Metagenomic Approaches as a Tool to Unravel Promising Biocatalysts from Natural Resources: Soil and Water
Abstract
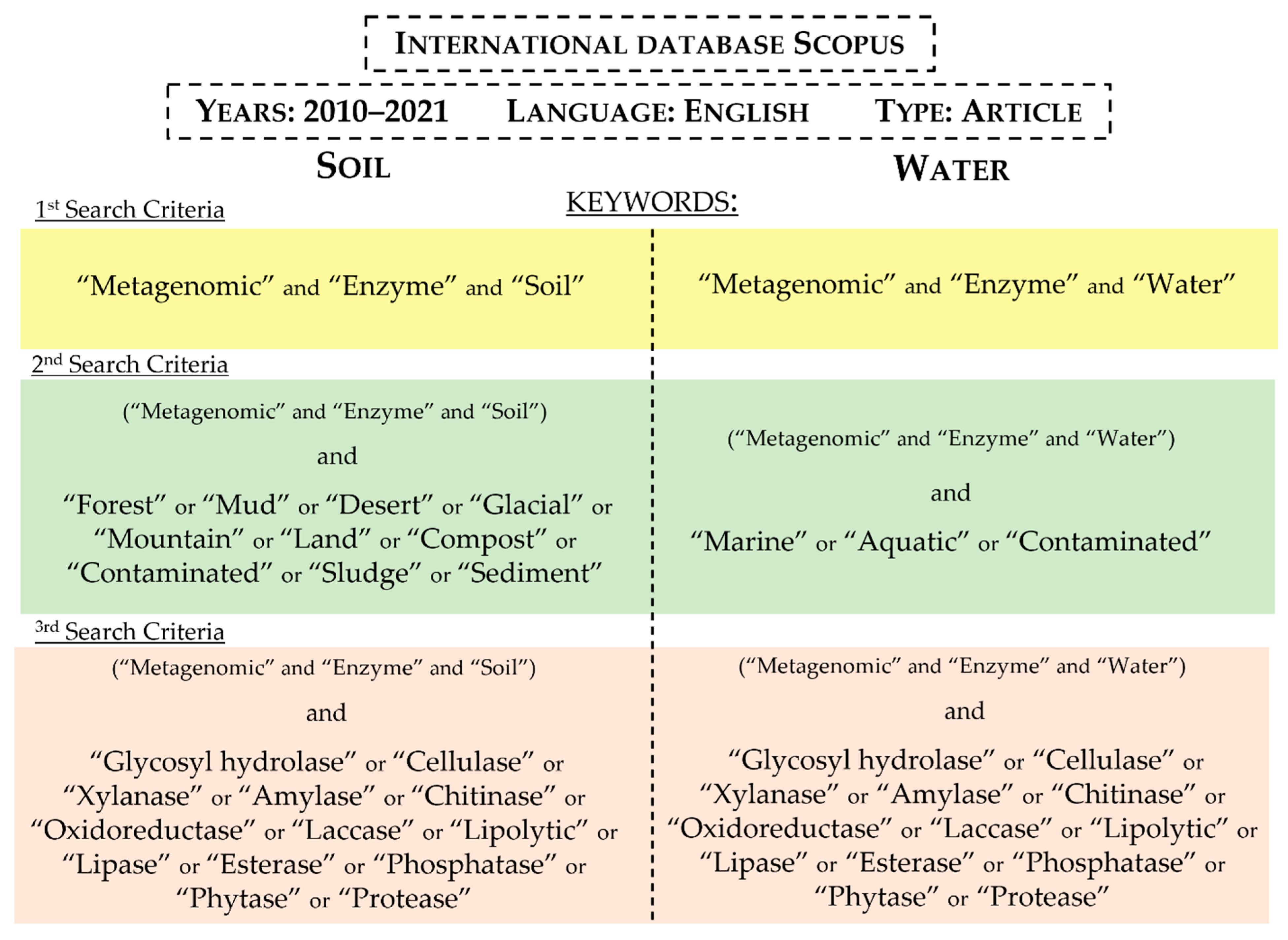
:1. Introduction
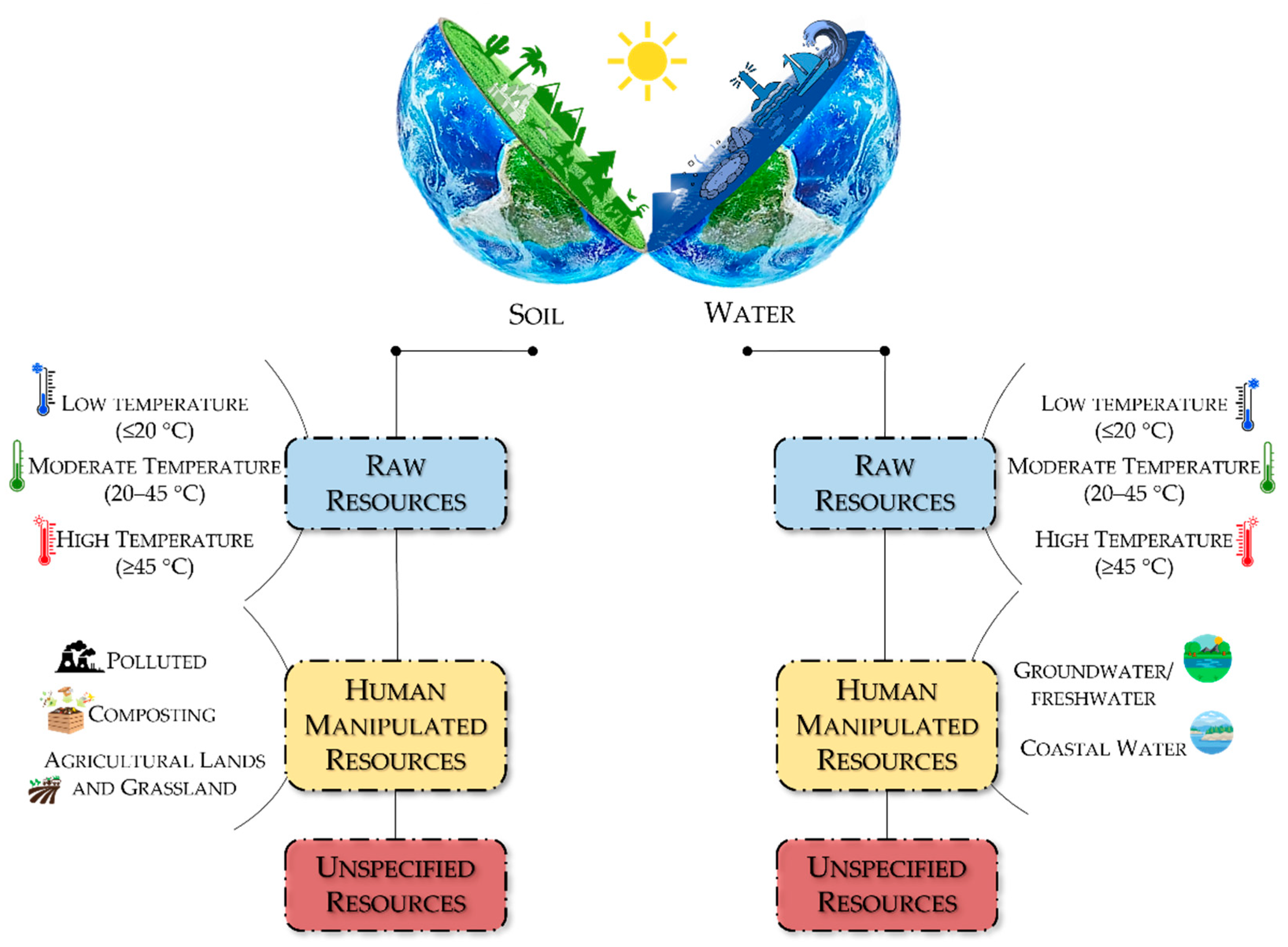
2. Soil
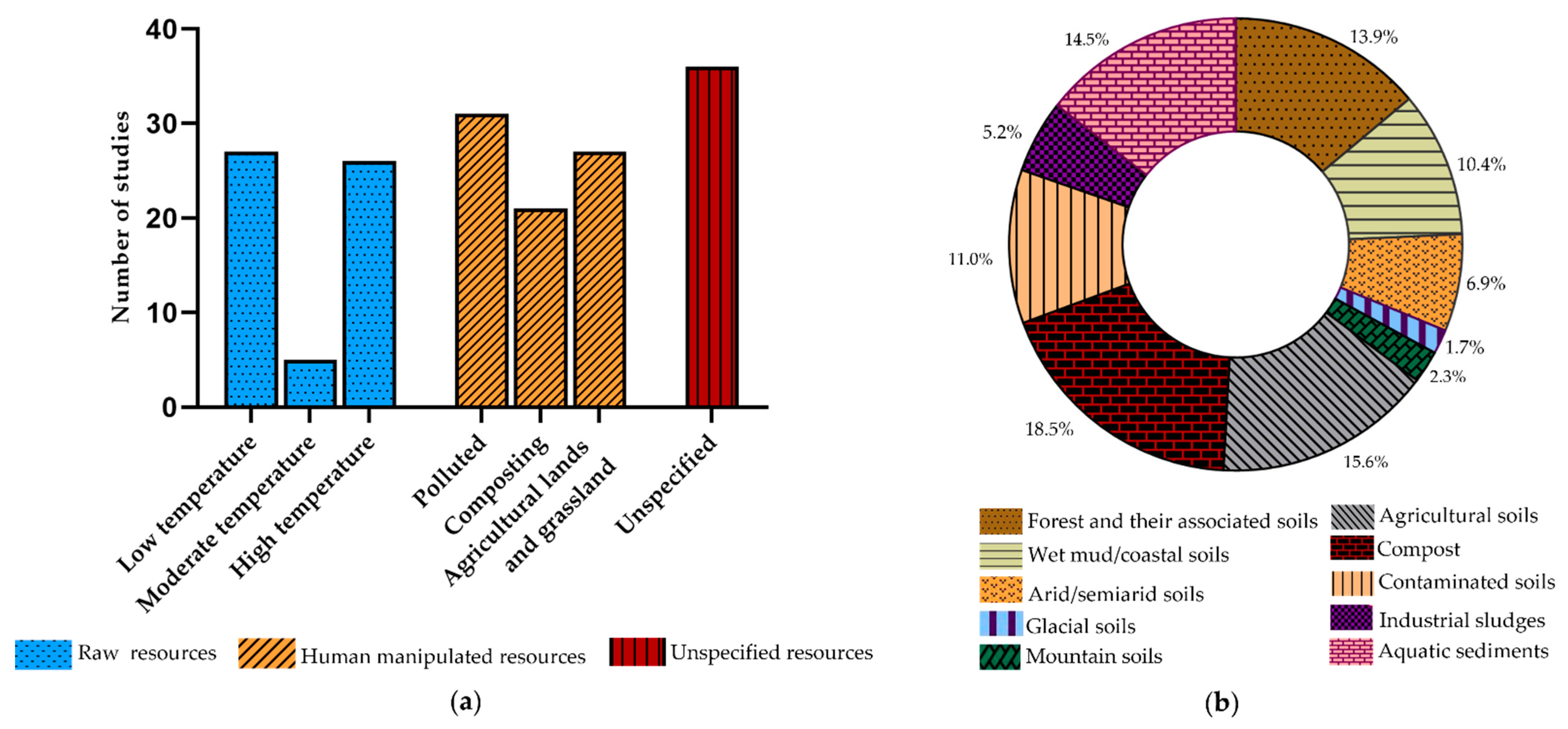
2.1. Raw Resources
2.1.1. Low-Temperature Environments
2.1.2. Moderate Temperature Environments
2.1.3. High-Temperature Environments
2.2. Human Manipulated Resources
2.2.1. Polluted Environments
2.2.2. Agricultural Lands and Grassland
2.2.3. Industrial Composting
2.3. Unspecified Resources
3. Water
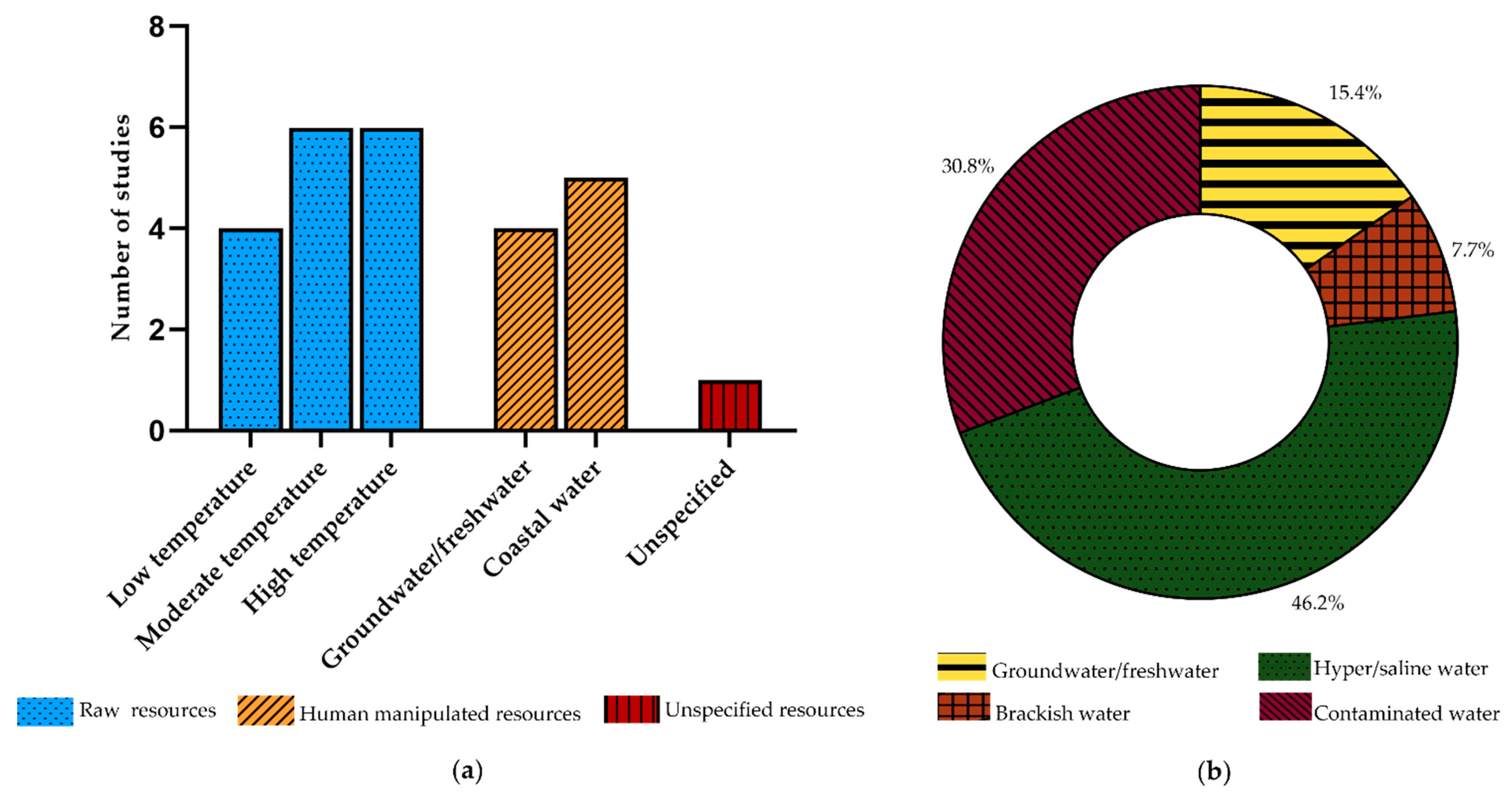
3.1. Raw Resources
3.1.1. Low-Temperature Environments
3.1.2. Moderate Temperature Environments
3.1.3. High-Temperature Environments
3.2. Human Manipulated Resources
3.2.1. Groundwater/Freshwater
3.2.2. Coastal Water
3.3. Unspecified Resource
4. Soil versus Water
5. General Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arocena, J.M.; Driscoll, K.G. Natural resources of the world. In Knowledge for Sustainable Development: An Insight into the Encyclopaedia of Life Support Systems; UNESCO/EOLSS: Zurich, Switzerland, 2002; pp. 261–290. [Google Scholar]
- Kattumuri, R. Sustaining natural resources in a changing environment: Evidence, policy and impact. Contemp. Soc. Sci. 2018, 13, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Lampert, A. Over-exploitation of natural resources is followed by inevitable declines in economic growth and discount rate. Nat. Commun. 2019, 10, 1419. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, S. Running out? Rethinking resource depletion. Extr. Ind. Soc. 2020, 7, 838–840. [Google Scholar] [CrossRef] [PubMed]
- Chettri, D.; Verma, A.K.; Verma, A.K. Innovations in CAZyme gene diversity and its modification for biorefinery applications. Biotechnol. Rep. 2020, 28, e00525. [Google Scholar] [CrossRef] [PubMed]
- Gudiña, E.J.; Amorim, C.; Braga, A.; Costa, Â.; Rodrigues, J.L.; Silvério, S.; Rodrigues, L.R. Biotech green approaches to unravel the potential of residues into valuable products. In Sustainable Green Chemical Processes and Their Allied Applications; Inamuddin, M., Asiri, A.M., Eds.; Springer: Cham, Switzerland, 2020; pp. 97–150. [Google Scholar]
- Berini, F.; Casciello, C.; Marcone, G.L.; Marinelli, F. Metagenomics: Novel enzymes from non-culturable microbes. FEMS Microbiol. Lett. 2017, 364, fnx211. [Google Scholar] [CrossRef] [PubMed]
- Costa, Â.M.A.; Santos, A.O.; Sousa, J.; Rodrigues, J.L.; Gudiña, E.J.; Silvério, S.C.; Rodrigues, L.R. Improved method for the extraction of high-quality DNA from lignocellulosic compost samples for metagenomic studies. Appl. Microbiol. Biotechnol. 2021, 105, 8881–8893. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.L.; Piel, J.; Sunagawa, S. A roadmap for metagenomic enzyme discovery. Nat. Prod. Rep. 2021, 38, 1994–2023. [Google Scholar] [CrossRef] [PubMed]
- Batista-García, R.A.; del Rayo Sánchez-Carbente, M.; Talia, P.; Jackson, S.A.; O’Leary, N.D.; Dobson, A.D.W.; Folch-Mallol, J.L. From lignocellulosic metagenomes to lignocellulolytic genes: Trends, challenges and future prospects. Biofuels Bioprod. Biorefining 2016, 10, 864–882. [Google Scholar] [CrossRef]
- Wang, H.; Hart, D.J.; An, Y. Functional metagenomic technologies for the discovery of novel enzymes for biomass degradation and biofuel production. BioEnergy Res. 2019, 12, 457–470. [Google Scholar] [CrossRef]
- Escuder-Rodríguez, J.-J.; DeCastro, M.-E.; Becerra, M.; Rodríguez-Belmonte, E.; González-Siso, M.-I. Advances of functional metagenomics in harnessing thermozymes. In Metagenomics: Perspectives, Methods, and Applications; Academic Press: Cambridge, MA, USA, 2018; pp. 289–307. [Google Scholar]
- Koorevaar, G.P.; Menelik, C.D. (Eds.) 1 Composition and physical properties of soils. In Developments in Soil Science; Elsevier: Amsterdam, The Netherlands, 1983; pp. 1–36. [Google Scholar]
- Bhattacharyya, T.; Pal, K.D. The Soil: A natural resource. In Soil Science: An Introduction; Rattan, R.K., Katyal, J.C., Dwivedi, B.S., Sarkar, A.K., Tapas Bhattacharyya, J.C., Tarafdar, S.K., Eds.; ISSS: New Delhi, India, 2015; pp. 1–19. [Google Scholar]
- Jindal, S. Microbes in soil and their Metagenomics. In Microbial Diversity, Interventions and Scope; Sharma, S.G., Neeta Raj Sharma, M.S., Eds.; Springer: Singapore, 2020; pp. 85–96. [Google Scholar]
- Wang, L.; D’Odorico, P. Decomposition and mineralization. In Reference Module in Earth Systems and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2013; pp. 280–285. [Google Scholar]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the extremes: Extremophiles and the limits of life in a planetary context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirete, S.; Morgante, V.; González-Pastor, J.E. Functional metagenomics of extreme environments. Curr. Opin. Biotechnol. 2016, 38, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Khan, F.G.; Qazi, G.N. Molecular cloning and characterization of amylase from soil metagenomic library derived from Northwestern Himalayas. Appl. Microbiol. Biotechnol. 2010, 86, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lei, Y.; Zhang, X.; Gao, Y.; Xiao, Y.; Peng, H. Identification and phylogenetic characterization of a new subfamily of α-amylase enzymes from marine microorganisms. Mar. Biotechnol. 2012, 14, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Vester, J.K.; Glaring, M.A.; Stougaard, P. Discovery of novel enzymes with industrial potential from a cold and alkaline environment by a combination of functional metagenomics and culturing. Microb. Cell Fact. 2014, 13, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Govil, T.; Capalash, N.; Sharma, P. Characterization of a glycoside hydrolase family 1 β-galactosidase from hot spring metagenome with transglycosylation activity. Appl. Biochem. Biotechnol. 2012, 168, 1681–1693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, H.; Li, C.J.; Ma, T.; Li, G.; Liu, Y.H. Metagenomic approach for the isolation of a thermostable β-galactosidase with high tolerance of galactose and glucose from soil samples of Turpan Basin. BMC Microbiol. 2013, 13, 237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, H.; Ozaki, M.; Kanaya, E.; Kim, J.J.; Angkawidjaja, C.; Koga, Y.; Kanaya, S. Structure and stability of metagenome-derived glycoside hydrolase family 12 cellulase (LC-CelA) a homolog of Cel12A from Rhodothermus marinus. FEBS Open Bio 2014, 4, 936–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sae-Lee, R.; Boonmee, A. Newly derived GH43 gene from compost metagenome showing dual xylanase and cellulase activities. Folia Microbiol. 2014, 59, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-F.; Chang, S.C.-Y.; Kuo, H.-W.; Tong, C.-G.; Yu, S.-M.; Ho, T.-H.D. A metagenomic approach for the identification and cloning of an endoglucanase from rice straw compost. Gene 2013, 519, 360–366. [Google Scholar] [CrossRef]
- Suleiman, M.; Schröder, C.; Klippel, B.; Schäfers, C.; Krüger, A.; Antranikian, G. Extremely thermoactive archaeal endoglucanase from a shallow marine hydrothermal vent from Vulcano Island. Appl. Microbiol. Biotechnol. 2019, 103, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Schröder, C.; Elleuche, S.; Blank, S.; Antranikian, G. Characterization of a heat-active archaeal β-glucosidase from a hydrothermal spring metagenome. Enzym. Microb. Technol. 2014, 57, 48–54. [Google Scholar] [CrossRef]
- Cao, L.C.; Wang, Z.J.; Ren, G.H.; Kong, W.; Li, L.; Xie, W.; Liu, Y.H. Engineering a novel glucose-tolerant β-glucosidase as supplementation to enhance the hydrolysis of sugarcane bagasse at high glucose concentration. Biotechnol. Biofuels 2015, 8, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuzawa, T.; Yaoi, K. Screening, identification, and characterization of a novel saccharide-stimulated β-glycosidase from a soil metagenomic library. Appl. Microbiol. Biotechnol. 2017, 101, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Klippel, B.; Sahm, K.; Basner, A.; Wiebusch, S.; John, P.; Lorenz, U.; Peters, A.; Abe, F.; Takahashi, K.; Kaiser, O.; et al. Carbohydrate-active enzymes identified by metagenomic analysis of deep-sea sediment bacteria. Extremophiles 2014, 18, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, G.; Kumar, J.; Sangwan, R.S.; Singh, S.P. Metagenomic analysis of geothermal water reservoir sites exploring carbohydrate-related thermozymes. Int. J. Biol. Macromol. 2018, 119, 882–895. [Google Scholar] [CrossRef] [PubMed]
- Colombo, L.T.; de Oliveira, M.N.V.; Carneiro, D.G.; de Souza, R.A.; Alvim, M.C.T.; dos Santos, J.C.; da Silva, C.C.; Vidigal, P.M.P.; da Silveira, W.B.; Passos, F.M.L. Applying functional metagenomics to search for novel lignocellulosic enzymes in a microbial consortium derived from a thermophilic composting phase of sugarcane bagasse and cow manure. Antonie Leeuwenhoek 2016, 109, 1217–1233. [Google Scholar] [CrossRef] [PubMed]
- Strazzulli, A.; Cobucci-Ponzano, B.; Iacono, R.; Giglio, R.; Maurelli, L.; Curci, N.; Schiano-di-Cola, C.; Santangelo, A.; Contursi, P.; Lombard, V.; et al. Discovery of hyperstable carbohydrate-active enzymes through metagenomics of extreme environments. FEBS J. 2020, 287, 1116–1137. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Kawarabayasi, Y.; Miyazaki, K.; Satyanarayana, T. Cloning, expression and characteristics of a novel alkalistable and thermostable xylanase encoding gene (Mxyl) retrieved from compost-soil metagenome. PLoS ONE 2013, 8, e52459. [Google Scholar] [CrossRef] [Green Version]
- Knapik, K.; Becerra, M.; González-Siso, M.I. Microbial diversity analysis and screening for novel xylanase enzymes from the sediment of the Lobios Hot Spring in Spain. Sci. Rep. 2019, 9, 11195. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa, T.; Kimura, N.; Suenaga, H.; Yaoi, K. Screening, identification, and characterization of α-xylosidase from a soil metagenome. J. Biosci. Bioeng. 2016, 122, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Yang, F.; Liu, X.; Wang, H. The discovery and characterization of a novel chitinase with dual catalytic domains from a Qinghai-Tibetan Plateau wetland soil metagenome. Int. J. Biol. Macromol. 2021, 188, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.P.; Heath, C.; Taylor, M.P.; Tuffin, M.; Cowan, D. A novel, extremely alkaliphilic and cold-active esterase from Antarctic desert soil. Extremophiles 2012, 16, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Martínez-martínez, M.; Alcaide, M.; Tchigvintsev, A.; Reva, O.; Polaina, J.; Bargiela, R.; Guazzaroni, M.-E.; Chicote, Á.; Canet, A.; Valero, F.; et al. Biochemical diversity of carboxyl esterases and lipases from Lake Arreo (Spain): A metagenomic approach. Appl. Environ. Microbiol. 2013, 79, 3553–3562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudan, A.K.; Vakhlu, J. Isolation of a thioesterase gene from the metagenome of a mountain peak, Apharwat, in the northwestern Himalayas. 3 Biotech 2013, 3, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Okano, H.; Hong, X.; Kanaya, E.; Angkawidjaja, C.; Kanaya, S. Structural and biochemical characterization of a metagenome-derived esterase with a long N-terminal extension. Protein Sci. 2015, 24, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Leis, B.; Angelov, A.; Mientus, M.; Li, H.; Pham, V.T.T.; Lauinger, B.; Bongen, P.; Pietruszka, J.; Gonçalves, L.G.; Sutherland, R. Identification of novel esterase-active enzymes from hot environments by use of the host bacterium Thermus thermophilus. Front. Microbiol. 2015, 6, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudan, A.K.; Vakhlu, J. Isolation and in silico characterization of novel esterase gene with β-lactamase fold isolated from metagenome of north western Himalayas. 3 Biotech 2015, 5, 553–559. [Google Scholar] [CrossRef] [Green Version]
- De Santi, C.; Ambrosino, L.; Tedesco, P.; de Pascale, D.; Zhai, L.; Zhou, C.; Xue, Y.; Ma, Y. Identification and characterization of a novel salt-tolerant esterase from a Tibetan glacier metagenomic library. Biotechnol. Prog. 2015, 31, 890–899. [Google Scholar] [CrossRef]
- Zarafeta, D.; Moschidi, D.; Ladoukakis, E.; Gavrilov, S.; Chrysina, E.D.; Chatziioannou, A.; Kublanov, I.; Skretas, G.; Kolisis, F.N. Metagenomic mining for thermostable esterolytic enzymes uncovers a new family of bacterial esterases. Sci. Rep. 2016, 6, 38886. [Google Scholar] [CrossRef] [Green Version]
- Petrovskaya, L.E.; Novototskaya-Vlasova, K.A.; Spirina, E.V.; Durdenko, E.V.; Lomakina, G.Y.; Zavialova, M.G.; Nikolaev, E.N.; Rivkina, E.M. Expression and characterization of a new esterase with GCSAG motif from a permafrost metagenomic library. FEMS Microbiol. Ecol. 2016, 92, fiw046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Santi, C.; Altermark, B.; Pierechod, M.M.; Ambrosino, L.; De Pascale, D.; Willassen, N.P. Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries. BMC Biochem. 2016, 17, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Wu, K.; Chen, L.; Fan, H.; Zhao, Z.; Gao, B.; Wang, H.; Wei, D. A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters Wenyuan. Microb. Cell Fact. 2016, 15, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popovic, A.; Hai, T.; Tchigvintsev, A.; Hajighasemi, M.; Nocek, B.; Khusnutdinova, A.N.; Brown, G.; Glinos, J.; Flick, R.; Skarina, T.; et al. Activity screening of environmental metagenomic libraries reveals novel carboxylesterase families. Sci. Rep. 2017, 7, 44103. [Google Scholar] [CrossRef] [PubMed]
- Li, P.-Y.; Yao, Q.-Q.; Wang, P.; Zhang, Y.; Li, Y.; Zhang, Y.-Q.; Hao, J.; Zhou, B.-C.; Chen, X.-L.; Shi, M.; et al. A novel subfamily esterase with a homoserine transacetylase-like fold but no transferase activity. Appl. Environ. Microbiol. 2017, 83, e00131-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayanath, G.; Mohandas, S.P.; Kachiprath, B.; Solomon, S.; Sajeevan, T.P.; Bright Singh, I.S.; Philip, R. A novel solvent tolerant esterase of GDSGG motif subfamily from solar saltern through metagenomic approach: Recombinant expression and characterization. Int. J. Biol. Macromol. 2018, 119, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fu, C.; Huang, Y.; Yin, Y.; Cheng, G.; Lei, F.; Lu, N.A.; Li, J.; Ashforth, E.J.; Zhang, L.; et al. Novel lipolytic genes from the microbial metagenomic library of the South China Sea marine sediment. FEMS Microbiol. Ecol. 2010, 72, 228–237. [Google Scholar] [CrossRef] [Green Version]
- Fu, C.; Hu, Y.; Xie, F.; Guo, H.; Ashforth, E.J.; Polyak, S.W.; Zhu, B.; Zhang, L. Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. Appl. Microbiol. Biotechnol. 2011, 90, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Zhang, X.; Shang, M.; Wang, X.; Wang, G.; Li, B.; Guan, G.; Li, Y.; Wang, Y. A novel esterase gene cloned from a metagenomic library from neritic sediments of the South China Sea. Microb. Cell Fact. 2011, 10, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, E.Y.; Kwon, M.A.; Lee, M.; Oh, J.Y.; Choi, J.E.; Lee, J.Y.; Song, B.K.; Hahm, D.H.; Song, J.K. Isolation and characterization of cold-active family VIII esterases from an arctic soil metagenome. Appl. Microbiol. Biotechnol. 2011, 90, 573–581. [Google Scholar] [CrossRef]
- Fan, X.; Liu, X.; Huang, R.; Liu, Y. Identification and characterization of a novel thermostable pyrethroid-hydrolyzing enzyme isolated through metagenomic approach. Microb. Cell Fact. 2012, 11, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Xu, X.; Huo, Y.; Wu, Y.; Zhu, X.; Zhang, X.; Wu, M. Identification and characterization of novel esterases from a deep-sea sediment metagenome. Arch. Microbiol. 2012, 194, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahoo, R.K.; Kumar, M.; Sukla, L.B.; Subudhi, E. Bioprospecting hot spring metagenome: Lipase for the production of biodiesel. Environ. Sci. Pollut. Res. 2017, 24, 3802–3809. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, R.K.; Das, A.; Sahoo, K.; Sahu, A.; Subudhi, E. Characterization of novel metagenomic–derived lipase from Indian hot spring. Int. Microbiol. 2020, 23, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, M.; Zhang, M.; Wang, C.; Liu, Y.; Fan, X.; Li, H. Characterization of a novel, cold-adapted, and thermostable laccase-like enzyme with high tolerance for organic solvents and salt and potent dye decolorization ability, derived from a marine metagenomic library. Front. Microbiol. 2018, 9, 2998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Li, C.; Pei, X.; Wang, Q.; Yin, X.; Xie, T. Isolation an aldehyde dehydrogenase gene from Metagenomics based on semi-nest touch-down PCR. Indian J. Microbiol. 2014, 54, 74–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, N.; Isotani, K.; Makino, Y.; Kato, M.; Kitayama, K.; Ishimota, T. PCR-based amplification and heterologous expression of Pseudomonas alcohol dehydrogenase genes from the soil metagenome for biocatalysis. Enzym. Microb. Technol. 2014, 55, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Kariya, S.; Kurokawa, J. Efficient PCR-based amplification of diverse alcohol dehydrogenase genes from metagenomes for improving biocatalysis: Screening of gene-specific amplicons from metagenomes. Appl. Environ. Microbiol. 2014, 80, 6280–6289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neveu, J.; Regeard, C.; Dubow, M.S. Isolation and characterization of two serine proteases from metagenomic libraries of the Gobi and Death Valley deserts. Appl. Microbiol. Biotechnol. 2011, 91, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Chopra, C.; Gupta, V.K.; Akhlaq, B.; Verma, V.; Rasool, S. Purification and characterization of CHpro1, a thermotolerant, alkali-stable and oxidation-resisting protease of Chumathang hotspring. Sci. Bull. 2015, 60, 1252–1260. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Zeng, Y.; Zheng, L.; Liu, W.; Lyu, Q. Discovery and characterization of a novel protease from the Antarctic soil. Process Biochem. 2021, 111, 270–277. [Google Scholar] [CrossRef]
- Kanaya, E.; Sakabe, T.; Nguyen, N.T.; Koikeda, S.; Koga, Y.; Takano, K.; Kanaya, S. Cloning of the RNase H genes from a metagenomic DNA library: Identification of a new type 1 RNase H without a typical active-site motif. J. Appl. Microbiol. 2010, 109, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.; Riyaz-Ul-Hassan, S.; Srivastava, N.; Johri, S. Molecular cloning of rhodanese gene from soil metagenome of cold desert of North-West Himalayas: Sequence and structural features of the rhodanese enzyme. 3 Biotech 2015, 5, 513–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva-Portela, R.C.B.; Carvalho, F.M.; Pereira, C.P.M.; De Souza-Pinto, N.C.; Modesti, M.; Fuchs, R.P.; Agnez-Lima, L.F. ExoMeg1: A new exonuclease from metagenomic library. Sci. Rep. 2016, 6, 19712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Jia, Z.; Li, S.; Li, Y.; You, Q.; Zhang, C.; Zheng, X.; Xiong, G.; Zhao, J.; Qi, C.; et al. Identification and characterization of a chitin deacetylase from a metagenomic library of deep-sea sediments of the Arctic Ocean. Gene 2016, 590, 79–84. [Google Scholar] [CrossRef]
- Agarwal, N.; Singh, S.P. A novel trehalose synthase for the production of trehalose and trehalulose. Microbiol. Spectr. 2021, 9, e01333-21. [Google Scholar] [CrossRef] [PubMed]
- Perfumo, A.; Banat, I.M.; Marchant, R. Going green and cold: Biosurfactants from low-temperature environments to biotechnology applications. Trends Biotechnol. 2018, 36, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Morton, B.; Blackmore, G. South China Sea. Mar. Pollut. Bull. 2001, 42, 1236–1263. [Google Scholar] [CrossRef] [PubMed]
- Pacchioni, R.G.; Carvalho, F.M.; Thompson, C.E.; Faustino, A.L.F.; Nicolini, F.; Pereira, T.S.; Silva, R.C.B.; Cantão, M.E.; Gerber, A.; Ana, T.R.; et al. Taxonomic and functional profiles of soil samples from Atlantic forest and Caatinga biomes in northeastern Brazil. Microbiologyopen 2014, 3, 299–315. [Google Scholar] [CrossRef] [PubMed]
- DeCastro, M.E.; Rodríguez-Belmonte, E.; González-Siso, M.I. Metagenomics of thermophiles with a focus on discovery of novel thermozymes. Front. Microbiol. 2016, 7, 1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, Ó.J.; Ospina, D.A.; Montoya, S. Compost supplementation with nutrients and microorganisms in composting process. Waste Manag. 2017, 69, 136–153. [Google Scholar] [CrossRef] [PubMed]
- Madhuri, R.J.; Saraswathi, M.; Gowthami, K.; Bhargavi, M.; Divya, Y.; Deepika, V. Recent approaches in the production of novel enzymes from environmental samples by enrichment culture and metagenomic approach. In Recent Developments in Applied Microbiology and Biochemistry; Buddolla, V., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 251–262. [Google Scholar]
- Wang, K.; Li, G.; Yu, S.Q.; Zhang, C.T.; Liu, Y.H. A novel metagenome-derived β-galactosidase: Gene cloning, overexpression, purification and characterization. Appl. Microbiol. Biotechnol. 2010, 88, 155–165. [Google Scholar] [CrossRef]
- Cheng, J.; Romantsov, T.; Engel, K.; Doxey, A.C.; Rose, D.R.; Neufeld, J.D.; Charles, T.C. Functional metagenomics reveals novel β-galactosidases not predictable from gene sequences. PLoS ONE 2017, 12, e0172545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allgaier, M.; Reddy, A.; Park, J.I.; Ivanova, N.; D’Haeseleer, P.; Lowry, S.; Sapra, R.; Hazen, T.C.; Simmons, B.A.; Vandergheynst, J.S.; et al. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS ONE 2010, 5, e8812. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, T.M.; Paiva, J.H.; Ruiz, D.M.; Cairo, J.P.L.F.; Pereira, I.O.; Paixão, D.A.A.; De Almeida, R.F.; Tonoli, C.C.C.; Ruller, R.; Santos, C.R.; et al. Structure and function of a novel cellulase 5 from sugarcane soil metagenome. PLoS ONE 2013, 8, e83635. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Li, S.X.; Luo, F.F.; Jin, K.; Wang, Q.; Hao, Z.Y.; Wu, L.L.; Zhao, G.C.; Ma, G.F.; Shen, P.H.; et al. Biochemical characterization of two novel β-glucosidase genes by metagenome expression cloning. Bioresour. Technol. 2011, 102, 3272–3278. [Google Scholar] [CrossRef]
- Liu, J.; Liu, W.D.; Zhao, X.L.; Shen, W.J.; Cao, H.; Cui, Z.L. Cloning and functional characterization of a novel endo-β-1,4-glucanase gene from a soil-derived metagenomic library. Appl. Microbiol. Biotechnol. 2011, 89, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Li, A.; Tian, C.; Zhou, Y.; Zhang, G.; Ma, Y. Identification and characterization of a new acid-stable endoglucanase from a metagenomic library. Protein Expr. Purif. 2014, 102, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Gulati, S.; Goyal, E.; Singh, S.; Kumar, K.; Nain, L.; Saxena, A.K. Construction and screening of metagenomic library derived from soil for β-1, 4-endoglucanase gene. Biocatal. Agric. Biotechnol. 2016, 5, 186–192. [Google Scholar] [CrossRef]
- Yang, C.; Xia, Y.; Qu, H.; Li, A.D.; Liu, R.; Wang, Y.; Zhang, T. Discovery of new cellulases from the metagenome by a metagenomics-guided strategy. Biotechnol. Biofuels 2016, 9, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meneses, C.; Silva, B.; Medeiros, B.; Serrato, R.; Johnston-Monje, D. A metagenomic advance for the cloning and characterization of a cellulase from red rice crop residues. Molecules 2016, 21, 831. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, A.C.; Ematsu, G.C.G.; Liberato, M.V.; Paixão, D.A.A.; Franco Cairo, J.P.L.; Mandelli, F.; Tramontina, R.; Gandin, C.A.; de Oliveira Neto, M.; Squina, F.M.; et al. Biochemical and biophysical properties of a metagenome-derived GH5 endoglucanase displaying an unconventional domain architecture. Int. J. Biol. Macromol. 2017, 99, 384–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.P.; Lee, H.W.; Na, H.B.; Lee, J.H.; Hong, Y.J.; Jeon, J.M.; Kwon, E.J.; Kim, S.K.; Kim, H. Characterization of truncated endo-β-1,4-glucanases from a compost metagenomic library and their saccharification potentials. Int. J. Biol. Macromol. 2018, 115, 554–562. [Google Scholar] [CrossRef]
- Wierzbicka-Woś, A.; Henneberger, R.; Batista-García, R.A.; Martínez-Ávila, L.; Jackson, S.A.; Kennedy, J.; Dobson, A.D.W. Biochemical characterization of a novel monospecific endo-β-1,4-glucanase belonging to GH family 5 from a rhizosphere metagenomic library. Front. Microbiol. 2019, 10, 1342. [Google Scholar] [CrossRef] [PubMed]
- Aymé, L.; Hébert, A.; Henrissat, B.; Lombard, V.; Franche, N.; Perret, S.; Jourdier, E.; Heiss-Blanquet, S. Characterization of three bacterial glycoside hydrolase family 9 endoglucanases with different modular architectures isolated from a compost metagenome. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129848. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, X.; Wei, W.; Xu, J.; Wang, W.; Xie, Z.; Zhang, Z.; Jiang, H.; Wang, Q.; Wei, C. A novel efficient β-glucanase from a paddy soil microbial metagenome with versatile activities. Biotechnol. Biofuels 2016, 9, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjort, K.; Bergström, M.; Adesina, M.F.; Jansson, J.K.; Smalla, K.; Sjöling, S. Chitinase genes revealed and compared in bacterial isolates, DNA extracts and a metagenomic library from a phytopathogen-suppressive soil. FEMS Microbiol. Ecol. 2010, 71, 197–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjort, K.; Presti, I.; Elväng, A.; Marinelli, F.; Sjöling, S. Bacterial chitinase with phytopathogen control capacity from suppressive soil revealed by functional metagenomics. Appl. Microbiol. Biotechnol. 2014, 98, 2819–2828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cretoiu, M.S.; Berini, F.; Kielak, A.M.; Marinelli, F.; van Elsas, J.D. A novel salt-tolerant chitobiosidase discovered by genetic screening of a metagenomic library derived from chitin-amended agricultural soil. Appl. Microbiol. Biotechnol. 2015, 99, 8199–8215. [Google Scholar] [CrossRef] [Green Version]
- Stöveken, J.; Singh, R.; Kolkenbrock, S.; Zakrzewski, M.; Wibberg, D.; Eikmeyer, F.G.; Pühler, A.; Schlüter, A.; Moerschbacher, B.M. Successful heterologous expression of a novel chitinase identified by sequence analyses of the metagenome from a chitin-enriched soil sample. J. Biotechnol. 2015, 201, 60–68. [Google Scholar] [CrossRef]
- Thimoteo, S.S.; Glogauer, A.; Faoro, H.; de Souza, E.M.; Huergo, L.F.; Moerschbacher, B.M.; Pedrosa, F.O. A broad pH range and processive chitinase from a metagenome library. Braz. J. Med. Biol. Res. 2017, 50, e5658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delavat, F.; Phalip, V.; Forster, A.; Plewniak, F.; Lett, M.C.; Liévremont, D. Amylases without known homologues discovered in an acid mine drainage: Significance and impact. Sci. Rep. 2012, 2, 354. [Google Scholar] [CrossRef] [PubMed]
- Soares, F.L.; Marcon, J.; Pereira e Silva, M.D.C.; Khakhum, N.; Cerdeira, L.T.; Ottoni, J.R.; Domingos, D.F.; Taketani, R.G.; De Oliveira, V.M.; Lima, A.O.D.S.; et al. A Novel Multifunctional β-N-Acetylhexosaminidase Revealed through Metagenomics of an Oil-Spilled Mangrove. Bioengineering 2017, 4, 62. [Google Scholar] [CrossRef] [Green Version]
- Jeong, Y.S.; Na, H.B.; Kim, S.K.; Kim, Y.H.; Kwon, E.J.; Kim, J.; Yun, H.D.; Lee, J.K.; Kim, H. Characterization of Xyn10J, a novel family 10 xylanase from a compost metagenomic library. Appl. Biochem. Biotechnol. 2012, 166, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, T.M.; Goldbeck, R.; dos Santos, C.R.; Paixão, D.A.A.; Gonçalves, T.A.; Franco Cairo, J.P.L.; Almeida, R.F.; de Oliveira Pereira, I.; Jackson, G.; Cota, J.; et al. Development and biotechnological application of a novel endoxylanase family GH10 identified from sugarcane soil metagenome. PLoS ONE 2013, 8, e70014. [Google Scholar] [CrossRef] [PubMed]
- Ellilä, S.; Bromann, P.; Nyyssönen, M.; Itävaara, M.; Koivula, A.; Paulin, L.; Kruus, K. Cloning of novel bacterial xylanases from lignocellulose-enriched compost metagenomic libraries. AMB Express 2019, 9, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Liang, J.; Li, Y.; Tian, L.; Wei, Y. Characterization of efficient xylanases from industrial-scale pulp and paper wastewater treatment microbiota. AMB Express 2021, 11, 19. [Google Scholar] [CrossRef]
- Ndata, K.; Nevondo, W.; Cekuse, B.; van Zyl, L.J.; Trindade, M. Characterization of a highly xylose tolerant β-xylosidase isolated from high temperature horse manure compost. BMC Biotechnol. 2021, 21, 61. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.J.; Jeong, Y.S.; Kim, Y.H.; Kim, S.K.; Na, H.B.; Kim, J.; Yun, H.D.; Kim, H. Construction of a metagenomic library from compost and screening of cellulase- and xylanase-positive clones. J. Appl. Biol. Chem. 2010, 53, 702–708. [Google Scholar] [CrossRef]
- Gladden, J.M.; Allgaier, M.; Miller, C.S.; Hazen, T.C.; VanderGheynst, J.S.; Hugenholtz, P.; Simmons, B.A.; Singer, S.W. Glycoside hydrolase activities of thermophilic bacterial consortia adapted to switchgrass. Appl. Environ. Microbiol. 2011, 77, 5804–5812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougherty, M.J.; D’haeseleer, P.; Hazen, T.C.; Simmons, B.A.; Adams, P.D.; Hadi, M.Z. Glycoside hydrolases from a targeted compost metagenome, activity-screening and functional characterization. BMC Biotechnol. 2012, 12, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanokratana, P.; Eurwilaichitr, L.; Pootanakit, K.; Champreda, V. Identification of glycosyl hydrolases from a metagenomic library of microflora in sugarcane bagasse collection site and their cooperative action on cellulose degradation. J. Biosci. Bioeng. 2015, 119, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Berlemont, R.; Pipers, D.; Delsaute, M.; Angiono, F.; Feller, G.; Galleni, M.; Power, P. Exploring the Antarctic soil metagenome as a source of novel cold-adapted enzymes and genetic mobile elements. Rev. Argent. Microbiol. 2011, 43, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Stroobants, A.; Portetelle, D.; Vandenbol, M. New carbohydrate-active enzymes identified by screening two metagenomic libraries derived from the soil of a winter wheat field. J. Appl. Microbiol. 2014, 117, 1045–1055. [Google Scholar] [CrossRef] [Green Version]
- Liaw, R.B.; Cheng, M.P.; Wu, M.C.; Lee, C.Y. Use of metagenomic approaches to isolate lipolytic genes from activated sludge. Bioresour. Technol. 2010, 101, 8323–8329. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Kwon, E.J.; Kim, S.K.; Jeong, Y.S.; Kim, J.; Yun, H.D.; Kim, H. Molecular cloning and characterization of a novel family VIII alkaline esterase from a compost metagenomic library. Biochem. Biophys. Res. Commun. 2010, 393, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Hong, K.S.; Malhotra, S.; Park, J.H.; Hwang, E.C.; Choi, H.K.; Kim, Y.S.; Tao, W.; Lee, S.W. A new esterase EstD2 isolated from plant rhizosphere soil metagenome. Appl. Microbiol. Biotechnol. 2010, 88, 1125–1134. [Google Scholar] [CrossRef]
- Sang, S.L.; Li, G.; Hu, X.P.; Liu, Y.H. Molecular cloning, overexpression and characterization of a novel feruloyl esterase from a soil metagenomic library. J. Mol. Microbiol. Biotechnol. 2011, 20, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.R.; Mercaldi, G.F.; Maester, T.C.; Balan, A.; De Macedo Lemos, E.G. Est16, a new esterase isolated from a metagenomic library of a microbial consortium specializing in diesel oil degradation. PLoS ONE 2015, 10, e0133723. [Google Scholar] [CrossRef]
- Ohlhoff, C.W.; Kirby, B.M.; Van Zyl, L.; Mutepfa, D.L.R.; Casanueva, A.; Huddy, R.J.; Bauer, R.; Cowan, D.A.; Tuffin, M. An unusual feruloyl esterase belonging to family VIII esterases and displaying a broad substrate range. J. Mol. Catal. B Enzym. 2015, 118, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.W.; Jung, W.K.; Kim, Y.H.; Ryu, B.H.; Doohun Kim, T.; Kim, J.; Kim, H. Characterization of a novel alkaline family viii esterase with S-enantiomer preference from a compost metagenomic library. J. Microbiol. Biotechnol. 2016, 26, 315–325. [Google Scholar] [CrossRef]
- Dukunde, A.; Schneider, D.; Lu, M.; Brady, S.; Daniel, R. A novel, versatile family IV carboxylesterase exhibits high stability and activity in a broad pH spectrum. Biotechnol. Lett. 2017, 39, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Dukunde, A.; Daniel, R. Biochemical profiles of two thermostable and organic solvent–tolerant esterases derived from a compost metagenome. Appl. Microbiol. Biotechnol. 2019, 103, 3421–3437. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Gui, L.; Yin, S. A novel esterase from a soil metagenomic library displaying a broad substrate range. AMB Express 2021, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Ding, L.; Zou, D.; Wang, L.; Tan, Y.; Guo, S.; Zhang, Y.; Xin, Z. Identification and characterization of a novel carboxylesterase EstQ7 from a soil metagenomic library. Arch. Microbiol. 2021, 203, 4113–4125. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Ding, L.; Zou, D.; Qiu, J.; Shao, Y.; Sun, S.; Li, L.; Xin, Z. Characterization of a novel carboxylesterase with catalytic activity toward di(2-ethylhexyl) phthalate from a soil metagenomic library. Sci. Total Environ. 2021, 785, 147260. [Google Scholar] [CrossRef]
- Park, J.E.; Jeong, G.S.; Lee, H.W.; Kim, H. Biochemical characterization of a family IV esterase with R-form enantioselectivity from a compost metagenomic library. Appl. Biol. Chem. 2021, 64, 81. [Google Scholar] [CrossRef]
- Park, J.E.; Jeong, G.S.; Lee, H.W.; Kim, H. Molecular characterization of novel family iv and viii esterases from a compost metagenomic library. Microorganisms 2021, 9, 1614. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Jeong, G.S.; Lee, H.W.; Kim, S.K.; Kim, J.; Kim, H. Characterization of a novel family iv esterase containing a predicted czco domain and a family v esterase with broad substrate specificity from an oil-polluted mud flat metagenomic library. Appl. Sci. 2021, 11, 5905. [Google Scholar] [CrossRef]
- O’Mahony, M.M.; Henneberger, R.; Selvin, J.; Kennedy, J.; Doohan, F.; Marchesi, J.R.; Dobson, A.D.W. Inhibition of the growth of Bacillus subtilis DSM10 by a newly discovered antibacterial protein from the soil metagenome. Bioengineered 2015, 6, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Glogauer, A.; Martini, V.P.; Faoro, H.; Couto, G.H.; Müller-Santos, M.; Monteiro, R.A.; Mitchell, D.A.; de Souza, E.M.; Pedrosa, F.O.; Krieger, N. Identification and characterization of a new true lipase isolated through metagenomic approach. Microb. Cell Fact. 2011, 10, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Liu, C.; Liu, L.; Jin, Q. Characterisation of a thermo-alkali-stable lipase from oil-contaminated soil using a metagenomic approach. Syst. Appl. Microbiol. 2013, 36, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Jeong, Y.S.; Jung, W.K.; Kim, S.K.; Lee, H.W.; Kahng, H.Y.; Kim, J.; Kim, H. Characterization of novel family IV esterase and family I.3 lipase from an oil-polluted mud flat metagenome. Mol. Biotechnol. 2015, 57, 781–792. [Google Scholar] [CrossRef]
- Yao, J.; Fan, X.J.; Lu, Y.; Liu, Y.H. Isolation and characterization of a novel tannase from a metagenomic library. J. Agric. Food Chem. 2011, 59, 3812–3818. [Google Scholar] [CrossRef]
- Nacke, H.; Will, C.; Herzog, S.; Nowka, B.; Engelhaupt, M.; Daniel, R. Identification of novel lipolytic genes and gene families by screening of metagenomic libraries derived from soil samples of the German Biodiversity Exploratories. FEMS Microbiol. Ecol. 2011, 78, 188–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stroobants, A.; Martin, R.; Roosens, L.; Portetelle, D.; Vandenbol, M. New lipolytic enzymes identified by screening two metagenomic libraries derived from the soil of a winter wheat field. Biotechnol. Agron. Soc. Environ. 2015, 19, 125–131. [Google Scholar]
- Kimura, N.; Kamagata, Y. A thermostable bilirubin-oxidizing enzyme from activated sludge isolated by a metagenomic approach. Microbes Environ. 2016, 31, 435–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, Q.; Liu, Y.; Deng, J.; Chen, G.; Yang, Y.; Shen, P.; Wu, B.; Jiang, C. A novel d-amino acid oxidase from a contaminated agricultural soil metagenome and its characterization. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2015, 107, 1615–1623. [Google Scholar] [CrossRef] [PubMed]
- Kimura, N.; Sakai, K.; Nakamura, K. Isolation and characterization of a 4-nitrotoluene-oxidizing enzyme from activated sludge by a metagenomic approach. Microbes Environ. 2010, 25, 133–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Singh Chauhan, N.; Thulasiram, H.V.; Taneja, V.; Sharma, R. Identification of two flavin monooxygenases from an effluent treatment plant sludge metagenomic library. Bioresour. Technol. 2010, 101, 8481–8484. [Google Scholar] [CrossRef] [PubMed]
- Chemerys, A.; Pelletier, E.; Cruaud, C.; Martin, F.; Violet, F.; Jouanneaua, Y. Characterization of novel polycyclic aromatic hydrocarbon dioxygenases from the bacterial metagenomic DNA of a contaminated soil. Appl. Environ. Microbiol. 2014, 80, 6591–6600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Yu, Y.; Zhou, R.; Sun, W.; Dai, C.; Wan, P.; Zhang, L.; Hao, D.; Ren, H. Cloning and characterisation of a novel 2,4-dichlorophenol hydroxylase from a metagenomic library derived from polychlorinated biphenyl-contaminated soil. Biotechnol. Lett. 2011, 33, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Mooij, M.J.; Barret, M.; Hegarty, P.M.; Harrington, C.; Dobson, A.D.W.; O’Gara, F. Identification of novel phytase genes from an agricultural soil-derived metagenome. J. Microbiol. Biotechnol. 2014, 24, 113–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, N.H.; Park, J.H.; Chung, E.; So, H.A.; Lee, M.H.; Kim, J.C.; Hwang, E.C.; Lee, S.W. Characterization of a soil metagenome-derived gene encoding wax ester synthase. J. Microbiol. Biotechnol. 2016, 26, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lin, M.; Zhang, Y.; Li, Y.; Xu, X.; Li, S.; Huang, H. Identification and characterization of a novel trehalose synthase gene derived from saline-alkali soil metagenomes. PLoS ONE 2013, 8, e77437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.D.; Guo, G.S.; Li, L.; Cao, L.C.; Tong, L.; Ren, G.H.; Liu, Y.H. Identification and characterization of an unusual glycosyltransferase-like enzyme with β-galactosidase activity from a soil metagenomic library. Enzym. Microb. Technol. 2014, 57, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Colin, P.Y.; Kintses, B.; Gielen, F.; Miton, C.M.; Fischer, G.; Mohamed, M.F.; Hyvönen, M.; Morgavi, D.P.; Janssen, D.B.; Hollfelder, F. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat. Commun. 2015, 6, 10008. [Google Scholar] [CrossRef] [PubMed]
- Ajibade, F.O.; Adelodun, B.; Lasisi, K.H.; Fadare, O.O.; Ajibade, T.F.; Nwogwu, N.A.; Sulaymon, I.D.; Ugya, A.Y.; Wang, H.C.; Wang, A. Chapter 25—Environmental pollution and their socioeconomic impacts. In Microbe Mediated Remediation of Environmental Contaminants; Kumar, A., Singh, V.K., Singh, P., Mishra, V.K., Eds.; Woodhead Publishing: Cambridge, UK, 2021; pp. 321–354. ISBN 9780128211991. [Google Scholar]
- Dindar, E.; Topaç Şağban, F.O.; Başkaya, H.S. Variations of soil enzyme activities in petroleum-hydrocarbon contaminated soil. Int. Biodeterior. Biodegrad. 2015, 105, 268–275. [Google Scholar] [CrossRef]
- Waldhauser, F.; Schaff, D.; Richards, P.G.; Kim, W.-Y. Lop Nor Revisited: Underground nuclear explosion locations, 1976–1996, from double-difference analysis of regional and teleseismic data. Bull. Seismol. Soc. Am. 2004, 94, 1879–1889. [Google Scholar] [CrossRef] [Green Version]
- Van Leeuwen, J.P.; Djukic, I.; Bloem, J.; Lehtinen, T.; Hemerik, L. Effects of land use on soil microbial biomass, activity and community structure at different soil depths in the Danube floodplain. Eur. J. Soil Biol. 2017, 79, 14–20. [Google Scholar] [CrossRef]
- Souza, R.C.; Hungria, M.; Cantão, M.E.; Vasconcelos, A.T.R.; Nogueira, M.A.; Vicente, V.A. Metagenomic analysis reveals microbial functional redundancies and specificities in a soil under different tillage and crop-management regimes. Appl. Soil Ecol. 2015, 86, 106–112. [Google Scholar] [CrossRef]
- Siles-Castellano, A.B.; López, M.J.; López-González, J.A.; Suárez-Estrella, F.; Jurado, M.M.; Estrella-González, M.J.; Moreno, J. Comparative analysis of phytotoxicity and compost quality in industrial composting facilities processing different organic wastes. J. Clean. Prod. 2020, 252, 119820. [Google Scholar] [CrossRef]
- Bergmann, J.C.; Costa, O.Y.A.; Gladden, J.M.; Singer, S.; Heins, R.; D’haeseleer, P.; Simmons, B.A.; Quirino, B.F. Discovery of two novel β-glucosidases from an Amazon soil metagenomic library. FEMS Microbiol. Lett. 2014, 351, 147–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couto, G.H.; Glogauer, A.; Faoro, H.; Chubatsu, L.S.; Souza, E.M.; Pedrosa, F.O. Isolation of a novel lipase from a metagenomic library derived from mangrove sediment from the south Brazilian coast. Genet. Mol. Res. 2010, 9, 514–523. [Google Scholar] [CrossRef]
- Ye, M.; Li, G.; Liang, W.Q.; Liu, Y.H. Molecular cloning and characterization of a novel metagenome-derived multicopper oxidase with alkaline laccase activity and highly soluble expression. Appl. Microbiol. Biotechnol. 2010, 87, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Jiang, Y.; Fan, X.J.; Liu, Y.H. Molecular cloning and characterization of a novel β-glucosidase with high hydrolyzing ability for soybean isoflavone glycosides and glucose-tolerance from soil metagenomic library. Bioresour. Technol. 2012, 123, 15–22. [Google Scholar] [CrossRef]
- Mai, Z.; Su, H.; Yang, J.; Huang, S.; Zhang, S. Cloning and characterization of a novel GH44 family endoglucanase from mangrove soil metagenomic library. Biotechnol. Lett. 2014, 36, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Mai, Z.; Su, H.; Zhang, S. Isolation and characterization of a glycosyl hydrolase family 16 β-agarase from a mangrove soil metagenomic library. Int. J. Mol. Sci. 2016, 17, 1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunterngsook, B.; Kanokratana, P.; Thongaram, T.; Tanapongpipat, S.; Uengwetwanit, T.; Rachdawong, S.; Vichitsoonthonkul, T.; Eurwilaichitr, L. Identification and characterization of lipolytic enzymes from a peat-swamp forest soil metagenome. Biosci. Biotechnol. Biochem. 2010, 74, 1848–1854. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.; Wu, X.; Xie, L.; Huang, Z.; Peng, W.; Gan, B. A novel phytase derived from an acidic peat-soil microbiome showing high stability under acidic plus pepsin conditions. J. Mol. Microbiol. Biotechnol. 2016, 26, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Berlemont, R.; Spee, O.; Delsaute, M.; Lara, Y.; Schuldes, J.; Simon, C.; Power, P.; Daniel, R.; Galleni, M. Novel organic solvent-tolerant esterase isolated by metagenomics: Insights into the lipase/esterase classification. Rev. Argent. Microbiol. 2013, 45, 3–12. [Google Scholar] [PubMed]
- Gu, X.; Wang, S.; Wang, S.; Zhao, L.X.; Cao, M.; Feng, Z. Identification and characterization of two novel esterases from a metagenomic library. Food Sci. Technol. Res. 2015, 21, 649–657. [Google Scholar] [CrossRef]
- Gomes-Pepe, E.S.; Sierra, E.G.M.; Pereira, M.R.; Castellane, T.C.L.; De Lemos, E.G.M. Bg10: A novel metagenomics alcohol-tolerant and glucose-stimulated gh1 β-glucosidase suitable for lactose-free milk preparation. PLoS ONE 2016, 11, e0167932. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Lee, M.H.; Yoon, M.Y.; Kim, J.C.; Malhotra, S.; Wu, J.; Hwang, E.C.; Lee, S.W. Characterization of two metagenome-derived esterases that reactivate chloramphenicol by counteracting chloramphenicol acetyltransferase. J. Microbiol. Biotechnol. 2011, 21, 1203–1210. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, X.; Liu, T.; Chen, S.; Liu, H.; Wang, H.; Li, K.; Song, Y.; Luo, X.; Zhao, J.; et al. The critical roles of exposed surface residues for the thermostability and halotolerance of a novel GH11 xylanase from the metagenomic library of a saline-alkaline soil. Int. J. Biol. Macromol. 2019, 133, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Istvan, P.; Souza, A.A.; Garay, A.V.; dos Santos, D.F.K.; de Oliveira, G.M.; Santana, R.H.; Lopes, F.A.C.; de Freitas, S.M.; Barbosa, J.A.R.G.; Krüger, R.H. Structural and functional characterization of a novel lipolytic enzyme from a Brazilian Cerrado soil metagenomic library. Biotechnol. Lett. 2018, 40, 1395–1406. [Google Scholar] [CrossRef]
- dos Santos, D.F.K.; Istvan, P.; Noronha, E.F.; Quirino, B.F.; Krüger, R.H. New dioxygenase from metagenomic library from Brazilian soil: Insights into antibiotic resistance and bioremediation. Biotechnol. Lett. 2015, 37, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, L.; Deng, Z.; Liu, S.; Yun, J.; Xiao, X.; Li, H. Screening, cloning, enzymatic properties of a novel thermostable cellulase enzyme, and its potential application on water hyacinth utilization. Int. Microbiol. 2021, 24, 337–349. [Google Scholar] [CrossRef]
- Erich, S.; Kuschel, B.; Schwarz, T.; Ewert, J.; Böhmer, N.; Niehaus, F.; Eck, J.; Lutz-Wahl, S.; Stressler, T.; Fischer, L. Novel high-performance metagenome β-galactosidases for lactose hydrolysis in the dairy industry. J. Biotechnol. 2015, 210, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Lezyk, M.; Jers, C.; Kjaerulff, L.; Gotfredsen, C.H.; Mikkelsen, M.D.; Mikkelsen, J.D. Novel α-L-fucosidases from a soil metagenome for production of fucosylated human milk oligosaccharides. PLoS ONE 2016, 11, e0147438. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Oh, D.B.; Kwon, O. Characterization of a lichenase isolated from soil metagenome. J. Microbiol. Biotechnol. 2014, 24, 1699–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathya, T.A.; Jacob, A.M.; Khan, M. Cloning and molecular modelling of pectin degrading glycosyl hydrolase of family 28 from soil metagenomic library. Mol. Biol. Rep. 2014, 41, 2645–2656. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Kamei, I.; Hirai, H.; Kondo, R. Identification of novel glycosyl hydrolases with cellulolytic activity against crystalline cellulose from metagenomic libraries constructed from bacterial enrichment cultures. SpringerPlus 2014, 3, 365. [Google Scholar] [CrossRef] [Green Version]
- Chai, S.; Zhang, X.; Jia, Z.; Xu, X.; Zhang, Y.; Wang, S.; Feng, Z. Identification and characterization of a novel bifunctional cellulase/hemicellulase from a soil metagenomic library. Appl. Microbiol. Biotechnol. 2020, 104, 7563–7572. [Google Scholar] [CrossRef] [PubMed]
- Faoro, H.; Glogauer, A.; Couto, G.H.; de Souza, E.M.; Rigo, L.U.; Cruz, L.M.; Monteiro, R.A.; de Oliveira Pedrosa, F. Characterization of a new Acidobacteria-derived moderately thermostable lipase from a Brazilian Atlantic Forest soil metagenome. FEMS Microbiol. Ecol. 2012, 81, 386–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.K.; Han, Y.J.; Hahm, M.S.; Park, S.Y.; Hwang, I.T. Isolation and characterization of a novel triolein selective lipase from soil environmental genes. Microbiol. Biotechnol. Lett. 2021, 48, 480–490. [Google Scholar] [CrossRef]
- Jin, P.; Pei, X.; Du, P.; Yin, X.; Xiong, X.; Wu, H.; Zhou, X.; Wang, Q. Overexpression and characterization of a new organic solvent-tolerant esterase derived from soil metagenomic DNA. Bioresour. Technol. 2012, 116, 234–240. [Google Scholar] [CrossRef]
- Biver, S.; Vandenbol, M. Characterization of three new carboxylic ester hydrolases isolated by functional screening of a forest soil metagenomic library. J. Ind. Microbiol. Biotechnol. 2013, 40, 191–200. [Google Scholar] [CrossRef]
- Jeon, J.H.; Lee, H.S.; Lee, J.H.; Koo, B.S.; Lee, C.M.; Lee, S.H.; Kang, S.G.; Lee, J.H. A novel family VIII carboxylesterase hydrolysing third- and fourth-generation cephalosporins. SpringerPlus 2016, 5, 525. [Google Scholar] [CrossRef] [Green Version]
- Park, J.M.; Won, S.M.; Kang, C.H.; Park, S.; Yoon, J.H. Characterization of a novel carboxylesterase belonging to family VIII hydrolyzing β-lactam antibiotics from a compost metagenomic library. Int. J. Biol. Macromol. 2020, 164, 4650–4661. [Google Scholar] [CrossRef]
- Nagayama, H.; Sugawara, T.; Endo, R.; Ono, A.; Kato, H.; Ohtsubo, Y.; Nagata, Y.; Tsuda, M. Isolation of oxygenase genes for indigo-forming activity from an artificially polluted soil metagenome by functional screening using Pseudomonas putida strains as hosts. Appl. Microbiol. Biotechnol. 2015, 99, 4453–4470. [Google Scholar] [CrossRef] [PubMed]
- Biver, S.; Portetelle, D.; Vandenbol, M. Characterization of a new oxidant-stable serine protease isolated by functional metagenomics. SpringerPlus 2013, 2, 410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, G.L.; Slingo, J.M.; Rignot, E.; Reager, J.T.; Hakuba, M.Z.; Durack, P.J.; Worden, J.; Rocca, R. Earth’s water reservoirs in a changing climate. Proc. R. Soc. A Math. Phys. Eng. Sci. 2020, 476, 20190458. [Google Scholar] [CrossRef] [Green Version]
- Parages, M.L.; Gutiérrez-Barranquero, J.A.; Reen, F.J.; Dobson, A.D.W.; O’Gara, F. Integrated (Meta) genomic and synthetic biology approaches to develop new biocatalysts. Mar. Drugs 2016, 14, 62. [Google Scholar] [CrossRef]
- Jiang, T.; Sun, S.; Chen, Y.; Qian, Y.; Guo, J.; Dai, R.; An, D. Microbial diversity characteristics and the influence of environmental factors in a large drinking-water source. Sci. Total Environ. 2021, 769, 144698. [Google Scholar] [CrossRef]
- Kamble, P.; Vavilala, S.L. Discovering novel enzymes from marine ecosystems: A metagenomic approach. Bot. Mar. 2018, 61, 161–175. [Google Scholar] [CrossRef]
- Fang, W.; Liu, J.; Hong, Y.; Peng, H.; Zhang, X.; Sun, B.; Xiao, Y. Cloning and characterization of a β -glucosidase from marine microbial metagenome with excellent glucose tolerance. J. Microbiol. Biotechnol. 2010, 20, 1351–1358. [Google Scholar] [CrossRef]
- Wierzbicka-Woś, A.; Bartasun, P.; Cieśliński, H.; Kur, J. Cloning and characterization of a novel cold-active glycoside hydrolase family 1 enzyme with β-glucosidase, β-fucosidase and β-galactosidase activities. BMC Biotechnol. 2013, 13, 22. [Google Scholar] [CrossRef] [Green Version]
- Toyama, D.; de Morais, M.A.B.; Ramos, F.C.; Zanphorlin, L.M.; Tonoli, C.C.C.; Balula, A.F.; de Miranda, F.P.; Almeida, V.M.; Marana, S.R.; Ruller, R.; et al. A novel β-glucosidase isolated from the microbial metagenome of Lake Poraquê (Amazon, Brazil). Biochim. Biophys. Acta Prot. Proteom. 2018, 1866, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, Y.M.; Ghazy, M.A.; Sayed, A.; Ouf, A.; El-Dorry, H.; Siam, R. Isolation and characterization of a heavy metal-resistant, thermophilic esterase from a Red Sea Brine Pool. Sci. Rep. 2013, 3, 3358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Z.; Li, J.; Wang, Q.; Fang, W.; Peng, H.; Zhang, X.; Xiao, Y. A novel esterase from a marine metagenomic library exhibiting salt tolerance ability. J. Microbiol. Biotechnol. 2014, 24, 771–780. [Google Scholar] [CrossRef] [PubMed]
- López-López, O.; Knapik, K.; Cerdán, M.E.; González-Siso, M.I. Metagenomics of an alkaline hot spring in Galicia (Spain): Microbial diversity analysis and screening for novel lipolytic enzymes. Front. Microbiol. 2015, 6, 1291. [Google Scholar] [CrossRef] [Green Version]
- Fang, Z.; Li, T.; Wang, Q.; Zhang, X.; Peng, H.; Fang, W.; Hong, Y.; Ge, H.; Xiao, Y. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl. Microbiol. Biotechnol. 2011, 89, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.; Ghazy, M.A.; Ferreira, A.J.S.; Setubal, J.C.; Chambergo, F.S.; Ouf, A.; Adel, M.; Dawe, A.S.; Archer, J.A.C.; Bajic, V.B.; et al. A novel mercuric reductase from the unique deep brine environment of Atlantis II in the Red Sea. J. Biol. Chem. 2014, 289, 1675–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badiea, E.A.; Sayed, A.A.; Maged, M.; Fouad, W.M.; Said, M.M.; Esmat, A.Y. A novel thermostable and halophilic thioredoxin reductase from the Red Sea Atlantis II hot brine pool. PLoS ONE 2019, 14, e0217565. [Google Scholar] [CrossRef] [PubMed]
- Witte, B.; John, D.; Wawrik, B.; Paul, J.H.; Dayan, D.; Robert Tabita, F. Functional prokaryotic rubisCO from an oceanic metagenomic library. Appl. Environ. Microbiol. 2010, 76, 2997–3003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; Wu, L.L.; Zhao, G.C.; Shen, P.H.; Jin, K.; Hao, Z.Y.; Li, S.X.; Ma, G.F.; Luo, F.F.; Hu, G.Q.; et al. Identification and characterization of a novel fumarase gene by metagenome expression cloning from marine microorganisms. Microb. Cell Fact. 2010, 9, 91. [Google Scholar] [CrossRef] [Green Version]
- Sonbol, S.A.; Ferreira, A.J.S.; Siam, R. Red Sea Atlantis II brine pool nitrilase with unique thermostability profile and heavy metal tolerance. BMC Biotechnol. 2016, 16, 14. [Google Scholar] [CrossRef] [Green Version]
- Elbehery, A.H.A.; Leak, D.J.; Siam, R. Novel thermostable antibiotic resistance enzymes from the Atlantis II Deep Red Sea brine pool. Microb. Biotechnol. 2017, 10, 189–202. [Google Scholar] [CrossRef]
- Sobat, M.; Asad, S.; Kabiri, M.; Mehrshad, M. Metagenomic discovery and functional validation of L-asparaginases with anti-leukemic effect from the Caspian Sea. iScience 2021, 24, 101973. [Google Scholar] [CrossRef]
- Shu, W.S.; Huang, L.N. Microbial diversity in extreme environments. Nat. Rev. Microbiol. 2021, 20, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Finore, I.; Romano, I.; Gioiello, A.; Lama, L.; Nicolaus, B. Microbial diversity in extreme marine habitats and their biomolecules. Microorganisms 2017, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Ojaveer, H.; Jaanus, A.; Mackenzie, B.R.; Martin, G.; Olenin, S.; Radziejewska, T.; Telesh, I.; Zettler, M.L.; Zaiko, A. Status of biodiversity in the Baltic Sea. PLoS ONE 2010, 5, e12467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sass, A.M.; Sass, H.; Coolen, M.J.L.; Cypionka, H.; Overmann, J. Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania Basin, Mediterranean Sea). Appl. Environ. Microbiol. 2001, 67, 5392–5402. [Google Scholar] [CrossRef] [Green Version]
- Borah, P.; Kumar, M.; Devi, P. Types of inorganic pollutants: Metals/metalloids, acids, and organic forms. In Inorganic Pollutants in Water; Devi, P., Singh, P., Kansal, S.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 17–31. ISBN 9780128189658. [Google Scholar]
- Ouyang, L.M.; Liu, J.Y.; Qiao, M.; Xu, J.H. Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl. Biochem. Biotechnol. 2013, 169, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.N.; Gupta, A.; Singh, V.S.; Mishra, R.; Kateriya, S.; Tripathi1, A.K. Identification and characterization of a novel phosphodiesterase from the metagenome of an Indian coalbed. PLoS ONE 2015, 10, e0118075. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Li, F.; Wang, L.; Zhu, Y.; Dong, Z.; Bai, L. Discovery and characterizaton of a novel lipase with transesterification activity from hot spring metagenomic library. Biotechnol. Rep. 2017, 14, 27–33. [Google Scholar] [CrossRef]
- Kotik, M.; Vanacek, P.; Kunka, A.; Prokop, Z.; Damborsky, J. Metagenome-derived haloalkane dehalogenases with novel catalytic properties. Appl. Microbiol. Biotechnol. 2017, 101, 6385–6397. [Google Scholar] [CrossRef]
- Apolinar, M.M.; Peña, Y.J.; Pérez-rueda, E.; Canto-canché, B.B.; Santos-briones, C.D.L.; Connor-sánchez, A.O. Identi fi cation and in silico characterization of two novel genes encoding peptidases S8 found by functional screening in a metagenomic library of Yucatán underground water. Gene 2016, 593, 154–161. [Google Scholar] [CrossRef]
- Aviv-Reuven, S.; Rosenfeld, A. Publication patterns’ changes due to the COVID-19 pandemic: A longitudinal and short-term scientometric analysis. Scientometrics 2021, 126, 6761–6784. [Google Scholar] [CrossRef]
- Lai, O.M.; Lee, Y.Y.; Phuah, E.T.; Akoh, C.C. Lipase/esterase: Properties and industrial applications. In Encyclopedia of Food Chemistry; Melton, L., Shahidi, P.V.F., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 158–167. ISBN 9780128140451. [Google Scholar]
- Linares-Pasten, J.; Andersson, M.; Karlsson, E. Thermostable glycoside hydrolases in biorefinery technologies. Curr. Biotechnol. 2014, 3, 26–44. [Google Scholar] [CrossRef]
- Liu, X.; Kokare, C. Microbial enzymes of use in industry. In Biotechnology of Microbial Enzymes; Brahmachari, G., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 267–298. [Google Scholar]
- Martínez, A.T.; Ruiz-Dueñas, F.J.; Camarero, S.; Serrano, A.; Linde, D.; Lund, H.; Vind, J.; Tovborg, M.; Herold-Majumdar, O.M.; Hofrichter, M.; et al. Oxidoreductases on their way to industrial biotransformations. Biotechnol. Adv. 2017, 35, 815–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alneyadi, A.H.; Rauf, M.A.; Ashraf, S.S. Oxidoreductases for the remediation of organic pollutants in water—A critical review. Crit. Rev. Biotechnol. 2018, 38, 971–988. [Google Scholar] [CrossRef] [PubMed]
- Jatuwong, K.; Suwannarach, N.; Kumla, J.; Penkhrue, W.; Kakumyan, P.; Lumyong, S. Bioprocess for production, characteristics, and biotechnological applications of fungal phytases. Front. Microbiol. 2020, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.F.; Shang, C.; Zelazny, L.W. Measurement of phytase activity in soil using a chromophoric tethered phytic acid probe. Soil Biol. Biochem. 2009, 41, 192–200. [Google Scholar] [CrossRef]
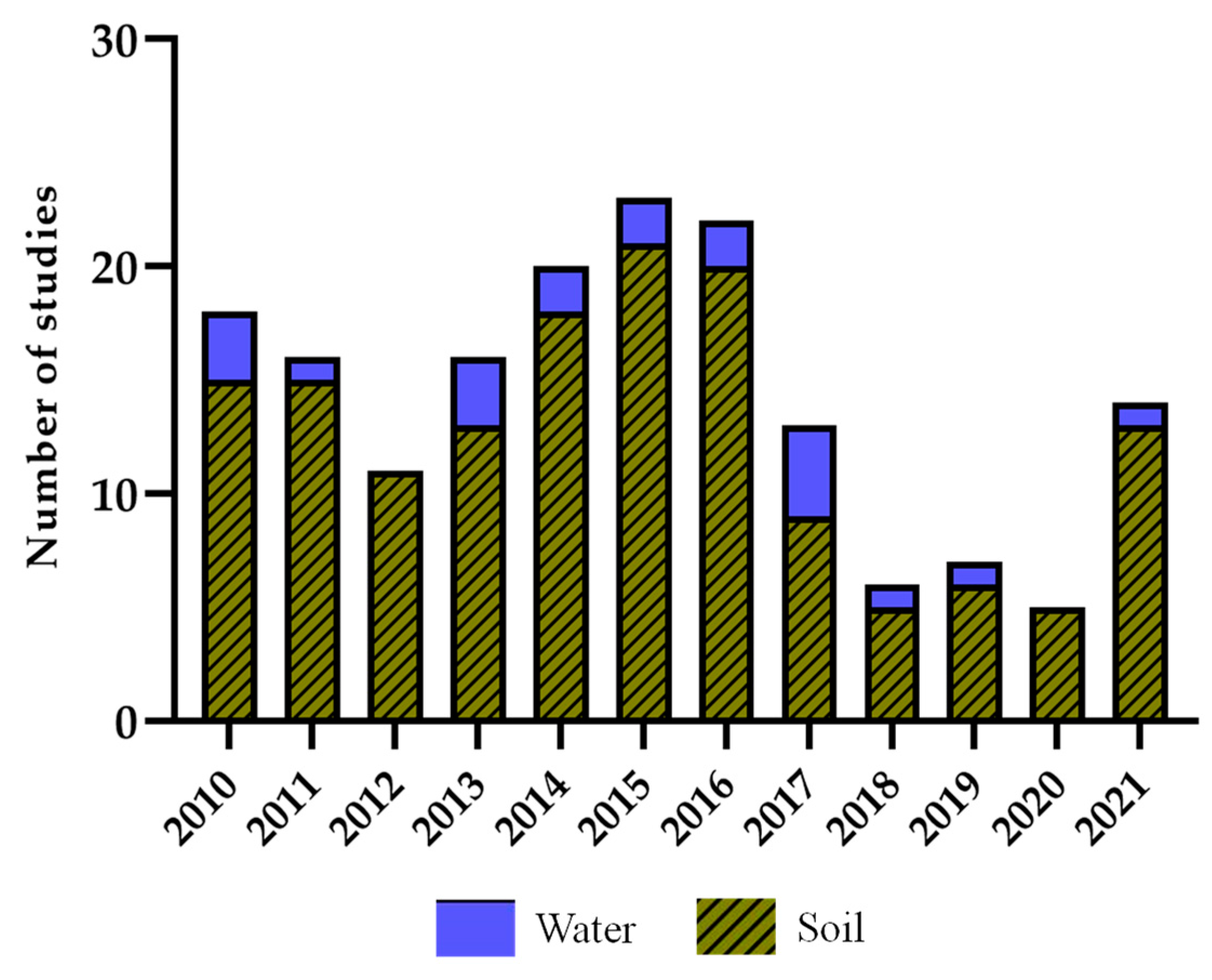

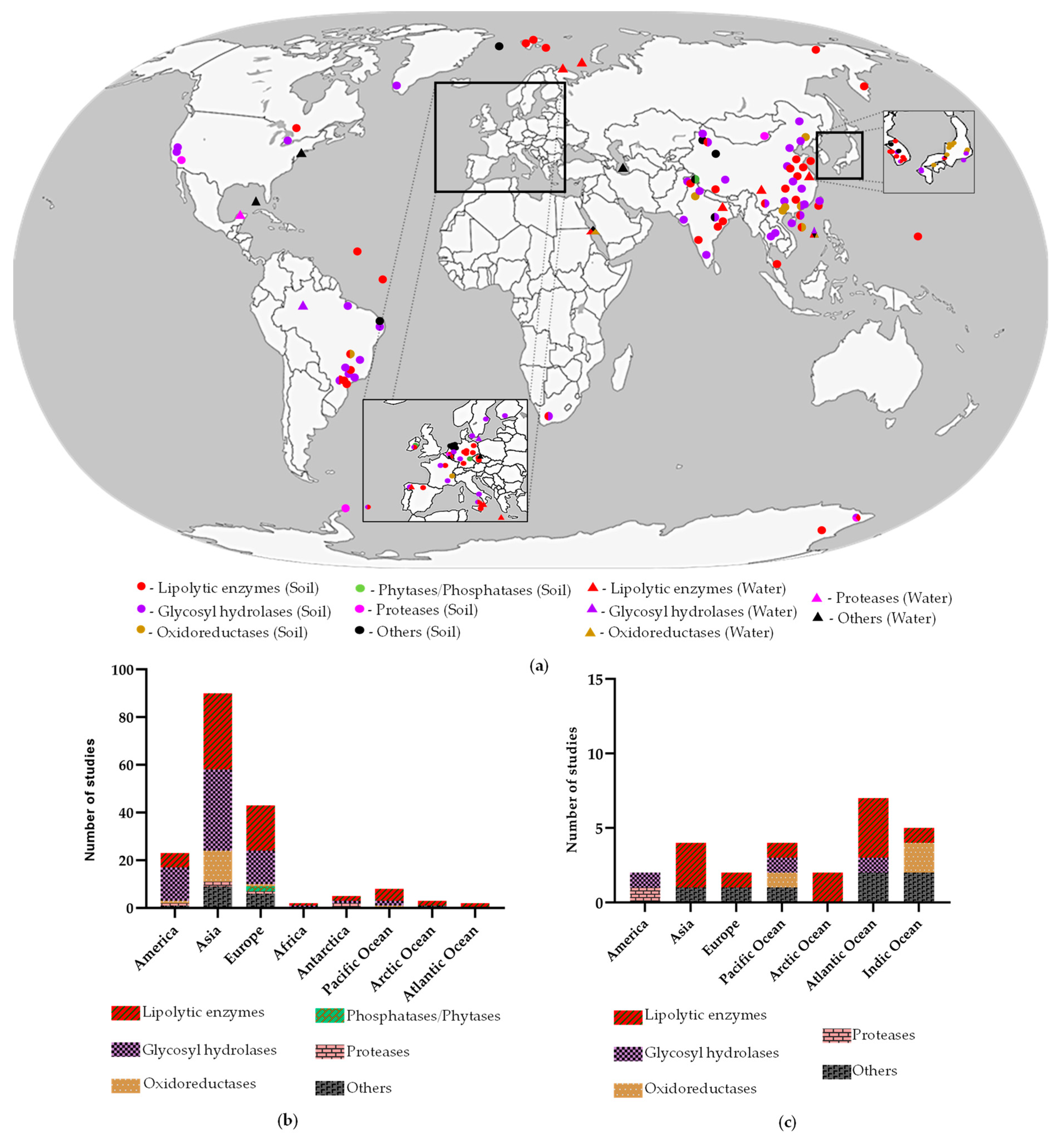

| Sample Source | Source Properties | Metagenomic Approach | Vector/ Host | No. Positive Clones/ Total Clones | Enzyme | No. Citations * | Reference | |
|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | |||||||
| Astaka region, Kargil, Northwestern Himalayas (India) | - | - | Functional | Cosmid/E. coli | 1/35,000 | Amylase | 42 | [19] |
| Deep-sea, South China Sea (Pacific Ocean) | - | - | Functional | Fosmid/E. coli | 1/20,000 | α-amylase | 32 | [20] |
| Ikaite tufa columns, Ikka Fjord (Greenland) | 10.4 | 4.0 | Sequence-based and functional | BAC/E. coli | 3/2843 2/2843 | α-amylase β-galactosidases | 70 | [21] |
| Hot sulfur springs, Nubra Valley, Leh, Northern Himalayas (India) | 7.5–8.5 | 60.0–80.0 | Functional | Plasmid/E. coli | 1/10,000 | β-d-galactosidase | 24 | [22] |
| Mountain of Flames, Turpan Basin, Xinjiang Uygur Autonomous Region (China) | - | 76.0 | Functional | Plasmid/E. coli | 1/8000 | β-galactosidase | 21 | [23] |
| Composting, EXPO Park, Osaka (Japan) | 7.5 | 67.0 | Functional | Fosmid/E. coli | 10/6000 | Cellulase | 15 | [24] |
| Farm-made composting, Khon Kaen (Thailand) | 4.2–6.8 | 30.0–50.0 | Functional | Fosmid/E. coli | 10/251 | Endocellulase | 13 | [25] |
| Composting, Tainan (Taiwan) | - | - | Functional | Lambda phage vector/E. coli | 1/2739 | Endoglucanase | 30 | [26] |
| Hydrothermal vent, Vulcano island (Italy) | 5.9 | 100.0 | Sequence-based | - | - | Endo-β-glucanase | 14 | [27] |
| Caldeirão hot spring, Furnas Valley, São Miguel Island, Azores (Portugal) | 6.0–7.0 | 60.0–70.0 | Functional | Plasmid/E. coli | - | β-glucosidase | 70 | [28] |
| Turpan Basin, Xinjiang Uygur Autonomous Region (China) | - | 82.0 | Functional | Plasmid/E. coli | 5/50,000 | β-glucosidase | 68 | [29] |
| Forest, National Institute of Advanced Industrial Science and Technology, Tsukuba (Japan) | - | 15.0 | Functional | Fosmid/E. coli | 1/50,000 | β-glycosidase | 22 | [30] |
| Suruga Bay, Shizuoka, Japan (Pacific Ocean) | - | - | Sequence-based | - | - | β-glucosidases Endomannanase Endoxylanases β-xylosidases | 17 | [31] |
| Tattapani thermal spring, Chhattisgarh (India) | 7.5–8.3 | 55.0–98.0 | Sequence-based | - | - | β-glucosidase Xylanase | 24 | [32] |
| Thermophilic composting, Viçosa and Urucânia, Minas Gerais (Brazil) | - | 55.0–65.0 | Functional | Fosmid/E. coli | 159/6720 9/6720 14/6720 | Cellulase Xylanase β-glucosidase | 11 | [33] |
| Solfatara-Pisciarelli hydrothermal pool, Agnano, Naples (Italy) | 1.5 | 92.0 | Sequence-based | - | - | β-mannanase/ β-1,3-glucosidase β-N-acetylglucosaminidase/β-glucosidase | 11 | [34] |
| Hot spring compost-soil, Fukuoka (Japan) | 3.0–4.5 | - | Functional | Plasmid/E. coli | - | Xylanase | 73 | [35] |
| Lobios hot spring, Ourense, Galicia (Spain) | 5.9 | 76.0 | Sequence-based and functional | Fosmid/E. coli | 1/150,000 | Xylanase | 15 | [36] |
| Forest, National Institute of Advanced Industrial Science and Technology, Tsukuba (Japan) | - | 15.0 | Functional | Fosmid/E. coli | 1/50,000 | α-xylosidase | 21 | [37] |
| Haiyan wetland, Qinghai-Tibetan Plateau (China) | - | - | Sequence-based | - | - | Chitinase | 1 | [38] |
| Cold desert, McMurdo Dry Valleys, South Victoria Land (Antarctica) | - | 3.0 | Functional | Fosmid/E. coli | -/10,000 | Esterase | 38 | [39] |
| Lake Arreo, Álava (Spain) | 8.0 | 6.9 | Functional | Fosmid/E. coli | 10/11,520 | Esterases/lipases | 39 | [40] |
| Apharwat mountain, Jammu and Kashmir, North-western Himalayas (India) | - | - | Functional | Plasmid/E. coli | 1/10,000 | Thioesterase | 1 | [41] |
| Composting, EXPO Park, Osaka (Japan) | 7.5 | 67.0 | Functional | Fosmid/E. coli | 19/6000 | Esterase | 9 | [42] |
| Hot spring, Furnas Valley, São Miguel Island, Azores (Portugal) | 2.0–7.0 | 60.0–62.0 | Sequence-based and functional | Fosmid/E. coli Fosmid/ T. thermophilus | 1/6048 5/6048 | Esterase | 50 | [43] |
| Apharwat mountain, Jammu and Kashmir, North-western Himalayas (India) | - | - | Functional | Plasmid/E. coli | 3/10,000 | Esterase | 3 | [44] |
| Karuola glacier, Tibetan Plateau (China) | - | - | Functional | Fosmid/E. coli | 5/10,000 | Esterase | 10 | [45] |
| Solnechny hot spring, Uzon Caldera, Kronotsky reserve, Kamchatka (Russia) | 5.8–6.0 | 61.0–64.0 | Sequence-based | - | - | Esterase | 39 | [46] |
| Permafrost, Kolyma Lowland, North-eastern Siberia (Russia) | - | - | Functional | Fosmid/E. coli | 7/5000 | Esterase | 30 | [47] |
| Deep-sea, Barents Sea (Arctic Ocean) | - | - | Functional | Fosmid/E. coli | 19/3884 | Esterase | 24 | [48] |
| Deep-sea, Svalbard e Jan Mayen (Arctic Ocean) | - | - | Functional | Fosmid/E. coli | 19/3884 | Esterase | ||
| Yellow Sea, China (Pacific Ocean) | - | - | Functional | Fosmid/E. coli | 34/40,000 | Esterase | 42 | [49] |
| Acidic pool, Vulcano island (Italy) | - | ≈25.0 | Functional | Fosmid/E. coli | 44/1920 | Carboxylesterases | 44 | [50] |
| Deep-sea (Atlantic Ocean) | - | - | Functional | Fosmid/E. coli | 6/17,000 | Esterase | 7 | [51] |
| Deep-sea (Atlantic Ocean) | - | - | Functional | Fosmid/E. coli | 6/17,000 | Esterase | ||
| Solar saltern, Ribandar, Goa (India) | 7.5 | 35.0 | Functional | Fosmid/E. coli | 1/5100 | Esterase | 14 | [52] |
| Qiongdongnan Basin, South China Sea (Pacific Ocean) | - | - | Functional | Fosmid/E. coli | 19/40,000 | Esterase | 68 | [53] |
| Qiongdongnan Basin, South China Sea (Pacific Ocean) | - | - | Functional | Fosmid/E. coli | 1/200,000 | Esterase | 52 | [54] |
| Deep-sea, South China Sea (Pacific Ocean) | - | - | Functional | Plasmid/E. coli | 15/60,000 | Esterase | 43 | [55] |
| Kongsfjorden seashore, Ny-Alesund, Svalbard Archipelago (Norway) | - | - | Functional | Fosmid/E. coli | 2/60,000 | Esterase | 61 | [56] |
| Turpan Basin, Xinjiang Uygur Autonomous Region (China) | - | 82.0 | Functional | Plasmid/E. coli | 3/21,000 | Pyrethroid- hydrolysing esterase | 49 | [57] |
| Deep-Sea (Pacific Ocean) | - | 1.5 | Functional | Fosmid/E. coli | 12/20,000 | Esterase | 43 | [58] |
| Composting, EXPO Park, Osaka (Japan) | 7.5 | 67.0 | Functional | Fosmid/E. coli | 19/6000 | Cutinase | 180 | [59] |
| Taptapani hot spring, Odisha (India) | 8.5 | 50.0 | Functional | Plasmid/E. coli | 7/13,298 | Lipase | 21 | [60] |
| Taptapani hot spring, Odisha (India) | 8.5 | 50.0 | Functional | Plasmid/E. coli | 7/13,000 | Lipase | 4 | [61] |
| Qiongdongnan Basin, South China Sea (Pacific Ocean) | - | - | Functional | Plasmid/E. coli | 1/60,000 | Laccase | 28 | [62] |
| Hot spring, Jiuqu, Guangxi (China) | - | 65.0–68.0 | Sequence-based | - | - | Aldehyde dehydrogenase | 6 | [63] |
| Ikahama hot spring, Tonami, Toyama (Japan) | - | - | Sequence-based | Plasmid/E. coli | 240/2800 | Alcohol dehydrogenases | 18 | [64] |
| Farm composting, Toyama (Japan) | - | 35.0–45.0 | Sequence-based and functional | Plasmid/E. coli | 1200/2000 | Alcohol dehydrogenases | 15 | [65] |
| Death Valley desert dune, California (USA) | - | - | Functional | Plasmid/E. coli | 1/30,000 | Serine proteases | 48 | [66] |
| Gobi Desert dune, Ulaanbadrakh (Mongolia) | Fosmid/E. coli | 16/17,000 | ||||||
| Chumathang hot spring, Ladakh region, North-western Himalayas (India) | - | - | Functional | Plasmid/E. coli | 1/9000 | Protease | 10 | [67] |
| Creek soil, Ardley Island, FILDES Peninsula (Antarctica) | - | - | Functional | Fosmid/E. coli | 7/- | Protease | 0 | [68] |
| Composting, EXPO Park, Osaka (Japan) | 7.0 | 50.0 | Functional | Plasmid/E. coli | 17/300,000 | RNase H | 10 | [69] |
| Ladakh region, North-western Himalayas (India) | - | - | Functional | Plasmid/E. coli | 1/8500 | Rhodanase | 4 | [70] |
| Caatinga biome, João Câmara, Rio Grande do Norte (Brazil) | - | 21.0–32.0 | Functional | Plasmid/E. coli | 1/2688 | Exonuclease | 12 | [71] |
| Deep-sea (Arctic Ocean) | - | - | Sequence-based | Plasmid/E. coli | 1/2750 | Chitin deacetylase | 11 | [72] |
| Tattapani thermal spring, Chhattisgarh (India) | 7.7 | 98.0 | Sequence-based | - | - | Trehalose synthase | 0 | [73] |
| Sample Source | Source Properties | Metagenomic Approach | Vector/ Host | No. Positive Clones/ Total Clones | Enzyme | No. Citations * | Reference | |
|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | |||||||
| Daqing oil field, Heilongjiang (China) | - | - | Functional | Plasmid/E. coli | 3/12,000 | β-galactosidase | 47 | [80] |
| Agricultural corn field, Elora, Ontario (Canada) | 7.8 | - | Functional | Cosmid/E. coli | 161/79,060 | β-galactosidase | 28 | [81] |
| Grover Soil Solutions compost facility, California (USA) | - | - | Sequence-based | - | - | Cellulase | 139 | [82] |
| Sugarcane land field, São Carlos (Brazil) | - | - | Functional | Plasmid/E. coli | 1/26,900 | Cellulase | 43 | [83] |
| Pollutants- contaminated stream ground surface, Guangxi (China) | 9.5 | - | Functional | Plasmid/E. coli | 2/30,000 | β-glucosidases | 40 | [84] |
| Yingtan Red Soil Ecological Station, Jiangxi (South China) | - | - | Functional | Plasmid/E. coli | 1/3024 | Endo-β-1,4- glucanase | 54 | [85] |
| Straw stook, Jiangxia, Wuhan (China) | - | - | Functional | Plasmid/E. coli | 1/24,000 | Endoglucanase | 12 | [86] |
| Agricultural fields irrigated with effluents of paper and pulp mill, Uttarakhand (India) | - | - | Functional | Fosmid/E. coli | 1/7500 | β-1,4- endoglucanase | 6 | [87] |
| Shek Wu Hui Sewage Treatment Works, Hong Kong (China) | - | 55.0 | Sequence-based | - | - | Endo-β-1,4- glucanases | 50 | [88] |
| Composting, State University of Paraíba composting cells, Campina Grande (Brazil) | - | 55.0–70.0 | Functional | Cosmid/E. coli | 1/10,000 | Endoglucanase | 16 | [89] |
| Sugarcane land field, São Carlos (Brazil) | - | - | Functional | Plasmid/E. coli | 1/26,900 | Endoglucanase | 15 | [90] |
| Yonghyeon Nonghyup compost factory, Sacheon (South Korea) | 8.9–9.2 | 40.0–73.0 | Functional | Fosmid/E. coli | 2/12,380 | Endo-β-1,4- glucanases | 10 | [91] |
| Grassland rhizosphere, Teagasc Oak Park research facility, Carlow (Ireland) | - | - | Functional | Fosmid/E. coli | 1/45,000 | Endo-β-1,4- glucanase | 14 | [92] |
| Municipal compost platform, Bailly (France) | - | 50.0 | Functional | Fosmid/E. coli | 74/48,000 | Endoglucanases | 2 | [93] |
| Paddy soil, Liaoning (China) | - | - | Functional | Fosmid/E. coli | -/25,000 | β-glucanase | 26 | [94] |
| University of Agricultural Sciences field, Uppsala (Sweden) | 6.9 | - | Sequence-based | Fosmid/E. coli | -/7800 | Chitinase | 64 | [95] |
| University of Agricultural Sciences field, Uppsala (Sweden) | 6.9 | - | Functional | Fosmid/E. coli | 1/7800 | Chitinase | 57 | [96] |
| Chitin-treated agricultural field, Experimental farm “Vredepeel” (The Netherlands) | 5.7 | - | Sequence-based and functional | Fosmid/E. coli | 5/145,000 | Chitinase | 17 | [97] |
| Chitin- contaminated soil, Mahtani Chitosan, Gujarat (India) | - | - | Sequence-based | - | - | Chitinase | 13 | [98] |
| Fat-contaminated soil, industrial wastewater treatment plant lagoon, Paraná (Brazil) | - | 30.0 | Functional | Fosmid/E. coli | 15/500,000 | Chitinase | 15 | [99] |
| Carnoulès acid- mine drainage, Gard (France) | 3.8 | 15.1 | Functional | Plasmid/E. coli | 28/80,000 | Amylases | 16 | [100] |
| Oil-contaminated mangrove site, Bertioga, São Paulo (Brazil) | - | - | Functional | Fosmid/E. coli | 1/12,960 | β-N- acetylhexosaminidase | 8 | [101] |
| Yonghyeon Nonghyup compost factory, Sacheon (South Korea) | 8.9–9.2 | 40.0–73.0 | Functional | Fosmid/E. coli | 5/12,380 | Xylanase | 30 | [102] |
| Sugarcane land field, São Carlos (Brazil) | - | - | Functional | Plasmid/E. coli | 1/26,900 | Endoxylanase | 20 | [103] |
| Ämmässuo composting plant (Finland) | - | - | Functional | Fosmid/E. coli Plasmid/E. coli | 21/43,000 18/40,000 1/760,000 | Xylanases | 8 | [104] |
| Jiaozuo Ruifeng Paper Co., Ltd., Jiaozuo, Henan province (China) | - | - | Sequence-based | - | - | Xylanase | 2 | [105] |
| Commercial compost production facility, Western Cape (South Africa) | - | 70.0 | Functional | Fosmid/E. coli | 26/20,000 | β-xylosidase | 0 | [106] |
| Yonghyeon Nonghyup compost factory, Sacheon (South Korea) | 8.9–9.2 | 40.0–73.0 | Functional | Fosmid/E. coli | 2/12,380 5/12,380 | Cellulase Xylanases | 16 | [107] |
| Grover Soil Solutions and the Jepson Prairie Organics compost factories, California (USA) | - | - | Sequence-based | - | - | Endoglucanase Xylanase | 76 | [108] |
| Grover Soil Solutions compost facility, California (USA) | - | - | Sequence- based | - | - | β-xylosidase/ α-arabinofuranosidase Endoxylanases α-fucosidase | 41 | [109] |
| Industrial bagasse collection site, Phu Khieo Bio-Energy, Chaiyapoom (Thailand) | - | 49.0–52.0 | Functional | Fosmid/E. coli | 7/100,000 | Endoglucanase Endoxylanase | 34 | [110] |
| Oil- contaminated soils, Ile des Petrels, Terre Adélie (Antarctica) | - | 12.0 | Functional | BAC/E. coli | 14/113,742 14/113,742 3/113,742 11/113,742 | Lipases/esterases Amylases Proteases Cellulases | 32 | [111] |
| Experimental field luvisoil, Gembloux (Belgium) | 6.5–7.0 | - | Functional | Yeast episomal shuttle vector/ E. coli | 7/500,000 2/500,000 | β-glycosidases Glycosyltransferases | 7 | [112] |
| Swine wastewater treatment facility, Tainan (Taiwan) | 7.6 | 30.0 | Sequence-based and functional | Plasmid/E. coli | 13/3818 | Esterases | 37 | [113] |
| Yonghyeon Nonghyup compost factory, Sacheon (South Korea) | 8.9–9.2 | 40.0–73.0 | Functional | Fosmid/E. coli | 19/23,400 | Esterase | 52 | [114] |
| Plants rhizosphere, Gyeongsang (South Korea) | - | - | Functional | Fosmid/E. coli | 14/142,900 | Esterase | 42 | [115] |
| Wheat field, Shouguang, Shandong (China) | - | - | Functional | Plasmid/E. coli | 6/50,000 | Feruloyl esterase | 18 | [116] |
| Petroleum hydrocarbons- contaminated region, Ribeirão Preto, São Paulo (Brazil) | - | - | Functional | Fosmid/E. coli | 30/4224 | Esterase | 21 | [117] |
| Commercial compost production facility, Western Cape (South Africa) | 6.1 | 70.0 | Sequence-based and functional | Fosmid/E. coli | 25/110,592 | Feruloyl esterase | 12 | [118] |
| Bioenergiezentrum GmbH compost facility, Göttingen (Germany) | - | 63.3 | Sequence-based and functional | Fosmid/E. coli Fosmid/ T. thermophilus | 1/1920 1/1920 | Esterase | 50 | [43] |
| Yonghyeon Nonghyup compost factory, Sacheon (South Korea) | 8.9–9.2 | 40.0–73.0 | Functional | Fosmid/E. coli | 18/23,400 | Esterase | 28 | [119] |
| Tembec Paper Mill, Temiscaming, Ontario (Canada) | - | 25.0–35.0 | Functional | Plasmid/E. coli | 1/53,500 | Carboxylesterases | 44 | [50] |
| Waste water treatment plant anaerobic digester, Evry (France) | - | 33.0 | Fosmid/E. coli | 254/47,616 | ||||
| Oil- contaminated harbour, Priolo Gargallo (Italy) | - | 15.0 | Plasmid/E. coli | 4/118,500 | ||||
| PAH contaminated soil, Michle (Czech Republic) | - | 20.0–25.0 | 20/99,900 | |||||
| PAH contaminated bioremediation site, Sobeslav (Czech Republic) | - | - | 5/114,000 | |||||
| Composting plant, Liemehna (Germany) | - | 30.0–50.0 | 6/60,000 | |||||
| Biodiversity Exploratories Schorfheide-Chorin forest and grassland site (Germany) | - | 8.0–8.5 | Functional | Plasmid/E. coli Fosmid/E. coli | 28/40,000–341,000 9/4600–300,000 | Esterase | 15 | [120] |
| Bioenergiezentrum GmbH compost facility, Göttingen (Germany) | - | 55.0 | Functional | Plasmid/E. coli | 279/675,200 | Esterases | 6 | [121] |
| Cotton field (China) | - | - | Functional | Plasmid/E. coli | 1/92,000 | Esterase | 0 | [122] |
| Cornfield, Shangqiu, Henan province (China) | - | - | Functional | Fosmid/E. coli | 1/30,000 | Carboxylesterase | 1 | [123] |
| Waste contaminated farmland, Nanjing, Jiangsu province (China) | - | - | Functional | Fosmid/E. coli | 9/60,000 | Carboxylesterase | 2 | [124] |
| Yonghyeon Nonghyup compost factory, Sacheon (South Korea) | 8.9–9.2 | 40.0–73.0 | Functional | Fosmid/E. coli | 19/23,400 | Esterase | 0 | [125] |
| Yonghyeon Nonghyup compost factory, Sacheon (South Korea) | 8.9–9.2 | 40.0–73.0 | Functional | Fosmid/E. coli | 19/23,400 | Esterases | 1 | [126] |
| Oil-contaminated Mud Flat, Taean (South Korea) | - | - | Functional | Plasmid/E. coli | 8/3000 | Esterases | 2 | [127] |
| Teagasc Oak Park research facility organic field trial site, Carlow (Ireland) | - | - | Functional | Fosmid/E. coli | 1/14,000 | Esterase/lipase | 9 | [128] |
| Fat-contaminated soil, Paraná (Brazil) | - | 30.0 | Functional | Fosmid/E. coli | 32/500,000 | Lipase | 129 | [129] |
| Oil-contaminated soil, Qingshan Branch oil field, Anhui (China) | - | - | Functional | Plasmid/E. coli | 6/20,000 | Lipase | 17 | [130] |
| Oil-contaminated mud flat, Taean (South Korea) | - | - | Functional | Plasmid/E. coli | 9/3000 | Lipase | 25 | [131] |
| Cotton field (China) | - | - | Functional | Plasmid/E. coli | 1/92,000 | Tannase | 28 | [132] |
| Biodiversity Exploratories Hainich-Dün forest and grassland site (Germany) | 6.5–8.0 | - | Functional | Plasmid/E. coli Fosmid/E. coli | 28/40,000–341,000 9/4600–300,000 | Lipolytic enzymes | [133] | |
| Biodiversity Exploratories Schwäbische Alb forest and grassland site (Germany) | 6.0–7.0 | 54 | ||||||
| Biodiversity Exploratories Schorfheide-Chorin forest and grassland site (Germany) | 8.0–8.5 | |||||||
| Experimental field luvisoil, Gembloux (Belgium) | 6.5–7.0 | - | Functional | Yeast episomal shuttle vector/E. coli | 19/420,000 | Lipolytic enzymes | 1 | [134] |
| Wastewater treatment facility (Japan) | - | - | Functional | Fosmid/E. coli | 1/100,000 | Bilirubin oxidase | 8 | [135] |
| Contaminated rice field, Nanning, Guangxi (China) | - | - | Sequence-based | Plasmid/E. coli | 1/32,000 | D-Amino acid oxidase | 7 | [136] |
| Coke plant wastewater treatment facility (Japan) | - | - | Functional | Fosmid/E. coli | 6/40,000 | Oxygenases | 19 | [137] |
| Pesticide industry effluent treatment plant (India) | - | - | Functional | Plasmid/E. coli | 2/40,000 | Flavin monooxygenases | 22 | [138] |
| PAH-contaminated wetland, Chambéry (France) | - | - | Sequence-based | - | - | Dioxygenases | 34 | [139] |
| Polychlorinated biphenyl-contaminated, Fushun, Liaoning (China) | 8.3 | - | Functional | Plasmid/E. coli | 1/- | 2,4-dichlorophenol hydroxylase | 23 | [140] |
| Compost-producing company, Toyama (Japan) | - | 50.0–80.0 | Sequence-based and functional | Plasmid/E. coli | 1200/2000 | Alcohol dehydrogenases | 15 | [65] |
| Farm, Knockbeg (Ireland) | - | - | Functional | Fosmid/E. coli | 28/14,400 | Phytases | 14 | [141] |
| Paddy field, Yuseong, Daejeon (South Korea) | - | - | Functional | Fosmid/E. coli | 157/326,200 | Wax ester synthase | 6 | [142] |
| Plants rhizosphere, Gyeongsang (South Korea) | ||||||||
| Lop Nur, Xinjiang Uigur Autonomous Region (China) | - | - | Functional | Plasmid/E. coli | 1/85,000 | Trehalose synthase | 34 | [143] |
| Cornfield, Turpan Basin, Xinjiang Uygur Autonomous Region (China) | - | - | Functional | Plasmid/E. coli | 1/700,000 | Glycosyltransferase | 14 | [144] |
| Municipal sewage plant, Garmerwolde (The Netherlands) | 6.4 | - | Functional | Plasmid/E. coli | 6/1,250,000 8/1,250,000 | Sulfatases Phosphotriesterases | 146 | [145] |
| Urban composting facility, Groningen (The Netherlands) | 7.9 | |||||||
| Local agricultural field (The Netherlands) | 8.2 | |||||||
| Sample Source | Source Properties | Metagenomic Approach | Vector/ Host | No. Positive Clones/ Total Clones | Enzyme | No. Citations * | Reference | |
|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | |||||||
| Shenzhen Mangrove Reserve, Guangdong (China) | - | - | Functional | Plasmid/E. coli | 1/30,000 | β-glucosidase | 64 | [155] |
| Amazon forest, Moju, Pará (Brazil) | 5.5 | - | Functional | Fosmid/E. coli | 5/97,500 | β-glucosidase | 18 | [152] |
| Eucalyptus sp. forest, UNESP campus, São Paulo (Brazil) | - | - | Sequence-based | - | - | β-glucosidase | 16 | [162] |
| Mangrove Reserve, Sanya, Hainan (China) | - | - | Functional | Fosmid/E. coli | 1/100,000 | Endo-β-1,4-glucanase | 24 | [156] |
| Junggar Basin, Xinjiang (China) | - | - | Functional | Plasmid/E. coli | 1/7200 | Cellulase | 4 | [167] |
| Garden, Zwingenberg (Germany) | - | - | Functional | Plasmid/E. coli | 6/1,335,000 | β-galactosidase | 39 | [168] |
| RGS 90 A/S environmental company, Copenhagen (Denmark) | - | - | Functional | Fosmid/E. coli | 7/100,000 | α-fucosidase | 43 | [169] |
| Forest, Daejeon (South Korea) | - | - | Functional | Plasmid/E. coli | 1/19,626 | Lichenase | 6 | [170] |
| Mangrove Reserve, Sanya, Hainan (China) | - | - | Functional | Fosmid/E. coli | 1/100,000 | β-agarase | 18 | [157] |
| Sathyamangalam forest, Erode, Tamilnadu (India) | - | - | Functional | Plasmid/E. coli | 9/2000 | Pectin degrading glycosyl hydrolase | 13 | [171] |
| Coastal saline area, Tianjin (China) | - | - | Sequence-based | Plasmid/E. coli | 12/- | Xylanase | 12 | [164] |
| Backyard, Kyushu University, Fukuoka (Japan) | - | - | Functional | Plasmid/E. coli | 5/150,000 1/150,000 | Cellulase Xylanase | 24 | [172] |
| Forest, Yunnan (China) | - | - | Functional | Cosmid/E. coli | 1/150,000 | Cellulase/ Hemicellulase | 8 | [173] |
| Mangrove forest, Pontal do Paraná (Brazil) | - | - | Functional | Fosmid/E. coli | 1/2400 | Lipase | 41 | [153] |
| Atlantic forest, Paraná (Brazil) | 3.7–4.4 | - | Functional | Fosmid/E. coli | 1/34,560 | Lipase | 25 | [174] |
| Cerrado sensu stricto area, Brazilian Institute of Geography and Statistics (Brazil) | 4.7 | - | Functional | Plasmid/E. coli | 3/6720 | Lipase | 3 | [165] |
| Mt. Jumbong reed marsh, Gangwon province (South Korea) | - | - | Functional | Plasmid/E. coli | 46/112,500 | Lipase | 0 | [175] |
| Sirinthon Peat-Swamp Forest, Narathiwat (Thailand) | 5.0 | - | Functional | Fosmid/E. coli | 6/15,000 | Esterase | 34 | [158] |
| Alluvium, Eulsukdo Island, Saha-Gu, Busan (South Korea) | - | - | Functional | Fosmid/E. coli | 50/45,300 | Esterases | 20 | [163] |
| Mount Fanjing, Tongren, Guizhou (China) | - | - | Functional | Fosmid/E. coli | 1/50,000 | Esterase | 35 | [176] |
| Göttingen beech forest, Georg-August University, Göttingen (Germany) | - | Functional | Plasmid/E. coli | 3/70,000 | Esterase | 11 | [160] | |
| Forest, Groenendaal (Belgium) | - | - | Functional | Plasmid/E. coli | 3/70,000 | Esterases | 40 | [177] |
| Chestnut grove surface, Yunnan (China) | - | - | Functional | Cosmid/E. coli | 2/100,000 | Esterases | 7 | [161] |
| Tree rhizosphere, Korea Expressway Corporation Arboretum, Jeonju (South Korea) | - | - | Functional | Cosmid/E. coli | 1/7968 | Carboxylesterase | 10 | [178] |
| Composting, Cheongyang (South Korea) | - | - | Functional | Fosmid/E. coli | 14/13,000 | Carboxylesterase | 4 | [179] |
| Shenzhen Mangrove Reserve, Guangdong (China) | - | - | Functional | Plasmid/E. coli | 1/8000 | Laccase | 69 | [154] |
| Lake Biwa, Otsu, Shiga (Japan) | - | - | Sequence-based | Plasmid/E. coli | 240/2800 | Alcohol dehydrogenases | 15 | [65] |
| Masukata Park, Uozu, Toyama (Japan) | ||||||||
| Cerrado sensu stricto area, Brazilian Institute of Geography and Statistics (Brazil) | 4.7 | - | Functional | Plasmid/E. coli Fosmid/E. coli | 3/150,000 3/65,000 | Dioxygenase | 12 | [166] |
| Forest, Ehime Research Institute of Agriculture, Forestry, and Fisheries, Matsuyama (Japan) | - | - | Functional | Cosmid/E. coli | 29/208,000 | Oxygenases | 35 | [180] |
| Schlöppnerbrunnen peatland site, Fichtelgebirge Mountains, Northeastern Bavaria (Germany) | 4.0 | - | Sequence-based | - | - | Phytase | 10 | [159] |
| Forest, Groenendaal (Belgium) | - | - | Functional | Plasmid/E. coli | 1/70,000 | Serine protease | 41 | [181] |
| Saline flats surface, Paesens-Moddergat (The Netherlands) | 8.0 | - | Functional | Plasmid/E. coli | 6/1,250,000 8/1,250,000 | Sulfatases Phosphotriesterases | 146 | [145] |
| Lauwersmeer lakeshore (The Netherlands) | 8.5 | |||||||
| Alluvium, Eulsukdo Island, Saha-Gu, Busan (South Korea) | - | - | Functional | Fosmid/E. coli | 158/326,200 | Wax ester synthase | 6 | [142] |
| Gwangneung forest, Korea National Arboretum, Gyeonggi (South Korea) | ||||||||
| Sample Source | Source Properties | Metagenomic Approach | Vector/ Host | No. Positive Clones/ Total Clones | Enzyme | No. Citations * | Reference | |
|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | |||||||
| Surface seawater, South China Sea (Pacific Ocean) | - | - | Functional | BAC/E. coli | 6/- | β-glucosidase | 95 | [186] |
| Baltic Sea, Kołobrzeg, Poland (Atlantic Ocean) | - | 0.8 | Functional | Plasmid/E. coli | 1/1100 | β-glucosidase | 47 | [187] |
| Lake Poraquê, Amazon (Brazil) | 7.1 | 29.8 | Sequence-based | - | - | β-glucosidase | 6 | [188] |
| Atlantis II brine pool, Red Sea (Indic Ocean) | 5.3 | 68.0 | Functional | Fosmid/E. coli | 5/10,656 | Esterase | 41 | [189] |
| Surface seawater, South China Sea (Pacific Ocean) | - | - | Sequence-based | - | - | Esterase | 25 | [190] |
| Lobios hot spring, Ourense, Galicia (Spain) | >8.2 | >76.0 | Sequence-based and functional | Fosmid/E. coli | 6/11,600 | Esterase | 36 | [191] |
| Urania, deep hypersaline anoxic basin interface, Mediterranean Sea (Atlantic Ocean) | - | 14.0 | Functional | Plasmid/E. coli | 41/90,800 | Carboxylesterases | 44 | [50] |
| Surface seawater, South China Sea (Pacific Ocean) | - | - | Sequence-based | BAC/E. coli | -/20,000 | Laccase | 93 | [192] |
| Atlantis II brine pool, Red Sea (Indic Ocean) | 5.4 | 68.1 | Sequence-based | - | - | Mercuric reductase | 30 | [193] |
| Atlantis II brine pool, Red Sea (Indic Ocean) | 5.7 | 68.4 | Sequence-based | - | - | Thioredoxin reductase | 4 | [194] |
| Seawater, Gulf of Mexico (Atlantic Ocean) | - | - | Sequence-based | BAC/E. coli | - | Ribulose 1,5-bisphosphate carboxylases/ oxygenases | 14 | [195] |
| Seawater, long-Term Ecosystem Observatory site, New Jersey, USA (Atlantic Ocean) | ||||||||
| Surface seawater, South China Sea (Pacific Ocean) | 8.2 | 15.0 | Sequence-based | Plasmid/E. coli | -/50,000 | Fumarase | 29 | [196] |
| Atlantis II brine pool, Red Sea (Indic Ocean) | 5.5 | 68.2 | Sequence-based | - | - | Nitrilase | 15 | [197] |
| Atlantis II brine pool, Red Sea (Indic Ocean) | 5.6 | 68.3 | Sequence-based | - | - | 3′-aminoglycoside phosphotransferase Beta-lactamase | 11 | [198] |
| Brackish water basin, Caspian Sea (Iran) | - | - | Sequence-based | - | - | L-asparaginases | 0 | [199] |
| Sample Source | Source Properties | Metagenomic Approach | Vector/ Host | No. Positive Clones/ Total Clones | Enzyme | No. Citations * | Reference | |
|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | |||||||
| Contaminated river, East China University of Science and Technology, Shanghai (China) | - | - | Functional | Fosmid/E. coli | 6/20,400 | Esterase | 19 | [205] |
| Coalbed water formation, Jharia coalfield, Jharkhand (India) | 7.2 | - | Functional | Fosmid/E. coli | 1/208 | Phosphodiesterase | 5 | [206] |
| Messina harbour, Mediterranean Sea, Italy (Atlantic Ocean) | - | 15.0 | Functional | Plasmid/E. coli | 18/24,000 | Carboxylesterases | [50] | |
| Messina harbour (Int II), Mediterranean Sea, Italy (Atlantic Ocean) | 15.0 | Fosmid/E. coli | 208/5760 | |||||
| Milazzo, Mediterranean Sea, Italy (Atlantic Ocean) | 18.0 | Plasmid/E. coli | 8/20,000 | 44 | ||||
| Oil-contaminated coastal water, Kolguev Island, Barents Sea, Russia (Arctic Ocean) | 3.0 | Plasmid/E. coli | 34/142,000 | |||||
| Oil-contaminated Murmansk Port, Barents Sea, Russia (Arctic Ocean) | 5.0 | Plasmid/E. coli | 43/108,000 | |||||
| Eryuan Niujie hot spring, Yunnan (China) | 7.0 | 58.0 | Functional | BAC/E. coli | 10/68,352 | Lipase | 11 | [207] |
| Oil industry products contaminated groudwater (Czech Republic) | 6.9 | 8.5 | Sequence-based | - | - | Haloalkane dehalogenases | 3 | [208] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, J.; Silvério, S.C.; Costa, A.M.A.; Rodrigues, L.R. Metagenomic Approaches as a Tool to Unravel Promising Biocatalysts from Natural Resources: Soil and Water. Catalysts 2022, 12, 385. https://doi.org/10.3390/catal12040385
Sousa J, Silvério SC, Costa AMA, Rodrigues LR. Metagenomic Approaches as a Tool to Unravel Promising Biocatalysts from Natural Resources: Soil and Water. Catalysts. 2022; 12(4):385. https://doi.org/10.3390/catal12040385
Chicago/Turabian StyleSousa, Joana, Sara C. Silvério, Angela M. A. Costa, and Ligia R. Rodrigues. 2022. "Metagenomic Approaches as a Tool to Unravel Promising Biocatalysts from Natural Resources: Soil and Water" Catalysts 12, no. 4: 385. https://doi.org/10.3390/catal12040385
APA StyleSousa, J., Silvério, S. C., Costa, A. M. A., & Rodrigues, L. R. (2022). Metagenomic Approaches as a Tool to Unravel Promising Biocatalysts from Natural Resources: Soil and Water. Catalysts, 12(4), 385. https://doi.org/10.3390/catal12040385










