Effects of Soil Surface Chemistry on Adsorption and Activity of Urease from a Crude Protein Extract: Implications for Biocementation Applications
Abstract
:1. Introduction
2. Results and Discussion
2.1. Surface Coverage
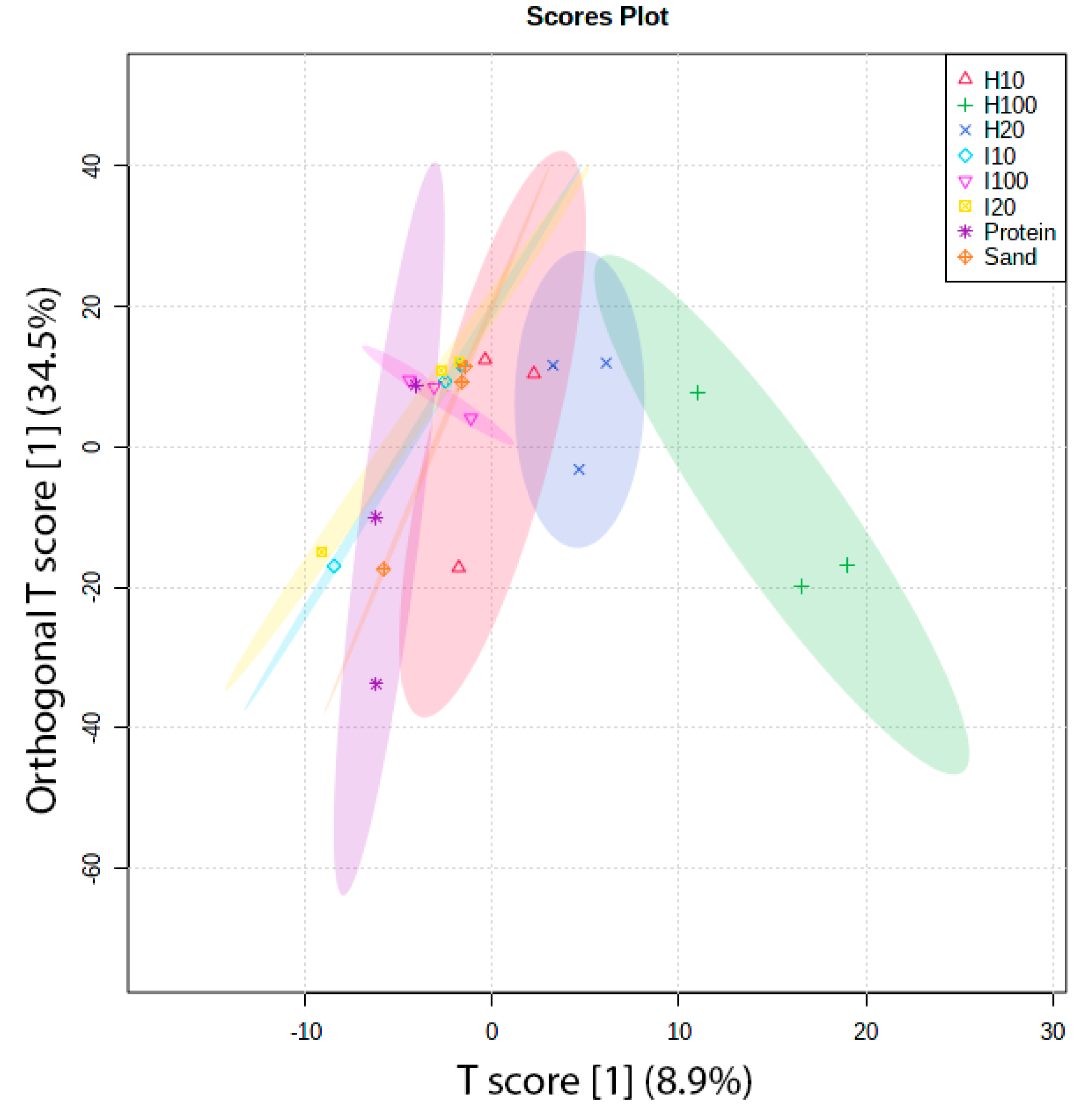
2.2. Protein Adsorption
2.3. Enzymatic Activity of Adsorbed Urease
3. Materials and Methods
3.1. Materials Used
3.2. Total Protein Extract
3.3. Soil Pretreatments
3.4. Soil Characterization
3.5. Adsorption Experiments
3.6. Urease Activity Measurements
3.7. Proteomics Analysis
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meyer, F.D.; Bang, S.; Min, S.; Stetler, L.D. Microbiologically-Induced Soil Stabilization: Application of Sporosarcina pasteurii for Fugitive Dust Control. In Proceedings of the Geo-Frontiers 2011: Advances in Geotechnical Engineering, Dallas, TX, USA, 13–16 March 2011; pp. 4002–4011. [Google Scholar] [CrossRef] [Green Version]
- Dhami, N.K.; Ereddy, M.S.; Mukherjee, A. Biomineralization of calcium carbonates and their engineered applications: A review. Front. Microbiol. 2013, 4, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whiffin, V.S.; van Paassen, L.; Harkes, M.P. Microbial Carbonate Precipitation as a Soil Improvement Technique. Geomicrobiol. J. 2007, 24, 417–423. [Google Scholar] [CrossRef]
- Van Paassen, L.A.; Ghose, R.; Van Der Linden, T.J.M.; Van Der Star, W.R.L.; Van Loosdrecht, M.C.M. Quantifying Biomediated Ground Improvement by Ureolysis: Large-Scale Biogrout Experiment. J. Geotech. Geoenviron. Eng. 2010, 136, 1721–1728. [Google Scholar] [CrossRef]
- DeJong, J.T.; Mortensen, B.M.; Martinez, B.C.; Nelson, D.C. Bio-mediated soil improvement. Ecol. Eng. 2010, 36, 197–210. [Google Scholar] [CrossRef]
- Cetin, B.; Aydilek, A.H.; Li, L. Trace Metal Leaching from Embankment Soils Amended with High-Carbon Fly Ash. J. Geotech. Geoenviron. Eng. 2014, 140, 1–13. [Google Scholar] [CrossRef]
- Komonweeraket, K.; Cetin, B.; Aydilek, A.H.; Benson, C.H.; Edil, T.B. Geochemical Analysis of Leached Elements from Fly Ash Stabilized Soils. J. Geotech. Geoenviron. Eng. 2015, 141, 04015012. [Google Scholar] [CrossRef]
- Komonweeraket, K.; Cetin, B.; Aydilek, A.H.; Benson, C.H.; Edil, T.B. Effects of pH on the leaching mechanisms of elements from fly ash mixed soils. Fuel 2015, 140, 788–802. [Google Scholar] [CrossRef]
- Qin, Y.; Cabral, J.M. Review Properties and Applications of Urease. Biocatal. Biotransform. 2002, 20, 1–14. [Google Scholar] [CrossRef]
- Vilar, R.P.; Ikuma, K. Adsorption of urease as part of a complex protein mixture onto soil and its implications for enzymatic activity. Biochem. Eng. J. 2021, 171, 108026. [Google Scholar] [CrossRef]
- Carmona, J.P.; Oliveira, P.V.; Lemos, L. Biostabilization of a Sandy Soil Using Enzymatic Calcium Carbonate Precipitation. Procedia Eng. 2016, 143, 1301–1308. [Google Scholar] [CrossRef] [Green Version]
- Kavazanjian, E.; Hamdan, N. Enzyme induced carbonate precipitation (eicp) columns for ground improvement. In Proceedings of the IFCEE 2015, Antonio, TX, USA, 17–21 March 2015; pp. 2252–2261. [Google Scholar] [CrossRef]
- Neupane, D.; Yasuhara, H.; Kinoshita, N.; Unno, T. Applicability of Enzymatic Calcium Carbonate Precipitation as a Soil-Strengthening Technique. J. Geotech. Geoenviron. Eng. 2013, 139, 2201–2211. [Google Scholar] [CrossRef]
- Yasuhara, H.; Neupane, D.; Hayashi, K.; Okamura, M. Experiments and predictions of physical properties of sand cemented by enzymatically-induced carbonate precipitation. Soils Found. 2012, 52, 539–549. [Google Scholar] [CrossRef] [Green Version]
- Larson, A.D.; Kalion, R.E. Purification and properties of bacterial urease. J. Bacteriol. 1954, 68, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, T.; Alleman, J.; Cetin, B.; Ikuma, K.; Choi, S.-G. Sand and silty-sand soil stabilization using bacterial enzyme–induced calcite precipitation (BEICP). Can. Geotech. J. 2019, 56, 808–822. [Google Scholar] [CrossRef]
- Hoang, T.; Alleman, J.; Cetin, B.; Choi, S.-G. Engineering Properties of Biocementation Coarse- and Fine-Grained Sand Catalyzed By Bacterial Cells and Bacterial Enzyme. J. Mater. Civ. Eng. 2020, 32, 04020030. [Google Scholar] [CrossRef]
- Fusi, P.; Ristori, G.; Calamai, L.; Stotzky, G. Adsorption and binding of protein on “clean” (homoionic) and “dirty” (coated with Fe oxyhydroxides) montmorillonite, illite and kaolinite. Soil Biol. Biochem. 1989, 21, 911–920. [Google Scholar] [CrossRef]
- Gianfreda, L.; Rao, M.A.; Violante, A. Invertase β-fructosidase)- Effects of montmorillonite, AL-hydroxide and AL(OH)x-montmorillonite on activity and kinetics properties. Soil Biol. Biochem. 1991, 23, 581–587. [Google Scholar] [CrossRef]
- Absolom, D.R.; Zingg, W.; Neumann, A.W. Protein adsorption to polymer particles: Role of surface properties. J. Biomed. Mater. Res. 1987, 21, 161–171. [Google Scholar] [CrossRef]
- Azioune, A.; Chehimi, M.M.; Miksa, B.; Basinska, T.; Slomkowski, S. Hydrophobic Protein−Polypyrrole Interactions: The Role of van der Waals and Lewis Acid−Base Forces As Determined by Contact Angle Measurements. Langmuir 2002, 18, 1150–1156. [Google Scholar] [CrossRef]
- Huang, Q.; Jiang, M.; Li, X. Adsorption and Properties of Urease Immobilized on Several Iron and Aluminum Oxides (Hydroxides) and Kaolinite. In Effect of Mineral-Organic-Microorganism Interactions on Soil and Freshwater Environments; Springer: Berlin/Heidelberg, Germany, 1999; pp. 167–168. [Google Scholar]
- Kim, J.; Somorjai, G.A. Molecular Packing of Lysozyme, Fibrinogen, and Bovine Serum Albumin on Hydrophilic and Hydrophobic Surfaces Studied by Infrared−Visible Sum Frequency Generation and Fluorescence Microscopy. J. Am. Chem. Soc. 2003, 125, 3150–3158. [Google Scholar] [CrossRef]
- Tilton, R.; Robertson, C.R.; Gast, A.P. Manipulation of hydrophobic interactions in protein adsorption. Langmuir 1991, 7, 2710–2718. [Google Scholar] [CrossRef]
- Tangpasuthadol, V.; Pongchaisirikul, N.; Hoven, V.P. Surface modification of chitosan films.: Effects of hydrophobicity on protein adsorption. Carbohydr. Res. 2003, 338, 937–942. [Google Scholar] [CrossRef]
- Wang, W.; Chen, L.; Zhang, Y.; Liu, G. Adsorption of bovine serum albumin and urease by biochar. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2017; Volume 61, pp. 8–13. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Su, T.; Thirtle, P.; Thomas, R.; Rennie, A.; Cubitt, R. The Denaturation of Lysozyme Layers Adsorbed at the Hydrophobic Solid/Liquid Surface Studied by Neutron Reflection. J. Colloid Interface Sci. 1998, 206, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Onweremadu, E.U. Hydrophobicity of soils formed over different lithologies. Malasyan J. Soil Sci. 2008, 12, 19–30. [Google Scholar]
- Hallett, P.; Baumgartl, T.; Young, I. Subcritical Water Repellency of Aggregates from a Range of Soil Management Practices. Soil Sci. Soc. Am. J. 2001, 65, 184–190. [Google Scholar] [CrossRef]
- Dekker, L.W.; Ritsema, C.J. How water moves in a water repellent sandy soil: 1. Potential and actual water repellency. Water Resour. Res. 1994, 30, 2507–2517. [Google Scholar] [CrossRef]
- Dekker, L.W.; Doerr, S.H.; Oostindie, K.; Ziogas, A.K.; Ritsema, C.J. Water Repellency and Critical Soil Water Content in a Dune Sand. Soil Sci. Soc. Am. J. 2001, 65, 1667–1674. [Google Scholar] [CrossRef]
- Jaramillo, D.F.; Dekker, L.W.; Ritsema, C.J.; Hendrickx, J.M.H. Soil water repellency in arid and humid climates. Soil Water Repel. Occur. Conseq. Amelior. 2003, 232, 93–98. [Google Scholar] [CrossRef]
- Roberts, F.; Carbon, B. Water repellence in sandy soils of South-Western Australia. II. Some chemical characteristics of the hydrophobic skins. Soil Res. 1972, 10, 35–42. [Google Scholar] [CrossRef]
- Jaramillo, D.F.; Herrón, F.E. Evaluacion de la repelencia al agua de algunos andisols de antioquia bajo cobertura de Pinus patula. Acta Agron. 1991, 4, 79–85. [Google Scholar]
- Debano, L.F.; Krammes, J.S. Water repellent soils and their relation to wildfire temperatures. Int. Assoc. Sci. Hydrol. Bull. 1966, 11, 14–19. [Google Scholar] [CrossRef]
- Doerr, S.; Shakesby, R.; Walsh, R. Soil water repellency: Its causes, characteristics and hydro-geomorphological significance. Earth-Sci. Rev. 2000, 51, 33–65. [Google Scholar] [CrossRef]
- McHale, G.; Newton, M.I.; Shirtcliffe, N.J. Water-repellent soil and its relationship to granularity, surface roughness and hydrophobicity: A materials science view. Eur. J. Soil Sci. 2005, 56, 445–452. [Google Scholar] [CrossRef] [Green Version]
- Osborn, J.F.; Pelishek, R.E.; Krammes, J.S.; Letey, J. Soil Wettability as a Factor in Erodibility. Soil Sci. Soc. Am. J. 1964, 28, 294–295. [Google Scholar] [CrossRef]
- Jungerius, P.; van der Meulen, F. Erosion processes in a dune landscape along the Dutch coast. Catena 1988, 15, 217–228. [Google Scholar] [CrossRef]
- Lowe, M.-A.; McGrath, G.; Leopold, M. The Impact of Soil Water Repellency and Slope upon Runoff and Erosion. Soil Tillage Res. 2021, 205, 104756. [Google Scholar] [CrossRef]
- Almajed, A.; Lemboye, K.; Arab, M.G.; Alnuaim, A. Mitigating wind erosion of sand using biopolymer-assisted EICP technique. Soils Found. 2020, 60, 356–371. [Google Scholar] [CrossRef]
- Chae, S.H.; Chung, H.; Nam, K. Evaluation of microbially Induced calcite precipitation (MICP) methods on different soil types for wind erosion control. Environ. Eng. Res. 2020, 26, 123–128. [Google Scholar] [CrossRef]
- Jiang, X.; Rutherford, C.; Cetin, B.; Ikuma, K. Reduction of Water Erosion Using Bacterial Enzyme Induced Calcite Precipitation (BEICP) for Sandy Soil. In Proceedings of the Geo-Congress 2020 Biogeotechnics, Reston, VA, USA; 2020; pp. 104–110. [Google Scholar] [CrossRef]
- Jiang, N.-J.; Soga, K. The applicability of microbially induced calcite precipitation (MICP) for internal erosion control in gravel–sand mixtures. Géotechnique 2017, 67, 42–53. [Google Scholar] [CrossRef] [Green Version]
- Gianfreda, L.; Rao, M.A.; Violante, A. Adsorption, activity and kinetic properties of urease on montmorillonite, aluminium hydroxide and AL(OH)x-montmorillonite complexes. Soil Biol. Biochem. 1992, 24, 51–58. [Google Scholar] [CrossRef]
- Artioli, Y. Adsorption. In Encyclopedia of Ecology; Elsevier: Amsterdam, The Netherlands, 2008; pp. 60–65. [Google Scholar]
- Hoeve, C.A.J.; DiMarzio, E.A.; Peyser, P. Adsorption of Polymer Molecules at Low Surface Coverage. J. Chem. Phys. 1965, 42, 2558–2563. [Google Scholar] [CrossRef]
- Larsericsdotter, H.; Oscarsson, S.; Buijs, J. Thermodynamic Analysis of Proteins Adsorbed on Silica Particles: Electrostatic Effects. J. Colloid Interface Sci. 2001, 237, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Norde, W.; Favier, J.P. Structure of adsorbed and desorbed proteins. Colloids Surfaces 1992, 64, 87–93. [Google Scholar] [CrossRef]
- Norde, W.; Giacomelli, C. BSA structural changes during homomolecular exchange between the adsorbed and the dissolved states. J. Biotechnol. 2000, 79, 259–268. [Google Scholar] [CrossRef]
- Norde, W.; Giacomelli, C.E. Conformational changes in proteins at interfaces: From solution to the interface, and back. Macromol. Symp. 1999, 145, 125–136. [Google Scholar] [CrossRef]
- Gianfreda, L.; Rao, M.A.; Violante, A. Formation and Activity of Urease-Tannate Complexes Affected by Aluminum, Iron, and Manganese. Soil Sci. Soc. Am. J. 1995, 59, 805–810. [Google Scholar] [CrossRef]
- Gianfreda, L.; De Cristofaro, A.; Rao, M.A.; Violante, A. Kinetic Behavior of Synthetic Organo-and Organo-Mineral-Urease Complexes. Soil Sci. Soc. Am. J. 1995, 59, 811–815. [Google Scholar] [CrossRef]
- He, S.; Feng, Y.; Ren, H.; Zhang, Y.; Gu, N.; Lin, X. The impact of iron oxide magnetic nanoparticles on the soil bacterial community. J. Soils Sediments 2011, 11, 1408–1417. [Google Scholar] [CrossRef]
- Rabe, M.; Verdes, D.; Seeger, S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011, 162, 87–106. [Google Scholar] [CrossRef] [Green Version]
- Bordbar, A.-K.; Sohrabi, N.; Hojjati, E. The estimation of the hydrophobic and electrostatic contributions to the free energy change upon cationic surfactants binding to Jack bean urease. Colloids Surf. B Biointerfaces 2004, 39, 171–175. [Google Scholar] [CrossRef]
- Tipping, E.; Jones, M.N.; Skinner, H.A. Enthalpy of interaction between some globular proteins and sodium n-dodecyl sulphate in aqueous solution. J. Chem. Soc. Faraday Trans. 1 Phys. Chem. Condens. Phases 1974, 70, 1306–1315. [Google Scholar] [CrossRef]
- Bordbar, A.-K. Thermodynamic Analysis for Cationic Surfactants Binding to Bovine Serum Albumin. J. Phys. Theor. Chem. 2006, 2, 197–204. [Google Scholar] [CrossRef]
- Takishima, K.; Suga, T.; Mamiya, G. The structure of jack bean urease. The complete amino acid sequence, limited proteolysis and reactive cysteine residues. JBIC J. Biol. Inorg. Chem. 1988, 175, 151–157. [Google Scholar] [CrossRef]
- Kuscu, I.S.K.; Cetin, M.; Yigit, N.; Savaci, G.; Sevik, H. Relationship between Enzyme Activity (Urease-Catalase) and Nutrient Element in Soil Use. Pol. J. Environ. Stud. 2018, 27, 2107–2112. [Google Scholar] [CrossRef]
- Christians, S.; Kaltwasser, H. Nickel-content of urease from Bacillus pasteurii. Arch. Microbiol. 1986, 145, 51–55. [Google Scholar] [CrossRef]
- Kuo, J.F.; Angeles, L. Further Investigation of the Surface Charge Properties of Oxide Surfaces in Oil-Bearing Sands and Sandstones. J. Colloid. 1987, 115, 9–16. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr. Protoc. Bioinform. 2011, 34. [Google Scholar] [CrossRef]
- Garwood, G.; Mortland, M.; Pinnavaia, T. Immobilization of glucose oxidase on montmorillonite clay: Hydrophobic and ionic modes of binding. J. Mol. Catal. 1983, 22, 153–163. [Google Scholar] [CrossRef]
- Quiquampoix, H.; Abadie, J.; Baron, M.H.; Leprince, F.; Matumoto-Pintro, P.T.; Ratcliffe, R.G.; Staunton, S. Mechanisms and Consequences of Protein Adsorption on Soil Mineral Surfaces. In Proteins at Interfaces II; ACS Symposium Series; American Chemical Society, ACS: Washington, DC, USA, 1995; pp. 321–333. Available online: https://pubs.acs.org/doi/abs/10.1021/bk-1995-0602.ch023 (accessed on 9 January 2022).
- Lahari, C.; Jasti, L.S.; Fadnavis, N.W.; Sontakke, K.; Ingavle, G.; Deokar, S.; Ponrathnam, S. Adsorption Induced Enzyme Denaturation: The Role of Polymer Hydrophobicity in Adsorption and Denaturation of α-Chymotrypsin on Allyl Glycidyl Ether (AGE)-Ethylene Glycol Dimethacrylate (EGDM) Copolymers. Langmuir 2010, 26, 1096–1106. [Google Scholar] [CrossRef]
- Hirai, M.; Kawai-Hirai, R.; Hirai, T.; Ueki, T. Structural change of jack bean urease induced by addition surfactants studied with synchrotron-radiation small-angle X-ray scattering. JBIC J. Biol. Inorg. Chem. 1993, 215, 55–61. [Google Scholar] [CrossRef]
- Contaxis, C.; Reithel, F. Studies on Protein Multimers: II. A Study of the Mechanism OF Urease Dissociation in 1, 2-Propanediol: Comparative Studies with Ethylene Glycol and Glycerol. J. Biol. Chem. 1971, 246, 677–685. [Google Scholar] [CrossRef]
- dos Santos, E.A.; Farina, M.; Soares, G.A.; Anselme, K. Surface energy of hydroxyapatite and β-tricalcium phosphate ceramics driving serum protein adsorption and osteoblast adhesion. J. Mater. Sci. Mater. Med. 2008, 19, 2307–2316. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Marchal, T.G.; Rouxhet, P.G. Influence of Substratum Surface Properties on the Organization of Adsorbed Collagen Films: In Situ Characterization by Atomic Force Microscopy. Langmuir 1999, 15, 2871–2878. [Google Scholar] [CrossRef]
- Benjamin, M.M.; Sletten, R.S.; Bailey, R.P.; Bennett, T. Sorption and filtration of metals using iron-oxide-coated sand. Water Res. 1996, 30, 2609–2620. [Google Scholar] [CrossRef]
- Olorunfemi, I. Soil Hydrophobicity: An Overview. J. Sci. Res. Rep. 2014, 3, 1003–1037. [Google Scholar] [CrossRef]
- Rahmatullah, M.; Boyde, T. Improvements in the determination of urea using diacetyl monoxime; methods with and without deproteinisation. Clin. Chim. Acta 1980, 107, 3–9. [Google Scholar] [CrossRef]
- Kandhavelu, J.; Demonte, N.L.; Namperumalsamy, V.P.; Prajna, L.; Thangavel, C.; Jayapal, J.M.; Kuppamuthu, D. Data set of Aspergillus flavus induced alterations in tear proteome: Understanding the pathogen-induced host response to fungal infection. Data Brief 2016, 9, 888–894. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto Vilar, R.; Ikuma, K. Effects of Soil Surface Chemistry on Adsorption and Activity of Urease from a Crude Protein Extract: Implications for Biocementation Applications. Catalysts 2022, 12, 230. https://doi.org/10.3390/catal12020230
Pinto Vilar R, Ikuma K. Effects of Soil Surface Chemistry on Adsorption and Activity of Urease from a Crude Protein Extract: Implications for Biocementation Applications. Catalysts. 2022; 12(2):230. https://doi.org/10.3390/catal12020230
Chicago/Turabian StylePinto Vilar, Rayla, and Kaoru Ikuma. 2022. "Effects of Soil Surface Chemistry on Adsorption and Activity of Urease from a Crude Protein Extract: Implications for Biocementation Applications" Catalysts 12, no. 2: 230. https://doi.org/10.3390/catal12020230
APA StylePinto Vilar, R., & Ikuma, K. (2022). Effects of Soil Surface Chemistry on Adsorption and Activity of Urease from a Crude Protein Extract: Implications for Biocementation Applications. Catalysts, 12(2), 230. https://doi.org/10.3390/catal12020230





