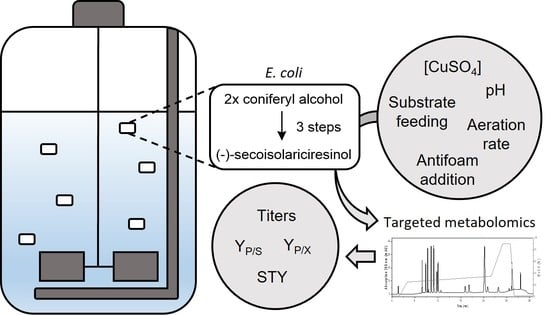
Heterologous Lignan Production in Stirred-Tank Reactors—Metabolomics-Assisted Bioprocess Development for an In Vivo Enzyme Cascade
Abstract
:1. Introduction
2. Results
2.1. Bioprocess Design
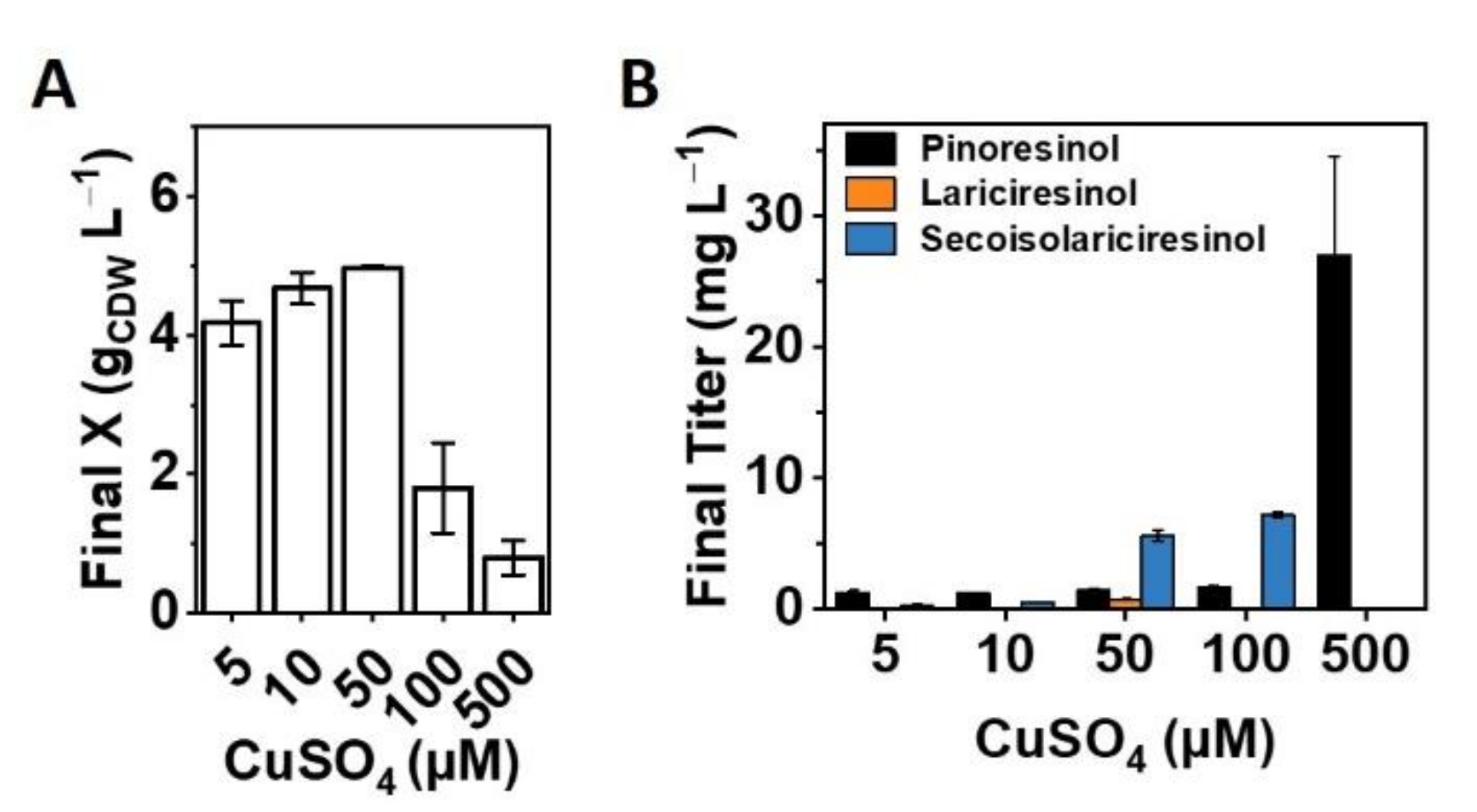
2.2. Optimizing the Copper Supply
2.3. Impact of Antifoam Addition and Oxygen Availability on Lignan Synthesis
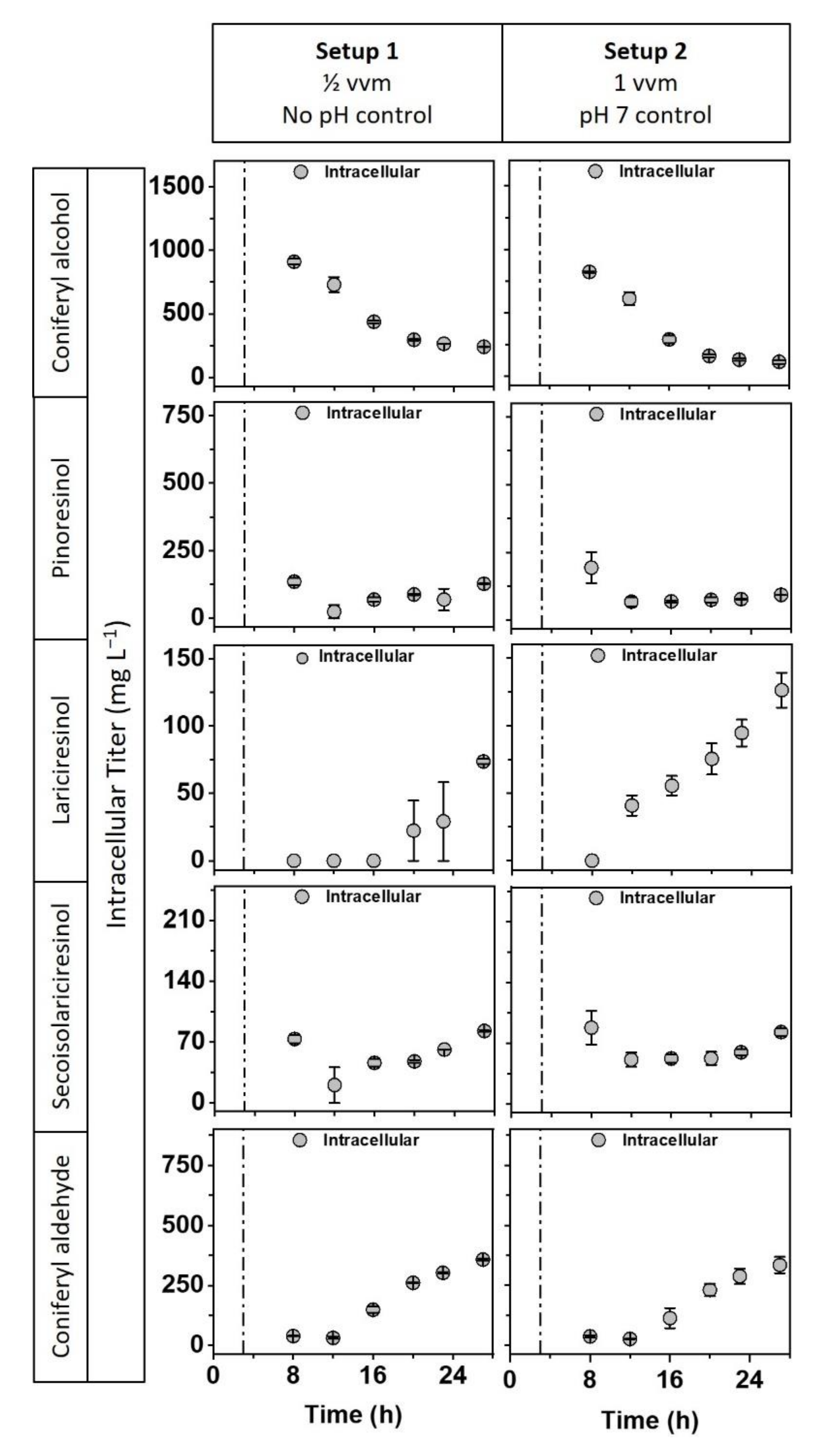
2.4. Transfer to Stirred-Tank Reactor with Regulated Conditions
2.5. Impact of Coniferyl Alcohol Feeding Strategies on Pathway Activity
2.6. Determination of Key Bioprocess Parameters
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, and Long-Term Storage
4.2. Medium and Chemicals
4.3. Fermentation Conditions
4.3.1. Batch Cultivation in Shake Flask
4.3.2. Batch Cultivation in Stirred-Tank Reactor
4.3.3. Determination of Biomass Growth
4.4. Analysis of Extra- and Intracellular Metabolites
4.4.1. Sampling
4.4.2. Extraction of Intracellular Metabolites
4.4.3. Quantification of Carbon Sources
4.4.4. Quantification of Phenylpropanoid and Lignan Metabolites
4.5. Calculation of Intracellular Metabolite Concentration and Key Bioprocess Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Satake, H.; Koyama, T.; Bahabadi, S.E.; Matsumoto, E.; Ono, E.; Murata, J. Essences in Metabolic Engineering of Lignan Biosynthesis. Metabolites 2015, 5, 270–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imran, M.; Ahmad, N.; Anjum, F.M.; Khan, M.K.; Mushtaq, Z.; Nadeem, M.; Hussain, S. Potential Protective Properties of Flax Lignan Secoisolariciresinol Diglucoside. Nutr. J. 2015, 14, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mark, R.; Lyu, X.; Lee, J.J.L.; Parra-Saldívar, R.; Chen, W.N. Sustainable Production of Natural Phenolics for Functional Food Applications. J. Funct. Foods 2019, 57, 233–254. [Google Scholar] [CrossRef]
- Nadeem, M.; Palni, L.M.S.; Purohit, A.N.; Pandey, H.; Nandi, S.K. Propagation and Conservation of Podophyllum Hexandrum Royle: An Important Medicinal Herb. Biol. Conserv. 2000, 92, 121–129. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, S.; Bornscheuer, U.T. Recent Advances in (Chemo)Enzymatic Cascades for Upgrading Bio-Based Resources. Chem. Commun. 2021, 57, 10661–10674. [Google Scholar] [CrossRef]
- An, C.; Maloney, K.M. Designing for Sustainability with Biocatalytic and Chemoenzymatic Cascade Processes. Curr. Opin. Green Sustain. Chem. 2022, 34, 100591. [Google Scholar] [CrossRef]
- Pickel, B.; Schaller, A. Dirigent Proteins: Molecular Characteristics and Potential Biotechnological Applications. Appl. Microbiol. Biotechnol. 2013, 97, 8427–8438. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Gang, D.R.; Davin, L.B.; Bedgar, D.L.; Chu, A.; Lewis, N.G. (+)-Pinoresinol/(+)-Lariciresinol Reductase from Forsythia Intermedia: Protein Purification, CDNA Cloning, Heterologous Expression and Comparison to Isoflavone Reductase. J. Biol. Chem. 1996, 271, 29473–29482. [Google Scholar] [CrossRef] [Green Version]
- Decembrino, D.; Ricklefs, E.; Wohlgemuth, S.; Girhard, M.; Schullehner, K.; Jach, G.; Urlacher, V.B. Assembly of Plant Enzymes in E. Coli for the Production of the Valuable (−)-Podophyllotoxin Precursor (−)-Pluviatolide. ACS Synth. Biol. 2020, 9, 3091–3103. [Google Scholar] [CrossRef]
- Lewis, N.G.; Davin, L.B.; Munasinghe, V.R.N.; Roberts, A.D. The Lignan Handbook; CRC Press: Boca Raton, FL, USA, 2022; ISBN 9781315152523. [Google Scholar]
- Ricklefs, E.; Girhard, M.; Urlacher, V.B. Three-Steps in One-Pot: Whole-Cell Biocatalytic Synthesis of Enantiopure (+)- and (−)-Pinoresinol via Kinetic Resolution. Microb. Cell Fact. 2016, 15, 1–11. [Google Scholar] [CrossRef]
- Decembrino, D.; Girhard, M.; Urlacher, V.B. Use of Copper as a Trigger for the in Vivo Activity of E. Coli Laccase CueO: A Simple Tool for Biosynthetic Purposes. ChemBioChem 2021, 22, 1470–1479. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.J.; Wei, Z.Y.; Lu, P.C.; Huang, P.L.; Lee, K.T. Bioconversion of Pinoresinol into Matairesinol by Use of Recombinant Escherichia Coli. Appl. Environ. Microbiol. 2014, 80, 2687–2692. [Google Scholar] [CrossRef] [Green Version]
- Cuppett, J.D. Evaluation of Copper Speciation and Water Quality Factors That Affect Aqueous Copper Tasting Response. Chem. Senses 2006, 31, 689–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Outten, F.W.; Huffman, D.L.; Hale, J.A.; O’Halloran, T.V. The Independent Cue and Cus Systems Confer Copper Tolerance during Aerobic and Anaerobic Growth in Escherichia Coli. J. Biol. Chem. 2001, 276, 30670–30677. [Google Scholar] [CrossRef] [Green Version]
- Singh, G.; Bhalla, A.; Kaur, P.; Capalash, N.; Sharma, P. Laccase from Prokaryotes: A New Source for an Old Enzyme. Rev. Environ. Sci. Biotechnol. 2011, 10, 309–326. [Google Scholar] [CrossRef]
- Hiraishi, T.; Tachibana, K.; Asakura, N.; Abe, H. Enhanced Expression of a Recombinant Multicopper Oxidase, CueO, from Escherichia Coli and Its Laccase Activity towards Aromatic Substrates. Polym. Degrad. Stab. 2019, 164, 1–8. [Google Scholar] [CrossRef]
- Solioz, M. Copper and Bacteria; SpringerBriefs in Molecular Science; Springer International Publishing: Cham, Switzerland, 2018; ISBN 978-3-319-94438-8. [Google Scholar]
- Oldiges, M.; Lütz, S.; Pflug, S.; Schroer, K.; Stein, N.; Wiendahl, C. Metabolomics: Current State and Evolving Methodologies and Tools. Appl. Microbiol. Biotechnol. 2007, 76, 495–511. [Google Scholar] [CrossRef]
- Steinmann, A.; Schullehner, K.; Kohl, A.; Dickmeis, C.; Finger, M.; Hubmann, G.; Jach, G.; Commandeur, U.; Girhard, M.; Urlacher, V.B.; et al. A Targeted Metabolomics Method for Extra- and Intracellular Metabolite Quantification Covering the Complete Monolignol and Lignan Synthesis Pathway. Metab. Eng. Commun. 2022, 15, e00205. [Google Scholar] [CrossRef]
- Riesenberg, D.; Schulz, V.; Knorre, W.A.; Pohl, H.D.; Korz, D.; Sanders, E.A.; Roß, A.; Deckwer, W.D. High Cell Density Cultivation of Escherichia Coli at Controlled Specific Growth Rate. J. Biotechnol. 1991, 20, 17–27. [Google Scholar] [CrossRef]
- Zhang, J.; Greasham, R. Chemically Defined Media for Commercial Fermentations. Appl. Microbiol. Biotechnol. 1999, 51, 407–421. [Google Scholar] [CrossRef]
- Meyer, H.; Minas, W.; Schmidhalter, D. Industrial-Scale Fermentation. In Industrial Biotechnology; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2016; pp. 1–53. [Google Scholar]
- Góralczyk-Bińkowska, A.; Jasińska, A.; Długoński, J. Characteristics and Use of Multicopper Oxidases Enzymes. Postępy Mikrobiol. Adv. Microbiol. 2019, 58, 7–18. [Google Scholar] [CrossRef] [Green Version]
- Vardar-Sukan, F. Foaming: Consequences, Prevention and Destruction. Biotechnol. Adv. 1998, 16, 913–948. [Google Scholar] [CrossRef]
- Phue, J.N.; Shiloach, J. Impact of Dissolved Oxygen Concentration on Acetate Accumulation and Physiology of E. Coli BL21, Evaluating Transcription Levels of Key Genes at Different Dissolved Oxygen Conditions. Metab. Eng. 2005, 7, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Changela, A.; Chen, K.; Xue, Y.; Holschen, J.; Outten, C.E.; O’Halloran, T.V.; Mondragón, A. Molecular Basis of Metal-Ion Selectivity and Zeptomolar Sensitivity by CueR. Science 2003, 301, 1383–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolle, P.; Hou, B.; Brüser, T. The Tat Substrate CueO Is Transported in an Incomplete Folding State. J. Biol. Chem. 2016, 291, 13520–13528. [Google Scholar] [CrossRef] [Green Version]
- Zeng, J.; Lin, X.; Zhang, J.; Li, X.; Wong, M.H. Oxidation of Polycyclic Aromatic Hydrocarbons by the Bacterial Laccase CueO from E. Coli. Appl. Microbiol. Biotechnol. 2011, 89, 1841–1849. [Google Scholar] [CrossRef]
- Lancefield, C.S.; Westwood, N.J. The Synthesis and Analysis of Advanced Lignin Model Polymers. Green Chem. 2015, 17, 4980–4990. [Google Scholar] [CrossRef] [Green Version]
- Freudenberg, K.; Harkin, J.M.; Reichert, M.; Fukuzumi, T. Die an Der Verholzung Beteiligten Enzyme. Die Dehydrierung Des Sinapinalkohols. Chem. Ber. 1958, 91, 581–590. [Google Scholar] [CrossRef]
- Halls, S.C.; Lewis, N.G. Secondary and Quaternary Structures of the (+)-Pinoresinol-Forming Dirigent Protein. Biochemistry 2002, 41, 9455–9461. [Google Scholar] [CrossRef]
- Ricklefs, E.; Girhard, M.; Koschorreck, K.; Smit, M.S.; Urlacher, V.B. Two-Step One-Pot Synthesis of Pinoresinol from Eugenol in an Enzymatic Cascade. ChemCatChem 2015, 7, 1857–1864. [Google Scholar] [CrossRef]
- Tarrago, L.; Modolo, C.; Yemloul, M.; Robert, V.; Rousselot-Pailley, P.; Tron, T. Controlling the Polymerization of Coniferyl Alcohol with Cyclodextrins. New J. Chem. 2018, 42, 11770–11775. [Google Scholar] [CrossRef] [Green Version]
- Rolf, J.; Julsing, M.K.; Rosenthal, K.; Lütz, S. A Gram-Scale Limonene Production Process with Engineered Escherichia Coli. Molecules 2020, 25, 1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Zhou, Y.J.; Ji, D.; Zhao, Z.K. An Accurate Method for Estimation of the Intracellular Aqueous Volume of Escherichia Coli Cells. J. Microbiol. Methods 2013, 93, 73–76. [Google Scholar] [CrossRef] [PubMed]



| Protein Synthesis | Biotransformation | ||||||
|---|---|---|---|---|---|---|---|
| E. coli Strain(s) | Medium | Conditions | Buffer | Conditions | Substrate | Product | Reference |
| C41(DE3) | 200 mL TB OD600 0.6: 0.5 mM IPTG 3 mM CuSO4 | 30 °C 140 rpm 21 h | 50 mM KPi buffer 10 + 10 mL pH 7.5 0.1 mM IPTG 20 g L−1 glucose | 70 gCWW L−1 Sequential coculture 24 h + 2/4/6 h 25 °C 140 rpm | Eugenol 1642 mg L−1 | Enantiopure (−)-pinoresinol 219 mg L−1 (24 + 2 h) | [11] |
| C41(DE3) | 50 mL TB OD600 0.6: 0.5 mM IPTG | 25 °C 120 rpm 48 h | 50 mM KPi buffer 10 mL pH 7.5 0.1 mM IPTG 5 mM CuSO4 500 mM glucose | 70 gCWW L−1 Monoculture 20 h 25 °C 200 rpm | Coniferyl alcohol 900 mg L−1 | (−)-Matairesinol (n.d.) | [12] |
| BL21(DE3) + C41(DE3) | 50 mL TB OD600 0.6: 0.5 mM IPTG BL21(DE3): 5 mM CuSO4 | BL21(DE3): 30 °C 140 rpm 21–22 h C41(DE3): 25 °C 120 rpm 48 h | 50 mM KPi buffer 5 + 5 mL pH 7.5 0.1 mM IPTG 500 mM glucose | 70 gCWW L−1 Coculture 20 h 25 °C 200 rpm | Coniferyl alcohol 900 mg L−1 | (−)-Matairesinol (n.q.) | [12] |
| BL21(DE3) + C41(DE3) | 50 mL TB OD600 0.6: 0.5 mM IPTG BL21(DE3): 5 mM CuSO4 | BL21(DE3): 30 °C 140 rpm 21–22 h C41(DE3): 25 °C 120 rpm 48 h | 50 mM KPi buffer 10 + 10 mL pH 7.5 0.1 mM IPTG 500 mM glucose | 70 gCWW L−1 Sequential Coculture 20 h + 4 h 25 °C 200 rpm | Coniferyl alcohol 900 mg L−1 | (−)-Matairesinol 89 mg L−1 | [12] |
| C41(DE3) | 50 mL TB OD600 0.6: 0.5 mM IPTG | 25 °C 120 rpm 48 h | 50 mM KPi buffer 500 µL pH 7.5 0.1 mM IPTG 500 mM glucose | 70 gCWW L−1 Monoculture 24 h 25 °C 1500 rpm | Pinoresinol (77% ee (+)) 72 mg L−1 | (−)-SILR (n.q.) | [9] |
| C41(DE3) | 50 mL TB OD600 0.6: 0.5 mM IPTG | 25 °C 120 rpm 48 h | 50 mM KPi buffer 500 µL pH 7.5 0.1 mM IPTG 500 mM glucose | 70 gCWW L−1 Monoculture 25 °C 1500 rpm | Pinoresinol (77% ee (+)) 72 mg L−1 | (−)-Matairesinol (n.q.) | [9] |
| C41(DE3) | 50 mL TB OD600 0.6: 0.5 mM IPTG 0.5 mM 5-Ala 0.1 mM FeSO4 | 25 °C 120 rpm 48 h | 50 mM KPi buffer 500 µL pH 7.5 0.1 mM IPTG 500 mM glucose | 70 gCWW L−1 Monoculture 24 h 25 °C 1500 rpm | (+)-Pinoresinol (≥96% ee) 72 mg L−1 | (−)-Pluviatolide (n.q.) | [9] |
| C41(DE3) | 50 mL TB OD600 0.6: 0.5 mM IPTG 0.5 mM 5-Ala 0.1 mM FeSO4 | 25 °C 120 rpm 48 h | 50 mM KPi buffer 10 mL pH 7.5 0.1 mM IPTG 500 mM glucose | 70 gCWW L−1 Monoculture 24 h 25 °C 180–250 rpm | (+)-Pinoresinol 180 mg L−1 | (−)-Pluviatolide 137 mg L−1 (250 rpm) | [9] |
| M15 | 100 mL LB OD600 0.6: 0.01 mM IPTG | 25 °C 130 rpm 9 h | 20 mM Tris buffer LB medium pH 8.0 | 2 × 109/1 × 1010 CFU Monoculture 3 h 22 °C vigorous shaking | (+)-Pinoresinol 18 mg L−1 | (−)-Matairesinol (n.q.) | [13] |
| Cultivation System | Experiment | t (h) | X (gCDW L−1) | cintra (mg L−1) | cextra (mg L−1) | ctotal (mg L−1) | YP/X (mg gCDW−1) | YP/S (mg g−1) | STY (mg L−1 h−1) |
|---|---|---|---|---|---|---|---|---|---|
| Shake flask | w/baffles w/o AF | 5 | 0.9 | 108.8 | 10.9 | 11.01 | 12.4 | 22.1 | 2.2 |
| 24 | 4.5 | 123.2 | 12.3 | 13.4 | 3.0 | 26.8 | 0.6 | ||
| w/baffles w/AF | 5 | 0.8 | 202.5 | 20.3 | 20.6 | 26.4 | 41.1 | 4.1 | |
| 24 | 4.8 | 134.0 | 13.4 | 14.6 | 3.0 | 29.3 | 0.6 | ||
| w/o baffles w/o AF | 5 | 0.8 | 118.6 | 11.9 | 12.0 | 15.7 | 24.1 | 2.4 | |
| 24 | 1.7 | 125.7 | 12.6 | 13.0 | 7.5 | 26.0 | 0.5 | ||
| STR | Setup 1 | 5 | 0.8 | 73.7 | 7.4 | 7.5 | 9.9 | 14.9 | 1.5 |
| 24 | 5.5 | 82.9 | 8.3 | 9.2 | 1.7 | 18.3 | 0.4 | ||
| Setup 2 | 5 | 0.7 | 88.0 | 8.8 | 8.9 | 12.6 | 17.8 | 1.8 | |
| 24 | 5.0 | 82.8 | 8.3 | 9.1 | 1.8 | 18.1 | 0.4 | ||
| F1 | 5 | 0.7 | 130.4 | 13.0 | 13.2 | 18.4 | 26.4 | 2.6 | |
| 24 | 5.2 | 90.6 | 9.1 | 10.0 | 1.9 | 19.9 | 0.4 | ||
| F2 | 5 | 1.2 | n.d. | - | - | - | - | - | |
| 24 | 5.4 | n.d. | - | - | - | - | - | ||
| F3 | 5 | 1.2 | n.d. | - | - | - | - | - | |
| 24 | 5.5 | n.d. | - | - | - | - | - | ||
| F4 | 5 | 1.2 | n.d. | - | - | - | - | - | |
| 24 | 5.4 | n.d. | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steinmann, A.; Finger, M.; Nowacki, C.; Decembrino, D.; Hubmann, G.; Girhard, M.; Urlacher, V.B.; Lütz, S. Heterologous Lignan Production in Stirred-Tank Reactors—Metabolomics-Assisted Bioprocess Development for an In Vivo Enzyme Cascade. Catalysts 2022, 12, 1473. https://doi.org/10.3390/catal12111473
Steinmann A, Finger M, Nowacki C, Decembrino D, Hubmann G, Girhard M, Urlacher VB, Lütz S. Heterologous Lignan Production in Stirred-Tank Reactors—Metabolomics-Assisted Bioprocess Development for an In Vivo Enzyme Cascade. Catalysts. 2022; 12(11):1473. https://doi.org/10.3390/catal12111473
Chicago/Turabian StyleSteinmann, Andrea, Maurice Finger, Christian Nowacki, Davide Decembrino, Georg Hubmann, Marco Girhard, Vlada B. Urlacher, and Stephan Lütz. 2022. "Heterologous Lignan Production in Stirred-Tank Reactors—Metabolomics-Assisted Bioprocess Development for an In Vivo Enzyme Cascade" Catalysts 12, no. 11: 1473. https://doi.org/10.3390/catal12111473