LuxAB-Based Microbial Cell Factories for the Sensing, Manufacturing and Transformation of Industrial Aldehydes
Abstract
:1. Introduction
2. Results
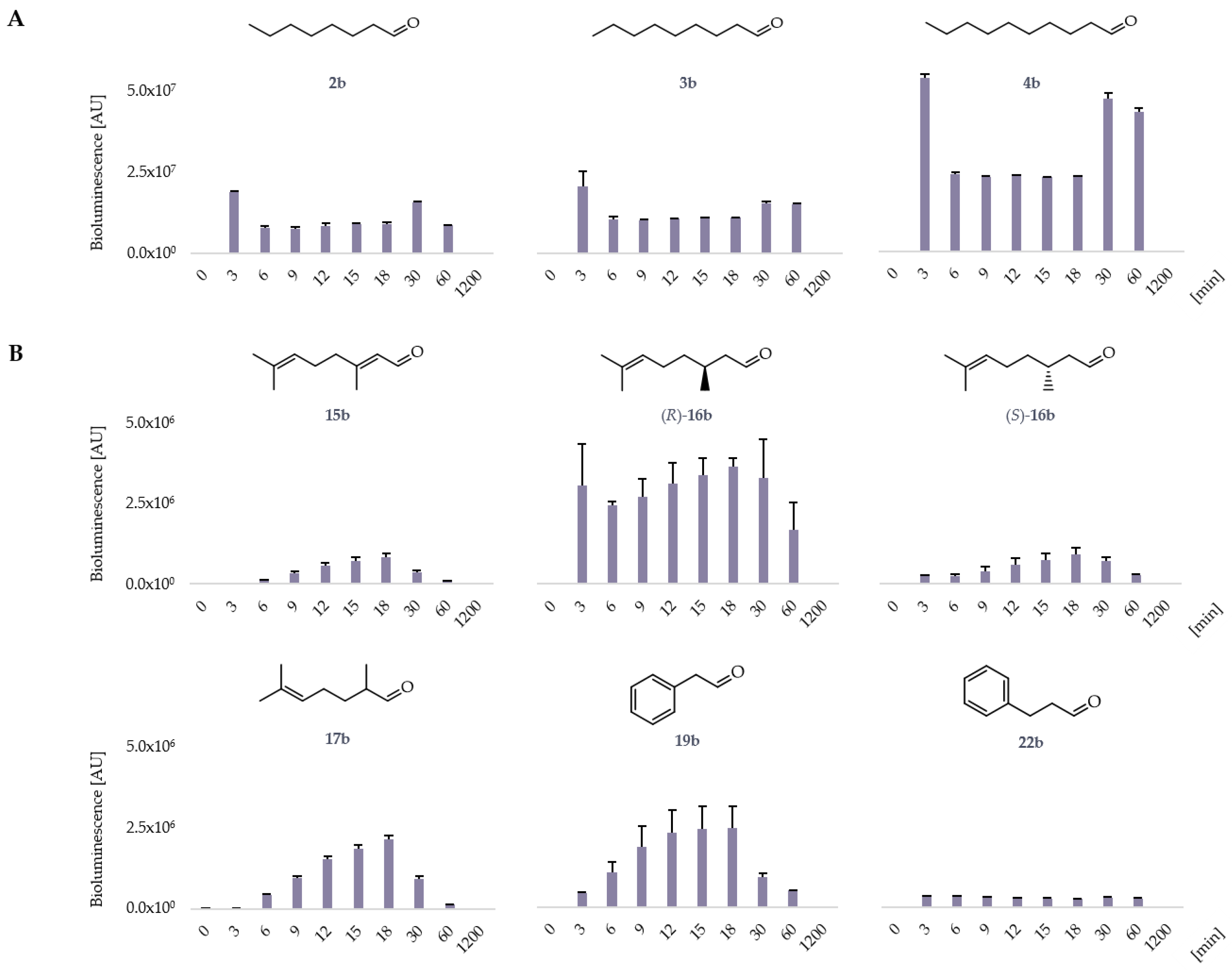
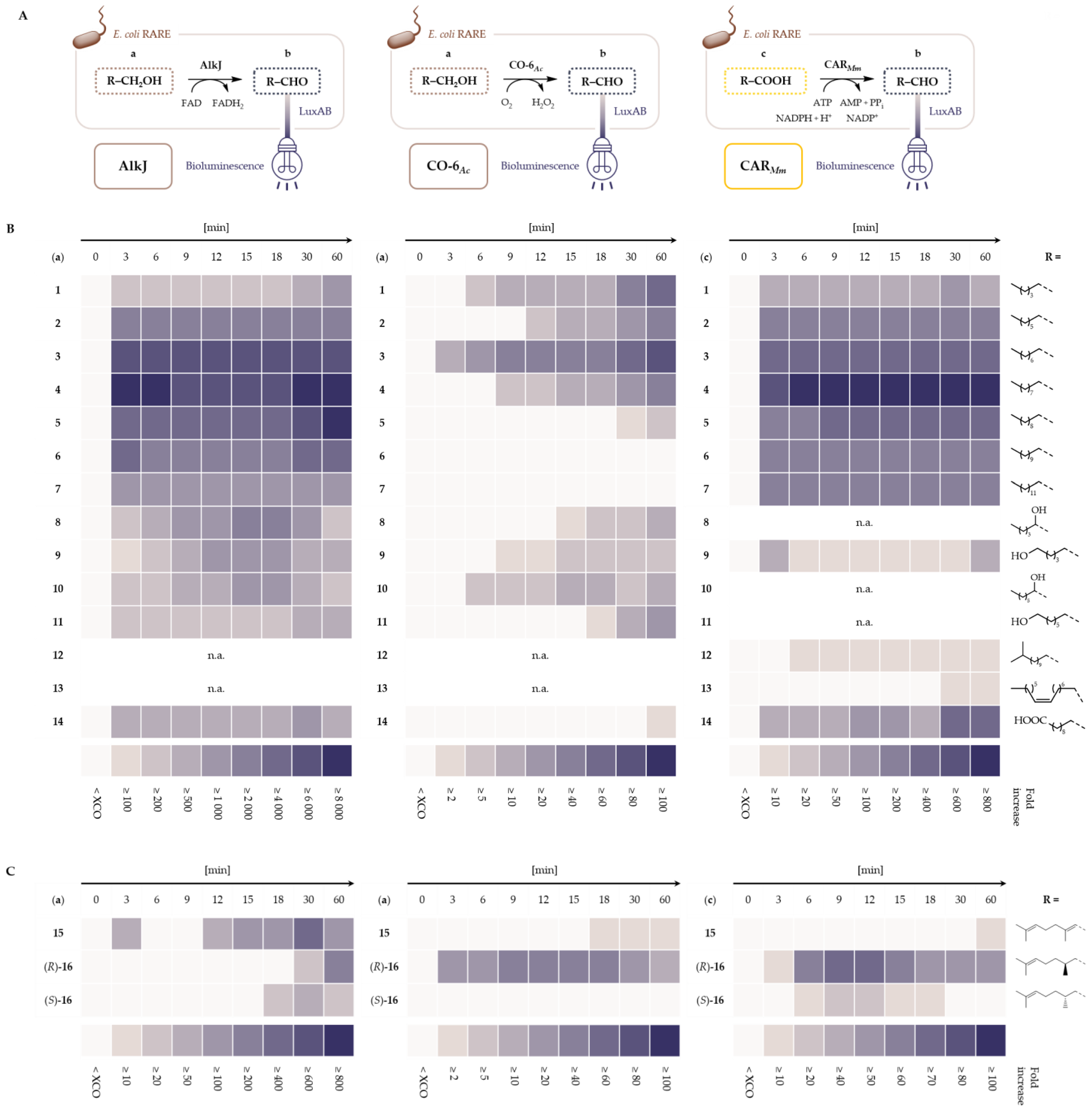
2.1. LuxAB Biosensor Assembly and High-Troughput (HT) Detection of Aldehydes In Vivo
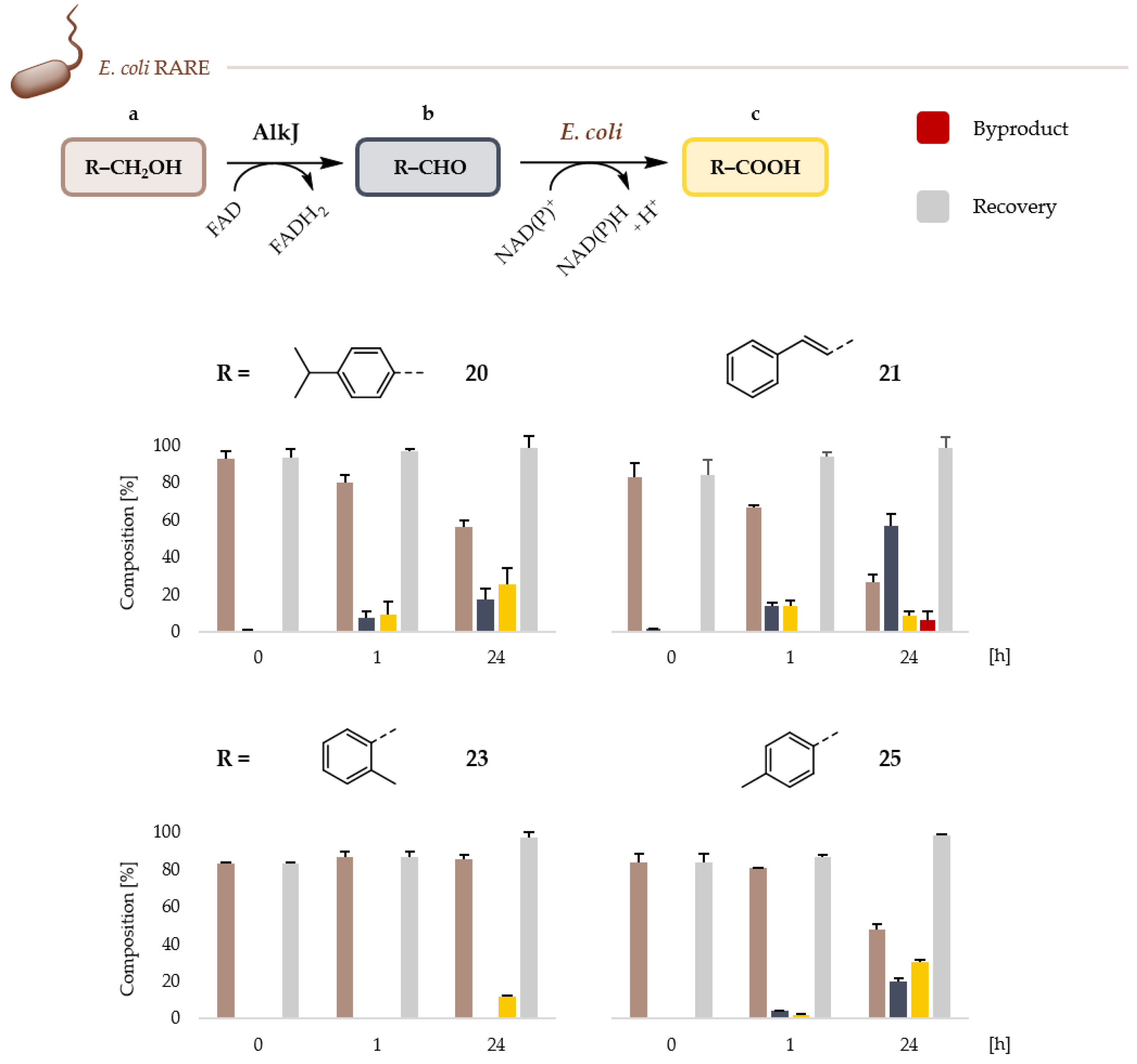
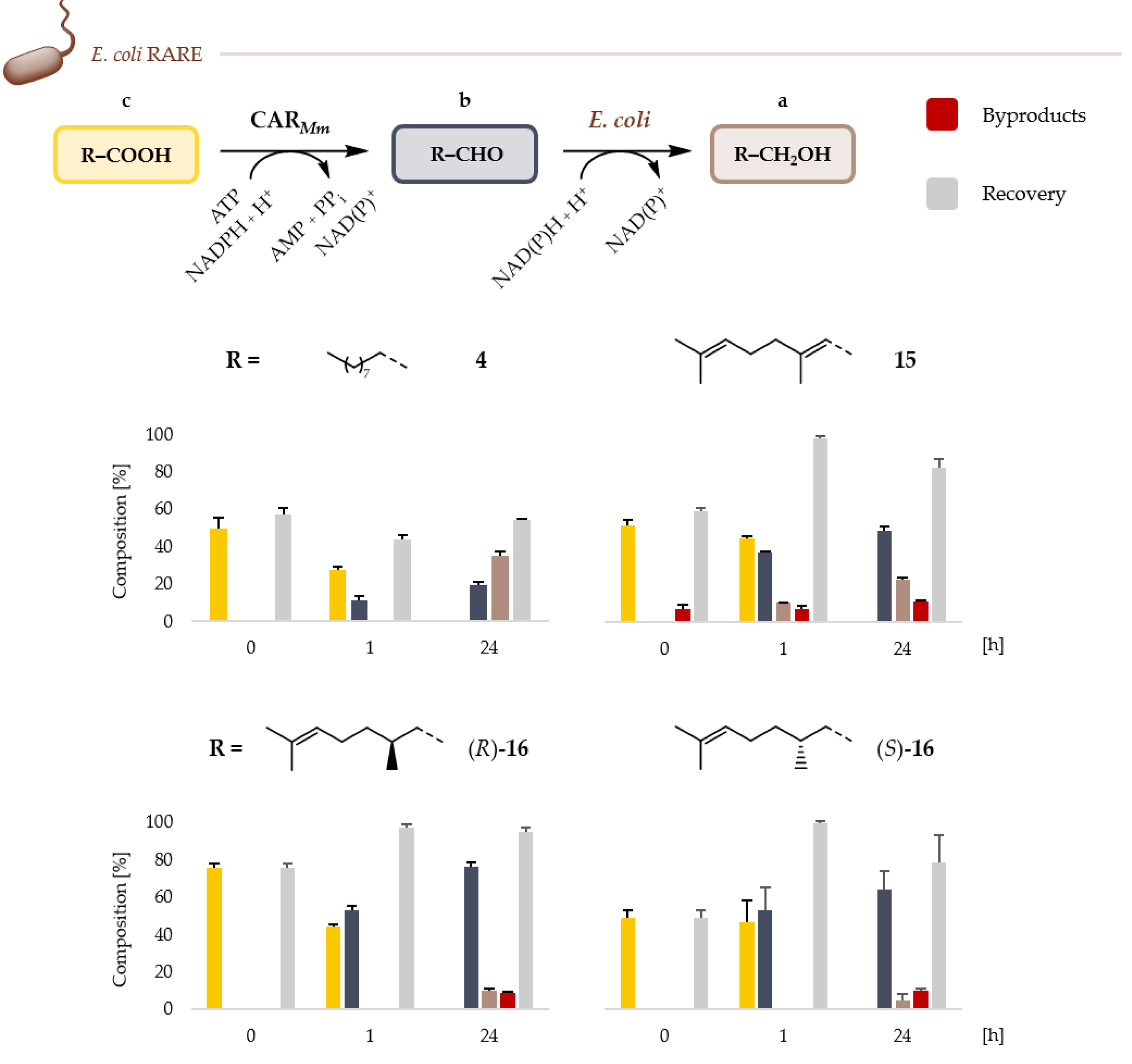
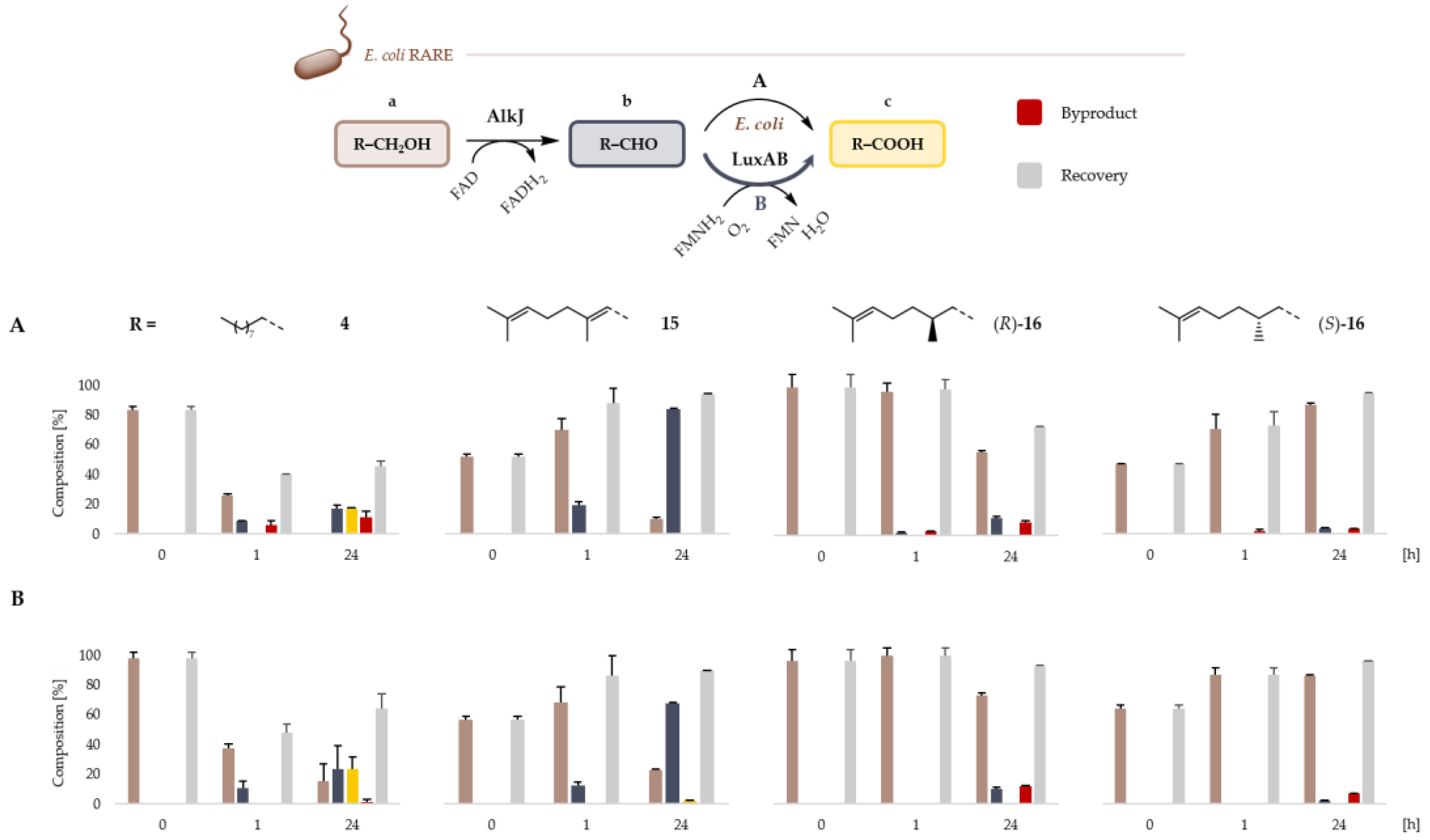
2.2. Microbial Cell Factories for the Production and Transformation of Value-Added Aldehydes
3. Discussion and Conclusions
4. Materials and Methods
4.1. General Information
4.2. Plasmid Assemblies by Sequence- and Ligation-Independent Cloning (SLIC) Techniques
4.3. Standard Conditions for Enzyme Production and the Preparation of Resting Cells (RCs)
4.4. LuxAB-Based HT Screening of Enzymes In Vivo
4.5. Whole-Cell Biotransformations and Gas Chromatographic (GC) Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dabirian, Y.; Gonçalves Teixeira, P.; Nielsen, J.; Siewers, V.; David, F. FadR-based biosensor-assisted screening for genes enhancing fatty acyl-CoA pools in Saccharomyces cerevisiae. ACS Synth. Biol. 2019, 8, 1788–1800. [Google Scholar] [CrossRef]
- Lehtinen, T.; Efimova, E.; Santala, S.; Santala, V. Improved fatty aldehyde and wax ester production by overexpression of fatty acyl-CoA reductases. Microb. Cell Fact. 2018, 17, 19. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Li, L.; Zhang, F.; Stephanopoulos, G.; Koffas, M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. USA 2014, 111, 11299–11304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunjapur, A.M.; Prather, K.L.J. Development of a Vanillate Biosensor for the Vanillin Biosynthesis Pathway in E. coli. ACS Synth. Biol. 2019, 8, 1958–1967. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Khatri, N.K.; Zsohár, A.; Kjærbølling, I.; Vogt, M.; Hammar, P.; Nielsen, C.F.; Marienhagen, J.; Sommer, M.O.A.; Joensson, H.N. Development of a bacterial biosensor for rapid screening of yeast p-coumaric acid production. ACS Synth. Biol. 2017, 6, 1860–1869. [Google Scholar] [CrossRef]
- Alvarez-Gonzalez, G.; Dixon, N. Genetically encoded biosensors for lignocellulose valorization. Biotechnol. Biofuels 2019, 12, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Reiter, M.A.; D’Espaux, L.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.; Harth, S.; Lin, W.; et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nat. Cell Biol. 2019, 567, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Meadows, A.L.; Hawkins, K.M.; Tsegaye, Y.; Antipov, E.; Kim, Y.; Raetz, L.; Dahl, R.H.; Tai, A.; Mahatdejkul-Meadows, T.; Xu, L.; et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nat. Cell Biol. 2016, 537, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Kunjapur, A.M.; Prather, K.L.J. Microbial Engineering for Aldehyde Synthesis. Appl. Environ. Microbiol. 2015, 81, 1892–1901. [Google Scholar] [CrossRef] [Green Version]
- Atsumi, S.; Hanai, T.; Liao, J. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nat. Cell Biol. 2008, 451, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Kaehne, F.; Buchhaupt, M.; Schrader, J. A recombinant α-dioxygenase from rice to produce fatty aldehydes using E. coli. Appl. Microbiol. Biotechnol. 2011, 90, 989–995. [Google Scholar] [CrossRef]
- Butler, N.; Kunjapur, A.M. Carboxylic acid reductases in metabolic engineering. J. Biotechnol. 2020, 307, 1–14. [Google Scholar] [CrossRef]
- Qu, G.; Guo, J.; Yang, D.; Sun, Z. Biocatalysis of carboxylic acid reductases: Phylogenesis, catalytic mechanism and potential applications. Green Chem. 2018, 20, 777–792. [Google Scholar] [CrossRef]
- Napora-Wijata, K.; Strohmeier, G.A.; Winkler, M. Biocatalytic reduction of carboxylic acids. Biotechnol. J. 2014, 9, 822–843. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.; Milker, S.; Wiesinger, T.; Winkler, M.; Mihovilovic, M.D.; Rudroff, F. In Vivo Synthesis of Polyhydroxylated Compounds from a “Hidden Reservoir” of Toxic Aldehyde Species. ChemCatChem 2017, 9, 2919–2923. [Google Scholar] [CrossRef]
- Wu, S.; Zhou, Y.; Wang, T.; Too, H.-P.; Wang, D.I.C.; Li, Z. Highly regio- and enantioselective multiple oxy- and amino-functionalizations of alkenes by modular cascade biocatalysis. Nat. Commun. 2016, 7, 11917. [Google Scholar] [CrossRef] [Green Version]
- Schrewe, M.; Julsing, M.K.; Lange, K.; Czarnotta, E.; Schmid, A.; Bühler, B. Reaction and catalyst engineering to exploit kinetically controlled whole-cell multistep biocatalysis for terminal FAME oxyfunctionalization. Biotechnol. Bioeng. 2014, 111, 1820–1830. [Google Scholar] [CrossRef]
- Rodriguez, G.M.; Atsumi, S. Isobutyraldehyde production from Escherichia coli by removing aldehyde reductase activity. Microb. Cell Fact. 2012, 11, 90. [Google Scholar] [CrossRef] [Green Version]
- He, A.; Li, T.; Daniels, L.; Fotheringham, I.; Rosazza, J.P.N. Nocardia sp. Carboxylic Acid Reductase: Cloning, Expression, and Characterization of a New Aldehyde Oxidoreductase Family. Appl. Environ. Microbiol. 2004, 70, 1874–1881. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, G.M.; Atsumi, S. Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab. Eng. 2014, 25, 227–237. [Google Scholar] [CrossRef] [Green Version]
- Kunjapur, A.M.; Tarasova, Y.; Prather, K.L.J. Synthesis and Accumulation of Aromatic Aldehydes in an Engineered Strain of Escherichia coli. J. Am. Chem. Soc. 2014, 136, 11644–11654. [Google Scholar] [CrossRef] [PubMed]
- Markel, U.; Essani, K.D.; Besirlioglu, V.; Schiffels, J.; Streit, W.R.; Schwaneberg, U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 2020, 49, 233–262. [Google Scholar] [CrossRef]
- Santos-Zavaleta, A.; Sánchez-Pérez, M.; Salgado, H.; Velázquez-Ramírez, D.A.; Gama-Castro, S.; Tierrafría, V.H.; Busby, S.J.W.; Aquino, P.; Fang, X.; Palsson, B.O.; et al. A unified resource for transcriptional regulation in Escherichia coli K-12 incorporating high-throughput-generated binding data into RegulonDB version 10.0. BMC Biol. 2018, 16, 91. [Google Scholar] [CrossRef]
- Jang, S.; Jang, S.; Xiu, Y.; Kang, T.J.; Lee, S.-H.; Koffas, M.A.G.; Jung, G.Y. Development of Artificial Riboswitches for Monitoring of Naringenin In Vivo. ACS Synth. Biol. 2017, 6, 2077–2085. [Google Scholar] [CrossRef] [PubMed]
- Lehtinen, T.; Santala, V.; Santala, S.M. Twin-layer biosensor for real-time monitoring of alkane metabolism. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Liu, Y.; Zhuang, Y.; Ding, D.; Xu, Y.; Sun, J.; Zhang, D. Biosensor-Based Evolution and Elucidation of a Biosynthetic Pathway in Escherichia coli. ACS Synth. Biol. 2017, 6, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Evans, T.; Zhang, F. Applications and advances of metabolite biosensors for metabolic engineering. Metab. Eng. 2015, 31, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brophy, J.A.; Voigt, C.A. Principles of genetic circuit design. Nat. Methods 2014, 11, 508–520. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, J.A.; McKee, A.E.; Keasling, J.D. High-Throughput Metabolic Engineering: Advances in Small-Molecule Screening and Selection. Annu. Rev. Biochem. 2010, 79, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Si, T.; Wang, M.; Zhao, H. Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening. ACS Synth. Biol. 2015, 4, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- DeLoache, W.C.; Russ, Z.N.; Narcross, L.; Gonzales, A.M.; Martin, V.J.J.; Dueber, J.E. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat. Chem. Biol. 2015, 11, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, J.A.; Shis, D.L.; Alikhani, A.; Keasling, J.D. Transcription Factor-Based Screens and Synthetic Selections for Microbial Small-Molecule Biosynthesis. ACS Synth. Biol. 2013, 2, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Schendzielorz, G.; Dippong, M.; Grünberger, A.; Kohlheyer, D.; Yoshida, A.; Binder, S.; Nishiyama, C.; Nishiyama, M.; Bott, M.; Eggeling, L. Taking Control over Control: Use of Product Sensing in Single Cells to Remove Flux Control at Key Enzymes in Biosynthesis Pathways. ACS Synth. Biol. 2014, 3, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; Rogers, J.K.; Taylor, N.D.; Church, G. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 17803–17808. [Google Scholar] [CrossRef] [Green Version]
- Chou, H.H.; Keasling, J. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013, 4, 2595. [Google Scholar] [CrossRef] [Green Version]
- Hossain, G.S.; Saini, M.; Miyake, R.; Ling, H.; Chang, M.W. Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms. Trends Biotechnol. 2020, 38, 797–810. [Google Scholar] [CrossRef]
- Volke, D.C.; Turlin, J.; Mol, V.; Nikel, P.I. Physical decoupling of XylS/Pm regulatory elements and conditional proteolysis enable precise control of gene expression in Pseudomonas putida. Microb. Biotechnol. 2019, 13, 222–232. [Google Scholar] [CrossRef] [Green Version]
- Rienzo, M.; Jackson, S.J.; Chao, L.K.; Leaf, T.; Schmidt, T.J.; Navidi, A.H.; Nadler, D.C.; Ohler, M.; Leavell, M.D. High-throughput screening for high-efficiency small-molecule biosynthesis. Metab. Eng. 2021, 63, 102–125. [Google Scholar] [CrossRef]
- Zhang, F.; Carothers, J.; Keasling, J. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 2012, 30, 354–359. [Google Scholar] [CrossRef]
- Doong, S.J.; Gupta, A.; Prather, K.L.J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc. Natl. Acad. Sci. USA 2018, 115, 2964–2969. [Google Scholar] [CrossRef] [Green Version]
- Calero, P.; Volke, D.C.; Lowe, P.T.; Gotfredsen, C.H.; O’Hagan, D.; Nikel, P.I. A fluoride-responsive genetic circuit enables in vivo biofluorination in engineered Pseudomonas putida. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Machado, L.F.M.; Currin, A.; Dixon, N. Directed evolution of the PcaV allosteric transcription factor to generate a biosensor for aromatic aldehydes. J. Biol. Eng. 2019, 13, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorentino, G.; Ronca, R.; Bartolucci, S. A novel E. coli biosensor for detecting aromatic aldehydes based on a responsive inducible archaeal promoter fused to the green fluorescent protein. Appl. Microbiol. Biotechnol. 2009, 82, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Frazão, C.R.; Maton, V.; François, J.M.; Walther, T. Development of a Metabolite Sensor for High-Throughput Detection of Aldehydes in Escherichia Coli. Front. Bioeng. Biotechnol. 2018, 6. [Google Scholar] [CrossRef]
- Colepicolo, P.; Cho, K.W.; Poinar, G.O.; Hastings, J.W. Growth and luminescence of the bacterium Xenorhabdus luminescens from a human wound. Appl. Environ. Microbiol. 1989, 55, 2601–2606. [Google Scholar] [CrossRef] [Green Version]
- Ressmann, A.; Schwendenwein, D.; Leonhartsbgerger, S.; Mihovilovic, M.D.; Bornscheuer, U.; Winkler, M.; Rudroff, F. Substrate-Independent High-Throughput Assay for the Quantification of Aldehydes. Adv. Synth. Catal. 2019, 361, 2538–2543. [Google Scholar] [CrossRef] [Green Version]
- Kitov, P.I.; Vinals, D.F.; Ng, S.; Tjhung, K.F.; Derda, R. Rapid, Hydrolytically Stable Modification of Aldehyde-Terminated Proteins and Phage Libraries. J. Am. Chem. Soc. 2014, 136, 8149–8152. [Google Scholar] [CrossRef] [PubMed]
- Santala, S.M.; Efimova, E.; Karp, M.; Santala, V. Real-Time monitoring of intracellular wax ester metabolism. Microb. Cell Fact. 2011, 10, 75. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Werling, U.; Edelmann, W. SLiCE: A novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012, 40, e55. [Google Scholar] [CrossRef] [Green Version]
- Wiesinger, T.; Bayer, T.; Milker, S.; Mihovilovic, M.D.; Rudroff, F. Cell factory design and optimization for the stereoselective synthesis of polyhydroxylated compounds. ChemBioChem 2018, 19, 361–368. [Google Scholar] [CrossRef]
- Heath, R.S.; Birmingham, W.R.; Thompson, M.P.; Taglieber, A.; Daviet, L.; Turner, N.J. An Engineered Alcohol Oxidase for the Oxidation of Primary Alcohols. ChemBioChem 2018, 20, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, W.; Thomas, A.; Cromar, H.; Gough, B.; Snajdrova, R.; Adams, J.P.; Littlechild, J.A.; Harmer, N.J. Characterization of Carboxylic Acid Reductases as Enzymes in the Toolbox for Synthetic Chemistry. ChemCatChem 2017, 9, 1005–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, M.K.; Turner, N.J.; Jones, P. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. USA 2013, 110, 87–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvat, M.; Winkler, M. In VivoReduction of Medium- to Long-Chain Fatty Acids by Carboxylic Acid Reductase (CAR) Enzymes: Limitations and Solutions. ChemCatChem 2020, 12, 5076–5090. [Google Scholar] [CrossRef]
- Del Solar, G.; Giraldo, R.; Ruiz-Echevarría, M.J.; Espinosa, M.; Díaz-Orejas, R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998, 62, 434–464. [Google Scholar] [CrossRef] [Green Version]
- Van Beilen, J.B.; Eggink, G.; Enequist, H.; Bos, R.; Witholt, B. DNA sequence determination and functional characterization of the OCT plasmid-encoded alkJKL genes of Pseudomonas oleovorans. Mol. Microbiol. 1992, 6, 3121–3136. [Google Scholar] [CrossRef]
- Juven, B.J.; Pierson, M.D. Antibacterial Effects of Hydrogen Peroxide and Methods for Its Detection and Quantitation†. J. Food Prot. 1996, 59, 1233–1241. [Google Scholar] [CrossRef]
- Milker, S.; Gonçalves, L.; Fink, M.J.; Rudroff, F. Escherichia coli Fails to Efficiently Maintain the Activity of an Important Flavin Monooxygenase in Recombinant Overexpression. Front. Microbiol. 2017, 8, 2201. [Google Scholar] [CrossRef] [Green Version]
- Mueller, N.J.; Stueckler, C.; Hauer, B.; Baudendistel, N.; Housden, H.; Bruce, N.C.; Faber, K. The Substrate Spectra of Pentaerythritol Tetranitrate Reductase, Morphinone Reductase, N-Ethylmaleimide Reductase and Estrogen-Binding Protein in the Asymmetric Bioreduction of Activated Alkenes. Adv. Synth. Catal. 2010, 352, 387–394. [Google Scholar] [CrossRef]
- Peters, C.; Kölzsch, R.; Kadow, M.; Skalden, L.; Rudroff, F.; Mihovilovic, M.D.; Bornscheuer, U.T. Identification, characterization and application of three enoate reductases from Pseudomonas putida in in vitro enzyme cascade reactions. ChemCatChem 2014, 6, 1021–1027. [Google Scholar] [CrossRef]
- Richardson, K.N.; Black, W.B.; Li, H. Aldehyde production in crude lysate- and whole cell-nased biotransformation using a noncanonical redox cofactor system. ACS Catal. 2020, 10, 889–903. [Google Scholar] [CrossRef]
- Williams, R.E.; Bruce, N.C. ‘New uses for an old enzyme’—The old yellow enzyme family of flavoenzymes. Microbiology 2002, 148, 1607–1614. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Thorpe, C.; Schulz, H. 2,4-Dienoyl-CoA reductase from Escherichia coli is a novel iron–sulfur flavoprotein that functions in fatty acid β-oxidation. Arch. Biochem. Biophys. 2000, 380, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Lee, J. Perspectives on Bioluminescence Mechanisms. Photochem. Photobiol. 2016, 93, 389–404. [Google Scholar] [CrossRef]
- Jimenez-Diaz, L.; Caballero, A.; Segura, A. Pathways for the Degradation of Fatty Acids in Bacteria. In Aerobic Utilization of Hydrocarbons, Oils and Lipids; Rojo, F., Ed.; Springer Internationl Publishing: Cham, Switzerland, 2017; pp. 1–23. [Google Scholar] [CrossRef]
- Janßen, H.J.; Steinbüchel, A. Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol. Biofuels 2014, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiltschi, B.; Cernava, T.; Dennig, A.; Casas, M.G.; Geier, M.; Gruber, S.; Haberbauer, M.; Heidinger, P.; Acero, E.H.; Kratzer, R.; et al. Enzymes revolutionize the bioproduction of value-added compounds: From enzyme discovery to special applications. Biotechnol. Adv. 2020, 40, 107520. [Google Scholar] [CrossRef]
- Li, C.; Zhang, R.; Wang, J.; Wilson, L.M.; Yan, Y. Protein Engineering for Improving and Diversifying Natural Product Biosynthesis. Trends Biotechnol. 2020, 38, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Fleiss, A.; Sarkisyan, K.S. A brief review of bioluminescent systems. Curr. Genet. 2019, 65, 877–882. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-K.; Kang, C.-S.; Lee, J.-K.; Kim, Y.-R.; Han, H.-Y.; Yun, H.K. Evaluation of Repellency Effect of Two Natural Aroma Mosquito Repellent Compounds, Citronella and Citronellal. Entomol. Res. 2005, 35, 117–120. [Google Scholar] [CrossRef]
- Nakahara, K.; Alzoreky, N.S.; Yoshihashi, T.; Nguyen, H.T.T.; Trakoontivakorn, G. Chemical Composition and Antifungal Activity of Essential Oil from Cymbopogon nardus (Citronella Grass). JARQ 2013, 37, 249–252. [Google Scholar] [CrossRef] [Green Version]
- Piancatelli, G.; Leonelli, F. Oxidation of nerol to neral with iodobenzene diacetate and TEMPO. Org. Synth. 2006, 83, 1. [Google Scholar] [CrossRef] [Green Version]
- Müller, G.C.; Junnila, A.; Kravchenko, V.D.; Revay, E.E.; Butler, J.; Orlova, O.B.; Weiss, R.W.; Schlein, Y. Ability of essential oil candles to repel biting insects in high and low biting pressure environments. J. Am. Mosq. Control Assoc. 2008, 24, 154–160. [Google Scholar] [CrossRef]
- Clark, B.; Powell, C.; Radford, T. The acid catalyzed cyclization of citral. Tetrahedron 1977, 33, 2187–2191. [Google Scholar] [CrossRef]
- Parker, G.; Smith, L.; Baxendale, I. Development of the industrial synthesis of vitamin A. Tetrahedron 2016, 72, 1645–1652. [Google Scholar] [CrossRef]
- Li, C.; Wen, A.; Shen, B.; Lu, J.; Huang, Y.; Chang, Y. FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011, 11, 92. [Google Scholar] [CrossRef] [Green Version]
- Julsing, M.K.; Schrewe, M.; Cornelissen, S.; Hermann, I.; Schmid, A.; Bühler, B. Outer Membrane Protein AlkL Boosts Biocatalytic Oxyfunctionalization of Hydrophobic Substrates in Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 5724–5733. [Google Scholar] [CrossRef] [Green Version]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayer, T.; Becker, A.; Terholsen, H.; Kim, I.J.; Menyes, I.; Buchwald, S.; Balke, K.; Santala, S.; Almo, S.C.; Bornscheuer, U.T. LuxAB-Based Microbial Cell Factories for the Sensing, Manufacturing and Transformation of Industrial Aldehydes. Catalysts 2021, 11, 953. https://doi.org/10.3390/catal11080953
Bayer T, Becker A, Terholsen H, Kim IJ, Menyes I, Buchwald S, Balke K, Santala S, Almo SC, Bornscheuer UT. LuxAB-Based Microbial Cell Factories for the Sensing, Manufacturing and Transformation of Industrial Aldehydes. Catalysts. 2021; 11(8):953. https://doi.org/10.3390/catal11080953
Chicago/Turabian StyleBayer, Thomas, Aileen Becker, Henrik Terholsen, In Jung Kim, Ina Menyes, Saskia Buchwald, Kathleen Balke, Suvi Santala, Steven C. Almo, and Uwe T. Bornscheuer. 2021. "LuxAB-Based Microbial Cell Factories for the Sensing, Manufacturing and Transformation of Industrial Aldehydes" Catalysts 11, no. 8: 953. https://doi.org/10.3390/catal11080953
APA StyleBayer, T., Becker, A., Terholsen, H., Kim, I. J., Menyes, I., Buchwald, S., Balke, K., Santala, S., Almo, S. C., & Bornscheuer, U. T. (2021). LuxAB-Based Microbial Cell Factories for the Sensing, Manufacturing and Transformation of Industrial Aldehydes. Catalysts, 11(8), 953. https://doi.org/10.3390/catal11080953









