Particle Recognition on Transmission Electron Microscopy Images Using Computer Vision and Deep Learning for Catalytic Applications
Abstract
:1. Introduction
2. Results and Discussion
2.1. Training
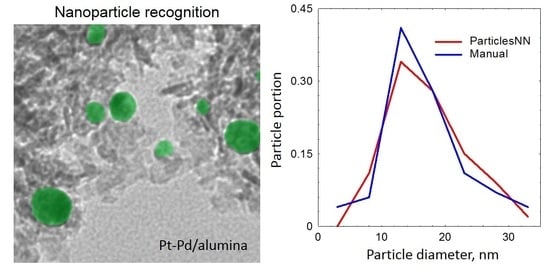
2.2. Comparison with Manual Analysis
3. Materials and Methods
3.1. TEM Data
3.2. Dataset
3.3. Neural Networks
3.4. Evaluation
3.5. Web Service
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nartova, A.V.; Kovtunova, L.K.; Khudorozhkov, A.K.; Shefer, K.I.; Shterk, G.V.; Kvon, R.I.; Bukhtiyarov, V.I. Influence of preparation conditions on catalytic activity and stability of platinum on alumina catalysts in methane oxidation. Appl. Catal. A Gen. 2018, 566, 174–180. [Google Scholar] [CrossRef]
- Hansen, T.W.; Delariva, A.T.; Challa, S.R.; Datye, A.K. Sintering of Catalytic Nanoparticles: Particle Migration or Ostwald Ripening? Acc. Chem. Res. 2013, 46, 1720–1730. [Google Scholar] [CrossRef]
- Akita, T.; Kohyama, M.; Haruta, A. Electron Microscopy Study of Gold Nanoparticles Deposited on Transition Metal Oxides. Acc. Chem. Res. 2013, 46, 1773–1782. [Google Scholar] [CrossRef] [PubMed]
- Hayden, B.E. Particle Size and Support Effects in Electrocatalysis. Acc. Chem. Res. 2013, 46, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, G.J.; Kiely, C.J. Strategies for the Synthesis of Supported Gold Palladium Nanoparticles with Controlled Morphology and Composition. Acc. Chem. Res. 2013, 46, 1759–1772. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Elam, J.W.; Stair, P. Synthesis and Stabilization of Supported Metal Catalysts by Atomic Layer Deposition. Acc. Chem. Res. 2013, 46, 1806–1815. [Google Scholar] [CrossRef]
- Okunev, A.G.; Mashukov, M.Y.; Nartova, A.V.; Matveev, A.V. Nanoparticle Recognition on Scanning Probe Microscopy Images Using Computer Vision and Deep Learning. Nanomaterials 2020, 10, 1285. [Google Scholar] [CrossRef] [PubMed]
- Russ, J.C. Computer—Assisted Microscopy: The Measurement and Analysis of Images; Plenum Press: New York, NY, USA, 1990; p. 451. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. Imagenet Classification with Deep Convolutional Neural Networks. In Proceedings of the Annual Conference on Neural Information Processing Systems (NIPS 2012), Lake Tahoe, NV, USA, 3–8 December 2012; pp. 1–9. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. SSD: Single Shot MultiboxDetector. In Computer Vision—ECCV 2016; Lecture Notes in Computer, Science; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer: Cham, Switzerland, 2016; Volume 9905, pp. 21–37. [Google Scholar]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the 2017 IEEE InternationalConference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Stringer, C.; Michaelos, M.; Pachitariu, M. Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 2021, 18, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Caicedo, J.; Goodman, A.; Karhohs, K.; Cimini, B.; Ackerman, J.; Haghighi, M.; Heng, C.; Becker, T.; Doan, M.; McQuin, C.; et al. Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Wu, P.; Hoeppner, D.J.; Metaxas, D. Pixel-wise neural cell instance segmentation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 373–377. [Google Scholar]
- Fu, G.; Sun, P.; Zhu, W.; Yang, J.; Cao, Y.; Ying-Yang, M.; Cao, Y.A. Deep-learning-based approach for fast and robust steel surface defects classification. Opt. Lasers Eng. 2019, 121, 397–405. [Google Scholar] [CrossRef]
- Poletaev, I.; Tokarev, M.P.; Pervunin, K.S. Bubble Patterns Recognition Using Neural Networks: Application to the Analysis of a Two-phase Bubbly Jet. Int. J. Multiph. Flow 2020, 126, 103194. [Google Scholar] [CrossRef]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.; Karhohs, K.; Doan, M.; Ding, L.; Rafelski, S.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.; Kausler, B.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2010, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Bannon, D.; Moen, E.; Schwartz, M.; Borba, E.; Kudo, T.; Greenwald, N.; Vijayakumar, V.; Chang, B.; Pao, E.; Osterman, E.; et al. DeepCell Kiosk: Scaling deep learning–enabled cellular image analysis with Kubernetes. Nat. Methods 2021, 18, 43–45. [Google Scholar] [CrossRef] [PubMed]
- Okunev, A.G.; Nartova, A.V.; Matveev, A.V. Recognition of Nanoparticles on Scanning Probe Microscopy Images Using Computer Vision and Deep Machine Learning. In Proceedings of the International Multi-Conference on Engineering, Computer and Information Sciences (SIBIRCON), Novosibirsk, Russia, 21–27 October 2019; pp. 940–943. [Google Scholar]
- Zhu, H.; Ge, W.; Liu, Z. Deep Learning-Based Classification of Weld Surface Defects. Appl. Sci. 2019, 9, 3312. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xu, K.; Xu, J. Periodic Surface Defect Detection in Steel Plates Based on Deep Learning. Appl. Sci. 2019, 9, 3127. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.; Zhou, H.; Dong, H. Using Deep Neural Network with Small Dataset to Predict Material Defects. Mater. Des. 2019, 162, 300–310. [Google Scholar] [CrossRef]
- Yang, T.; Xiao, L.; Gong, B.; Huang, L. Surface Defect Recognition of Varistor Based on Deep Convolutional Neural Networks. In Optoelectronic Imaging and Multimedia Technology VI, Proceedings of the SPIE/COS PHOTONICS ASIA, Hangzhou, China, 20–23 October 2019; Dai, Q., Shimura, T., Zheng, Z., Eds.; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 11187, p. 1118718. [Google Scholar]
- Ziatdinov, M.; Dyck, O.; Maksov, A.; Li, X.; Sang, X.; Xiao, K.; Unocic, R.; Vasudevan, R.; Jesse, S.; Kalinin, S.V. Deep Learning of Atomically Resolved Scanning Transmission Electron Microscopy Images: Chemical Identification and Tracking Local Transformations. ACS Nano 2017, 11, 12742–12752. [Google Scholar] [CrossRef] [Green Version]
- Modarres, M.H.; Aversa, R.; Cozzini, S.; Ciancio, R.; Leto, A.; Brandino, G.P. Neural Network for Nanoscience Scanning Electron Microscope Image Recognition. Sci. Rep. 2017, 7, 13282. [Google Scholar] [CrossRef] [Green Version]
- Qu, E.Z.; Jimenez, A.M.; Kumar, S.K.; Zhang, K. Quantifying Nanoparticle Assembly States in a Polymer Matrix Through Deep Learning. Macromolecules 2021, 54, 3034–3040. [Google Scholar] [CrossRef]
- Monchot, P.; Coquelin, L.; Guerroudj, K.; Feltin, N.; Delvallée, A.; Crouzier, L.; Fischer, N. Deep Learning Based Instance Segmentation of TitaniumDioxide Particles in the Form of Agglomerates in ScanningElectron Microscopy. Nanomaterials 2021, 11, 968. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Huang, J.Z.; Li, X.; Ding, Y. Robust Nanoparticles Detection from Noisy Background by Fusing Complementary Image Information. IEEE Trans. Image Process. 2016, 25, 5713–5726. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Ding, Y. Automating material image analysis for material discovery. MRS Commun. 2019, 9, 545–555. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Chen, H.; Wang, H.; Wei, D.; Wu, Y.; Fan, K. Detection of Nano-particles Based on Machine Vision. In Proceedings of the 2019 IEEE International Conference on Manipulation, Manufacturing and Measurement on the Nanoscale (3M-NANO), Zhenjiang, China, 4–8 August 2019; pp. 189–192. [Google Scholar]
- Oktay, A.B.; Gurses, A. Automatic detection, localization and segmentation of nano-particles with deep learning in microscopy images. Micron 2019, 120, 113–119. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Q.; Xiao, Z.; Wu, J.; Liu, Y. Spherical Nanoparticle Parameter Measurement Method based on Mask R-CNN Segmentation and Edge Fitting. In Proceedings of the 8th International Conference on Computing and Pattern Recognition (ICCPR’19), Beijing, China, 23–25 October 2019; pp. 205–212. [Google Scholar]
- Horcas, I.; Fernandez, R.; Gomez-Rodriguez, J.M.; Colchero, J.; Gomez-Herrero, J.; Baro, A.M. WSXM: A Software for Scanning Probe Microscopy and a Tool for Nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef]
- Schoonjans, F. Digimizer Manual: Easy-to-Use Image Analysis Software; 2019; 107p, ISBN 9781706417149. [Google Scholar]
- Wada, K. Labelme: Image Polygonal Annotation with Python. 2016. Available online: https://github.com/wkentaro/labelme (accessed on 1 June 2020).
- Lin, T.-Y.; Maire, M.; Belongie, S.; Bourdev, L.; Girshick, R.; Hays, J.; Perona, P.; Ramanan, D.; Zitnick, C.L.; Dollár, P. Microsoft COCO: Common Objects in Context. Lect. Notes Comput. Sci. 2014, 8693, 740–755. [Google Scholar]
- Cai, Z.; Vasconcelos, N. Cascade R-CNN: Delving into High Quality Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 6154–6162. [Google Scholar]
- Chen, K.; Wang, J.; Pang, J.; Cao, Y.; Xiong, Y.; Li, X.; Sun, S.; Feng, W.; Liu, Z.; Xu, J.; et al. MMDetection: Open MMLab Detection Toolbox and Benchmark. arXiv 2019, arXiv:1906.07155. [Google Scholar]
- Everingham, M.; Van Gool, L.; Williams, C.K.I.; Winn, J.; Zisserman, A. The Pascal Visual Object Classes (voc) Challenge. Int. J. Comput. Vis. 2010, 88, 303–338. [Google Scholar] [CrossRef] [Green Version]
- COCO API—Dataset. Available online: https://github.com/cocodataset/cocoapi (accessed on 1 June 2020).







| Number of Images | ‘Face’ | ‘Bottom’ | |
|---|---|---|---|
| Training | 26 | 1030 | 235 |
| Test | 5 | 130 | 33 |
| Total | 31 | 1160 | 268 |
| Particle Count | ||||||||
|---|---|---|---|---|---|---|---|---|
| Image No | ‘Face’ GT | ‘Bottom’ GT | ‘Face’ FN | ‘Bottom’ FN | ‘Face’ TP | ‘Face’ FP | ‘Bottom’ TP | ‘Bottom’ FP |
| 1 | 42 | 4 | 22 | 10 | 43 | 1 | 3 | 6 |
| 2 | 6 | 1 | 1 | 0 | 6 | 3 | 1 | 2 |
| 3 | 8 | 5 | 1 | 1 | 8 | 8 | 5 | 8 |
| 4 | 19 | 3 | 5 | 19 | 4 | 3 | 4 | |
| 5 | 16 | 0 | 1 | 0 | 16 | 1 | 0 | 5 |
| Total | 91 | 13 | 25 | 16 | 92 | 17 | 12 | 25 |
| Precision | Recall | ||
|---|---|---|---|
| ‘Bottom’ | ‘Face’ | ‘Bottom’ | ‘Face’ |
| 0.32 | 0.84 | 0.43 | 0.79 |
| Total | 0.71 | Total | 0.72 |
| Method of Particle Size Determining | Number of Particles | Mean Particle Size, nm | Standard Error Of Mean, nm |
|---|---|---|---|
| Manually | 54 | 17.2 | 1.8 |
| ParticlesNN | 53 * | 17.6 | 1.6 |
| Particle Size (pix) | ||
|---|---|---|
| Particle 1 | Particle 2 | |
| ParticlesNN, d | 74.5 | 79.8 |
| ImagJ | ||
| D1 | 84.4 | 94.9 |
| D2 | 62.9 | 66.7 |
| Dmean | 73.7 | 80.8 |
| D3 | 74.2 | 79.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nartova, A.V.; Mashukov, M.Y.; Astakhov, R.R.; Kudinov, V.Y.; Matveev, A.V.; Okunev, A.G. Particle Recognition on Transmission Electron Microscopy Images Using Computer Vision and Deep Learning for Catalytic Applications. Catalysts 2022, 12, 135. https://doi.org/10.3390/catal12020135
Nartova AV, Mashukov MY, Astakhov RR, Kudinov VY, Matveev AV, Okunev AG. Particle Recognition on Transmission Electron Microscopy Images Using Computer Vision and Deep Learning for Catalytic Applications. Catalysts. 2022; 12(2):135. https://doi.org/10.3390/catal12020135
Chicago/Turabian StyleNartova, Anna V., Mikhail Yu. Mashukov, Ruslan R. Astakhov, Vitalii Yu. Kudinov, Andrey V. Matveev, and Alexey G. Okunev. 2022. "Particle Recognition on Transmission Electron Microscopy Images Using Computer Vision and Deep Learning for Catalytic Applications" Catalysts 12, no. 2: 135. https://doi.org/10.3390/catal12020135