software
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandey, A. Solid-state fermentation. Biochem. Eng. J. 2003, 13, 81–84. [Google Scholar] [CrossRef]
- Kapilan, R. Solid state fermentation for microbial products: A review. Appl. Sci. Res. 2015, 7, 21–25. [Google Scholar]
- Pandey, A.; Soccol, C.R.; Mitchell, D. New developments in solid state fermentation: I-bioprocesses and products. Process. Biochem. 2000, 35, 1153–1169. [Google Scholar] [CrossRef]
- Singhania, R.R.; Patel, A.K.; Soccol, C.R.; Pandey, A. Recent advances in solid-state fermentation. Biochem. Eng. J. 2009, 44, 13–18. [Google Scholar] [CrossRef]
- Rodrı, S. Application of solid-state fermentation to food industry—A review. J. Food Eng. 2006, 76, 291–302. [Google Scholar]
- Ramachandran, S.; Singh, S.K.; Larroche, C.; Soccol, C.R.; Pandey, A. Oil cakes and their biotechnological applications—A review. Bioresour. Technol. 2007, 98, 2000–2009. [Google Scholar] [CrossRef]
- Soccol, C.R.; da Costa, E.S.F.; Letti, L.A.J.; Karp, S.G.; Wojciechowski, A.L.; de Souza Vandenberghe, L.P. Recent developments and innovations in solid state fermentation. Biotechnol. Res. Innov. 2017, 1, 52–71. [Google Scholar] [CrossRef]
- Kannan, T.R.; Kanagaraj, C. Molecular characteristic of α-amylase enzymes producing from Bacillus lichenformis (JQ946317) using solid state fermentation. Biocatal. Agric. Biotechnol. 2019, 20, 1–6. [Google Scholar] [CrossRef]
- Liguori, R.; Amore, A.; Faraco, V. Waste valorization by biotechnological conversion into added value products. Appl. Microbiol. Biotechnol. 2013, 97, 6129–6147. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M. Recent Advances in Solid-State Fermentation Applications for the Food Industry. Curr. Biochem. Eng. 2013, 1, 2–8. [Google Scholar] [CrossRef]
- Rodríguez Madrera, R.; Pando Bedriñana, R.; Suárez Valles, B. Production and characterization of aroma compounds from apple pomace by solid-state fermentation with selected yeasts. LWT Food Sci. Technol. 2015, 64, 1342–1353. [Google Scholar] [CrossRef]
- Raghavarao, K.S.M.S.; Ranganathan, T.V.; Karanth, N.G. Some engineering aspects of solid-state fermentation. Biochem. Eng. J. 2003, 13, 127–135. [Google Scholar] [CrossRef]
- Castilho, L.R.; Polato, C.M.S.; Baruque, E.A.; Sant’Anna, G.L.; Freire, D.M.G. Economic analysis of lipase production by Penicillium restrictum in solid-state and submerged fermentations. Biochem. Eng. J. 2000, 4, 239–247. [Google Scholar] [CrossRef]
- Boratyński, F.; Szczepańska, E.; Grudniewska, A.; Skalny, B.; Olejniczak, T. A novel approach for microbial synthesis of enantiomerically pure whisky lactones based on solid-state fermentation. Molecules 2018, 23, 659. [Google Scholar] [CrossRef]
- Chreptowicz, K.; Wielechowska, M.; Główczyk-Zubek, J.; Rybak, E.; Mierzejewska, J. Production of natural 2-phenylethanol: From biotransformation to purified product. Food Bioprod. Process. 2016, 100, 275–281. [Google Scholar] [CrossRef]
- Nagy, V.; Toke, E.R.; Keong, L.C.; Szatzker, G.; Ibrahim, D.; Omar, I.C.; Szakács, G.; Poppe, L. Kinetic resolutions with novel, highly enantioselective fungal lipases produced by solid state fermentation. J. Mol. Catal. B Enzym. 2006, 39, 141–148. [Google Scholar] [CrossRef]
- Marie, L.; Gori, K.; Agerlin, M.; Jespersen, L.; Arneborg, N. Flavour compound production by Yarrowia lipolytica, Saccharomyces cerevisiae and Debaryomyces hansenii in a cheese-surface model. Int. Dairy J. 2011, 21, 970–978. [Google Scholar]
- Marco, B.A.D.; Rechelo, B.S.; Tótoli, E.G.; Kogawa, A.C.; Regina, H.; Salgado, N. Evolution of green chemistry and its multidimensional impacts: A review. Saudi Pharm. J. 2019, 27, 1–8. [Google Scholar] [CrossRef]
- Braga, A.; Belo, I. Biotechnological production of gamma-decalactone, a peach like aroma, by Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2016, 32, 169. [Google Scholar] [CrossRef]
- Ashok, A.; Doriya, K.; Rao, D.R.M.; Kumar, D.S. Design of solid state bioreactor for industrial applications: An overview to conventional bioreactors. Biocatal. Agric. Biotechnol. 2017, 9, 11–18. [Google Scholar] [CrossRef]
- Rayhane, H.; Josiane, M.; Gregoria, M.; Yiannis, K.; Nathalie, D.; Ahmed, M.; Sevastianos, R. From flasks to single used bioreactor: Scale-up of solid state fermentation process for metabolites and conidia production by Trichoderma asperellum. J. Environ. Manage. 2019, 252, 109496. [Google Scholar] [CrossRef]
- Maga, J.A. Oak lactones in alcoholic beverages. Food Rev. Int. 1996, 12, 105–130. [Google Scholar] [CrossRef]
- Günther, C.; Mosandl, A. Stereoisomere Aromastoffe, XII. 3-Methyl-4-octanolid—“Quercuslacton, Whiskylacton”–Struktur und Eigenschaften der Stereoisomeren. Liebigs Ann. der Chemie 1986, 1986, 2112–2122. [Google Scholar] [CrossRef]
- Abbott, N.; Puech, J.L.; Bayonove, C.; Baumes, R. Determination of the Aroma Threshold of the cis and trans Racemic Forms of β-Methyl-γ-Octalactone by Gas Chromatography-Sniffing Analysis. Am. J. Enol. Vitic. 1995, 46, 292–294. [Google Scholar]
- Suzukt, Y.; Mori, W.; Ishizone, H.; Naito, K.; Honda, T. Concise enantiospecific syntheses of (+)-Eldanolide and (−)-cis-whisky lactone. Tetrahedron Lett. 1992, 33, 4931–4932. [Google Scholar] [CrossRef]
- Ito, K.; Yoshitake, M.; Katsuki, T. Chiral bipyrindine and biquinoline ligands: Their asymmetric synthesis and application to the synthesis of trans-whisky lactone. Tetrahedron 1996, 52, 3905–3920. [Google Scholar] [CrossRef]
- Jiang, X.; Fu, C.; Ma, S. A concise synthesis of (−)- and (+)-trans-whisky lactones. Eur. J. Org. Chem. 2010, 2010, 687–693. [Google Scholar] [CrossRef]
- Pisani, L.; Superchi, S.; D’Elia, A.; Scafato, P.; Rosini, C. Synthetic approach toward cis-disubstituted γ- and δ-lactones through enantioselective dialkylzinc addition to aldehydes: Application to the synthesis of optically active flavors and fragrances. Tetrahedron 2012, 68, 5779–5784. [Google Scholar] [CrossRef]
- Armstrong, A.; Ashraff, C.; Chung, H.; Murtagh, L. Oxidative rearrangement of 2-alkoxy-3,4-dihydro-2H-pyrans: Stereocontrolled synthesis of 4,5-cis-disubstituted tetrahydrofuranones including whisky and cognac lactones and crobarbatic acid. Tetrahedron 2009, 65, 4490–4504. [Google Scholar] [CrossRef]
- Xie, H.; Lu, J.; Gul, Y.; Gao, L.; Song, Z. (HMe2SiCH2)2: A useful reagent for B(C6F5)3-catalyzed reduction–lactonization of keto acids: Concise syntheses of (–)-cis-whisky and (–)-cis-cognac lactones. Synlett 2017, 28, 2453–2459. [Google Scholar]
- Boratyński, F.; Smuga, M.; Wawrzeńczyk, C. Lactones 42. Stereoselective enzymatic/microbial synthesis of optically active isomers of whisky lactone. Food Chem. 2013, 141, 419–427. [Google Scholar] [CrossRef]
- Boratyński, F.; Szczepańska, E.; Grudniewska, A.; Olejniczak, T. Microbial kinetic resolution of aroma compounds using solid-state fermentation. Catalysts 2018, 8, 28. [Google Scholar] [CrossRef]
- Boratyński, F.; Dancewicz, K.; Paprocka, M.; Gabryś, B.; Wawrzeńczyk, C. Chemo-enzymatic synthesis of optically active γ- and δ-decalactones and their effect on aphid probing, feeding and settling behavior. PLoS ONE 2016, 11, e0146160. [Google Scholar] [CrossRef]
- Boratyński, F.; Szczepańska, E.; De Simeis, D.; Serra, S.; Brenna, E. Bacterial biotransformation of oleic acid: New findings on the formation of γ-dodecalactone and 10-ketostearic acid in the culture of Micrococcus luteus. Molecules 2020, 25, 3024. [Google Scholar] [CrossRef]
- Boratyński, F.; Pannek, J.; Walczak, P.; Janik-Polanowicz, A.; Huszcza, E.; Szczepańska, E.; Martin-Rojas, E.; Olejniczak, T. Microbial alcohol dehydrogenase screening for enantiopure lactone synthesis: Down-stream process from microtiter plate to bench bioreactor. Process. Biochem. 2014, 49, 1637–1646. [Google Scholar] [CrossRef]
- Graminha, E.B.N.; Gonçalves, A.Z.L.; Pirota, R.D.P.B.; Balsalobre, M.A.A.; Da Silva, R.; Gomes, E. Enzyme production by solid-state fermentation: Application to animal nutrition. Anim. Feed Sci. Technol. 2008, 144, 1–22. [Google Scholar] [CrossRef]
- Gutierrez, C.; Rubilar, M.; Jara, C.; Verdugo, M.; Sineiro, J.; Shene, C. Flaxseed and Flaxseed cake as a source of compounds for food industry. J. Soil Sci. Plant. Nutr. 2009, 10, 454–463. [Google Scholar] [CrossRef]
- Stasiniewicz, T.; Niwińska, B.; Strzetelski, J.; Kowalczyk, J.; Maciaszek, K.; Bilik, K. Nutritive value of evening primrose (Oenothera paradoxa) cake for ruminants. J. Anim. Feed Sci. 1998, 7, 187–195. [Google Scholar] [CrossRef]
- Liu, J.; Du, C.; Beaman, H.; Monroe, M. Characterization of phenolic acid antimicrobial and antioxidant structure-property relationships. Pharmaceutics 2020, 21, 419. [Google Scholar] [CrossRef]
- Wilkinson, K.L.; Elsey, G.M.; Prager, R.H.; Tanaka, T.; Sefon, M.A. Precursors to oak lactone. Part 2: Synthesis, separation and cleavage of several β-D-glucopyranosides of 3-methyl-4-hydroxyoctanoic acid. Tetrahedron 2004, 60, 6091–6100. [Google Scholar] [CrossRef]




| Strain | Oil Cake | Time [days] | Conv. 1a [%] | Products | |||
|---|---|---|---|---|---|---|---|
| Trans 2a–b [%] | ee [%] | Cis 2c–d [%] | ee [%] | ||||
| Gordonia bronchialis PCM2167 | linseed | 3 | 100 | 83 (±0.7) | 33 (+)-(4S,5R)-2a | 17 (±1.5) | >99 (−)-(4S,5S)-2c |
| 7 | 100 | 81 (±1.2) | 33 (+)-(4S,5R)-2a | 19 (±1.3) | >99 (−)-(4S,5S)-2c | ||
| rapeseed | 3 | 9 (±0.9) | 7 (±0.4) | 32 (+)-(4S,5R)-2a | 2 (±0.1) | 0 | |
| 7 | 12 (±1.1) | 10 (±0.8) | 18 (+)-(4S,5R)-2a | 2 (±0.1) | 0 | ||
| Rhodococcus erythropolis DSM44534 | linseed | 3 | 100 | 97 (±1.2) | 20 (−)-(4R,5S)-2b | 3 (±0.2) | >99 (+)-(4R,5R)-2d |
| 7 | 100 | 90 (±0.9) | 70 (−)-(4R,5S)-2b | 10 (±0.5) | >99 (+)-(4R,5R)-2d | ||
| rapeseed | 3 | 15 (±0.7) | 8 (±0.7) | nd* | 7 (±0.2) | nd | |
| 7 | 43 (±2.1) | 9 (±0.8) | nd | 34 (±1.2) | nd | ||
| Rhodococcus rhodochrous PCM909 | linseed | 3 | 100 | 92 (±0.7) | 20 (+)-(4S,5R)-2a | 8 (±0.3) | >99 (+)-(4R,5R)-2d |
| 7 | 100 | 90 (±0.9) | 42 (+)-(4S,5R)-2a | 10 (±0.4) | >99 (+)-(4R,5R)-2d | ||
| rapeseed | 3 | 0 | 0 | 0 | 0 | 0 | |
| 7 | 0 | 0 | 0 | 0 | 0 | ||
| Rhodococcus ruber PCM2166 | linseed | 3 | 100 | 92 (±1.3) | 37 (+)-(4S,5R)-2a | 8 (±0.1) | >99 (−)-(4S,5S)-2c |
| 7 | 100 | 86 (±0.6) | 33 (+)-(4S,5R)-2a | 14 (±0.8) | 65 (−)-(4S,5S)-2c | ||
| rapeseed | 3 | 0 | 0 | 0 | 0 | 0 | |
| 7 | 84 (±0.6) | 67 (±1.2) | 5 (+)-(4S,5R)-2a | 17 (±1.2) | 0 | ||
| Strain | Oil Cake | Time [days] | Conv. 1b [%] | Products | |||
|---|---|---|---|---|---|---|---|
| Trans 2a–b [%] | ee [%] | Cis 2c–d [%] | ee [%] | ||||
| Gordonia bronchialis PCM2167 | linseed | 3 | 100 | 19 (±0.7) | >99 (+)-(4S,5R)-2a | 81 (±0.7) | 93 (+)-(4R,5R)-2d |
| 7 | 100 | 21 (±1.3) | >99 (+)-(4S,5R)-2a | 79 (±1.7) | 85 (+)-(4R,5R)-2d | ||
| rapeseed | 3 | 0 | 0 | 0 | 0 | 0 | |
| 7 | 12 (±0.9) | 10 | 0 | 2 (±0.4) | 82 (+)-(4R,5R)-2d | ||
| Rhodococcus erythropolis DSM44534 | linseed | 3 | 100 | 27 (±0.9) | 37 (+)-(4S,5R)-2a | 73 (±1.0) | 83 (+)-(4R,5R)-2d |
| 7 | 100 | 23 (±0.6) | 78 (+)-(4S,5R)-2a | 77 (±1.1) | 86 (+)-(4R,5R)-2d | ||
| rapeseed | 3 | 18 (±0.7) | 4 (±0.1) | 0 | 14 (±0.3) | 0 | |
| 7 | 57 (±1.5) | 9 (±0.3) | 0 | 48 (±1.6) | 4 (+)-(4R,5R)-2d | ||
| Rhodococcus rhodochrous PCM909 | linseed | 3 | 100 | 25 (±0.9) | >99 (+)-(4S,5R)-2a | 75 (±1.2) | 65 (+)-(4R,5R)-2d |
| 7 | 100 | 31 (±0.4) | >99 (+)-(4S,5R)-2a | 69 (±0.7) | 67 (+)-(4R,5R)-2d | ||
| rapeseed | 3 | 0 | 0 | 0 | 0 | 0 | |
| 7 | 0 | 0 | 0 | 0 | 0 | ||
| Rhodococcus ruber PCM2166 | linseed | 3 | 100 | 37 (±0.9) | >99 (+)-(4S,5R)-2a | 63 (±1.6) | 79 (+)-(4R,5R)-2d |
| 7 | 100 | 37 (±1.2) | >99 (+)-(4S,5R)-2a | 63 (±0.8) | 83 (+)-(4R,5R)-2d | ||
| rapeseed | 3 | 0 | 0 | 0 | 0 | 0 | |
| 7 | 22 (±0.7) | 6 (±0.3) | 0 | 16 (±0.9) | 0 | ||
| Strain | Time [days] | Conv. 1a–1b [%] | Products | |||
|---|---|---|---|---|---|---|
| Trans 2a–b [%] | ee (+)-(4S,5R)-2a[%] | Cis 2c–d [%] | ee (+)-(4R,5R)-2d[%] | |||
| Gordonia bronchialis PCM2167 | 3 | 100 | 23 (±0.9) | 55 | 77 (±1.2) | 90 |
| 7 | 100 | 45 (±0.7) | 52 | 55 (±0.6) | 70 | |
| Rhodococcus erythropolis DSM44534 | 3 | 100 | 47 (±1.1) | 32 | 53 (±1.4) | 77 |
| 7 | 100 | 47 (±0.4) | 35 | 53 (±0.7) | 79 | |
| Rhodococcus rhodochrous PCM909 | 3 | 100 | 39 (±0.8) | 70 | 61 (±1.1) | 60 |
| 7 | 100 | 47 (±1.7) | 76 | 53 (±1.9) | 74 | |
| Rhodococcus ruber PCM2166 | 3 | 100 | 58 (±1.4) | 30 | 42 (±1.1) | 32 |
| 7 | 100 | 60 (±1.7) | 37 | 40 (±0.3) | 35 | |
| Strain | Time [days] | Conv. 1a [%] | Products | |||
|---|---|---|---|---|---|---|
| Trans 2a–b [%] | ee [%] | Cis 2c–d [%] | ee [%] | |||
| Dietzia sp. DSM44016 | 3 | 100 | 80 (±1.1) | 31 (−)-(4R,5S)-2b | 20 (±0.6) | 84 (+)-(4R,5R)-2d |
| 7 | 100 | 82 (±0.8) | 44 (−)-(4R,5S)-2b | 18 (±0.7) | 85 (+)-(4R,5R)-2d | |
| Gordonia rubripertincta PCM2144 | 3 | 100 | 100 | 78 (+)-(4S,5R)-2a | 0 | 0 |
| 7 | 100 | 100 | 62 (+)-(4S,5R)-2a | 0 | 0 | |
| Micrococcus luteus PCM525 | 3 | 20 (±0.5) | 20 (±0.7) | 12 (+)-(4S,5R)-2a | 0 | 0 |
| 7 | 25 (±0.9) | 25 (±0.2) | 15 (+)-(4S,5R)-2a | 0 | 0 | |
| Rhodococcus coprophilus PCM2174 | 3 | 100 | 95 (±0.6) | 7 (+)-(4S,5R)-2a | 5 (±0.1) | 20 (+)-(4R,5R)-2d |
| 7 | 100 | 93 (±0.5) | 3 (+)-(4S,5R)-2a | 7 (±0.7) | 30 (+)-(4R,5R)-2d | |
| Rhodococcus erythropolis PCM2150 | 3 | 100 | 80 (±1.1) | 0 | 20 (±0.9) | 99 (−)-(4S,5S)-2c |
| 7 | 100 | 82 (±0.8) | 27 (+)-(4S,5R)-2a | 18 (±0.7) | 50 (−)-(4S,5S)-2c | |
| Rhodococcus ruber PCM2171 | 3 | 30 (±0.2) | 30 (±0.6) | 30 (+)-(4S,5R)-2a | 0 | 0 |
| 7 | 80 (±1.1) | 80 (±1.5) | 9 (+)-(4S,5R)-2a | 0 | 0 | |
| Rhodococcus ruber PCM2216 | 3 | 0 | 0 | 0 | 0 | 0 |
| 7 | 35 (±0.9) | 35 (±0.3) | 10 (+)-(4S,5R)-2a | 0 | 0 | |
| Streptomyces griseus subsp. griseus PCM2331 | 3 | 100 | 78 (±1.5) | 50 (+)-(4S,5R)-2a | 22 (±0.7) | 8 (+)-(4R,5R)-2d |
| 7 | 100 | 85 (±1.2) | 50 (+)-(4S,5R)-2a | 15 (±0.4) | 25 (+)-(4R,5R)-2d | |
| Strain | Time [days] | Conv. 1b [%] | Products | |||
|---|---|---|---|---|---|---|
| Trans 2a–b [%] | ee [%] | Cis 2c–d [%] | ee [%] | |||
| Dietzia sp. DSM44016 | 3 | 100 | 27 (±0.7) | 77 (+)-(4S,5R)-2a | 73 (±1.2) | 75 (+)-(4R,5R)-2d |
| 7 | 100 | 23 (±0.9) | 50 (+)-(4S,5R)-2a | 77 (±0.4) | 79 (+)-(4R,5R)-2d | |
| Gordonia rubripertincta PCM2144 | 3 | 100 | 40 (±0.3) | 77 (+)-(4S,5R)-2a | 60 (±1.2) | 32 (−)-(4S,5S)-2c |
| 7 | 100 | 16 (±0.5) | 62 (+)-(4S,5R)-2a | 84 (±2.1) | 30 (−)-(4S,5S)-2c | |
| Micrococcus luteus PCM525 | 3 | 30 (±0.4) | 0 | 0 | 30 (±0.9) | 20 (+)-(4R,5R)-2d |
| 7 | 35 (±1.3) | 0 | 0 | 35 (±1.1) | 29 (+)-(4R,5R)-2d | |
| Rhodococcus coprophilus PCM2174 | 3 | 100 | 15 (±0.4) | 60 (+)-(4S,5R)-2a | 85 (±1.2) | 33 (+)-(4R,5R)-2d |
| 7 | 100 | 30 (±0.9) | 75 (+)-(4S,5R)-2a | 70 (±0.7) | 33 (+)-(4R,5R)-2d | |
| Rhodococcus erythropolis PCM2150 | 3 | 100 | 0 | 0 | 100 | 5 (+)-(4R,5R)-2d |
| 7 | 100 | 0 | 0 | 100 | 2 (+)-(4R,5R)-2d | |
| Rhodococcus ruber PCM2171 | 3 | 30 (±0.2) | 0 | 0 | 30 (±1.7) | 10 (−)-(4S,5S)-2c |
| 7 | 60 (±1.3) | 0 | 0 | 60 (±1.8) | 0 | |
| Rhodococcus ruber PCM2216 | 3 | 35 (±0.7) | 0 | 0 | 35 (±1.1) | 30 (+)-(4R,5R)-2c |
| 7 | 35 (±0.4) | 0 | 0 | 35 (±1.3) | 0 | |
| Streptomyces griseus subsp. griseus PCM2331 | 3 | 10 (±0.2) | 5 | 0 | 5 (±0.3) | 60 (+)-(4R,5R)-2d |
| 7 | 27 (±1.2) | 16 | 0 | 11 (±0.3) | 60 (+)-(4R,5R)-2d | |
| Strain | Time [days] | Conv. 1b [%] | Products | |||
|---|---|---|---|---|---|---|
| Trans 2a | ee | Cis 2d | ee | |||
| [%] | [%] | [%] | [%] | |||
| Gordonia rubripertincta PCM2144 | 4 | 25 (±0.6) | 25 (±0.9) | 55 (+)-(4S,5R)-2a | 0 | 0 |
| 7 | 100 | 100 | 66 (+)-(4S,5R)-2a | 0 | 0 | |
| Rhodococcus erythropolis DSM44534 | 4 | 15 (±0.3) | 3 (±0.2) | 65 (+)-(4S,5R)-2a | 12 (±0.5) | 54 (+)-(4R,5R)-2d |
| 7 | 80 (±1.2) | 22 (±0.6) | 80(+)-(4S,5R)-2a | 58 (±0.5) | 66 (+)-(4R,5R)-2d | |
| Strain | Time [days] | Conv. 1a [%] | Products | |||
|---|---|---|---|---|---|---|
| Trans 2a | ee | Cis 2c | ee | |||
| [%] | [%] | [%] | [%] | |||
| Gordonia rubripertincta PCM2144 | 4 | 54 (±1.1) | 18 (±0.4) | 51 (+)-(4S,5R)-2a | 36 (±0.9) | 27 (−)-(4S,5S)-2c |
| 7 | 90 (±1.5) | 21 (±0.7) | 64 (+)-(4S,5R)-2a | 69 (±1.1) | 25 (−)-(4S,5S)-2c | |
| Rhodococcus erythropolis PCM2150 | 4 | 56 (±0.3) | 45 (±0.2) | 0 | 11 (±0.4) | 68 (−)-(4S,5S)-2c |
| 7 | 100 | 77 (±1.3) | 35 (+)-(4S,5R)-2a | 23 (±0.8) | 66 (−)-(4S,5S)-2c | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernik, D.; Pannek, J.; Szczepańska, E.; Olejniczak, T.; Boratyński, F. Bacterial Whole Cells Synthesis of Whisky Lactones in a Solid-State Fermentation Bioreactor Prototype. Catalysts 2021, 11, 320. https://doi.org/10.3390/catal11030320
Hernik D, Pannek J, Szczepańska E, Olejniczak T, Boratyński F. Bacterial Whole Cells Synthesis of Whisky Lactones in a Solid-State Fermentation Bioreactor Prototype. Catalysts. 2021; 11(3):320. https://doi.org/10.3390/catal11030320
Chicago/Turabian StyleHernik, Dawid, Jakub Pannek, Ewa Szczepańska, Teresa Olejniczak, and Filip Boratyński. 2021. "Bacterial Whole Cells Synthesis of Whisky Lactones in a Solid-State Fermentation Bioreactor Prototype" Catalysts 11, no. 3: 320. https://doi.org/10.3390/catal11030320
APA StyleHernik, D., Pannek, J., Szczepańska, E., Olejniczak, T., & Boratyński, F. (2021). Bacterial Whole Cells Synthesis of Whisky Lactones in a Solid-State Fermentation Bioreactor Prototype. Catalysts, 11(3), 320. https://doi.org/10.3390/catal11030320