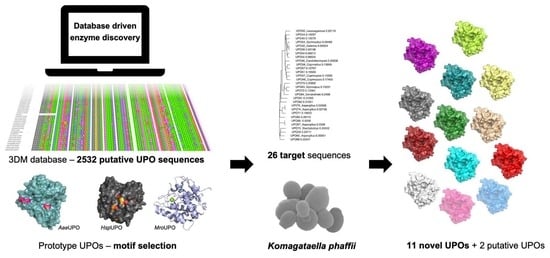
Discovery and Heterologous Expression of Unspecific Peroxygenases
Abstract
:1. Introduction
2. Results
2.1. Target Choice and Sequence Comparisons
2.2. Cloning
2.3. Screening of K. phaffi Culture Supernatants for UPO Activity and Rescreening
2.4. Shake Flask Cultivation of the Most Promising UPO Secreting K. phaffii Clones
3. Discussion
4. Materials and Methods
4.1. General
4.2. Target Choice and Sequence Comparison
4.3. Cloning of Putative UPOs and Strain Construction
4.4. Heterologous Production of Putative UPOs
4.4.1. Deep-Well Plate Cultivation (DWP)
4.4.2. Shake Flask Cultivation
4.5. Determination of UPO Activity
4.5.1. ABTS Assay
4.5.2. Naphthalene Assay
4.5.3. The 2,6- DMP Assay
4.5.4. NBD Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chakrabarty, S.; Wang, Y.; Perkins, J.C.; Narayan, A.R.H. Scalable Biocatalytic C-H Oxyfunctionalization Reactions. Chem. Soc. Rev. 2020, 49, 8137–8155. [Google Scholar] [CrossRef] [PubMed]
- Newhouse, T.; Baran, P.S. If C-H Bonds Could Talk: Selective C-H Bond Oxidation. Angew. Chemie Int. Ed. 2011, 50, 3362–3374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Van Beilen, J.B.; Duetz, W.A.; Schmid, A.; De Raadt, A.; Griengl, H.; Witholt, B. Oxidative Biotransformations Using Oxygenases. Curr. Opin. Chem. Biol. 2002, 6, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Ortiz De Montellano, P.R. Hydrocarbon Hydroxylation by Cytochrome P450 Enzymes. Chem. Rev. 2010, 110, 932–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollmann, F.; Arends, I.W.C.E.; Buehler, K.; Schallmey, A.; Bühler, B. Enzyme-Mediated Oxidations for the Chemist. Green Chem. 2011, 13, 226–265. [Google Scholar] [CrossRef]
- Ullrich, R.; Nueske, J.; Scheibner, K.; Spantzel, J.; Hofrichter, M. Novel Haloperoxidase from the Agaric Basidiomycete. Appl. Environ. Microbiol. 2004, 70, 4575–4581. [Google Scholar] [CrossRef] [Green Version]
- Kinner, A.; Rosenthal, K.; Lütz, S. Identification and Expression of New Unspecific Peroxygenases—Recent Advances, Challenges and Opportunities. Front. Bioeng. Biotechnol. 2021, 9, 705630. [Google Scholar] [CrossRef]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Liers, C.; Scheibner, K.; Kimani, V.W.; Ullrich, R. Fungal Peroxygenases: A Phylogenetically Old Superfamily of Heme Enzymes with Promiscuity for Oxygen Transfer Reactions. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 369–403. ISBN 978-3-030-29541-7. [Google Scholar]
- Hobisch, M.; Holtmann, D.; Gomez de Santos, P.; Alcalde, M.; Hollmann, F.; Kara, S. Recent Developments in the Use of Peroxygenases—Exploring Their High Potential in Selective Oxyfunctionalisations. Biotechnol. Adv. 2021, 51, 107615. [Google Scholar] [CrossRef]
- Hobisch, M.; De Santis, P.; Serban, S.; Basso, A.; Byström, E.; Kara, S. Peroxygenase-Driven Ethylbenzene Hydroxylation in a Rotating Bed Reactor. Org. Proc. Res. Dev. 2022, 26, 2761–2765. [Google Scholar] [CrossRef]
- Wang, Y.; Lan, D.; Durrani, R.; Hollmann, F. Peroxygenases En Route to Becoming Dream Catalysts. What Are the Opportunities and Challenges? Curr. Opin. Chem. Biol. 2017, 37, 1–9. [Google Scholar] [CrossRef]
- Rotilio, L.; Swoboda, A.; Ebner, K.; Rinnofner, C.; Glieder, A.; Kroutil, W.; Mattevi, A. Structural and Biochemical Studies Enlighten the Unspecific Peroxygenase from Hypoxylon Sp. EC38 as an Efficient Oxidative Biocatalyst. ACS Catal. 2021, 11, 11511–11525. [Google Scholar] [CrossRef]
- Van Rantwijk, F.; Sheldon, R.A. Selective Oxygen Transfer Catalysed by Heme Peroxidases: Synthetic and Mechanistic Aspects. Curr. Opin. Biotechnol. 2000, 11, 554–564. [Google Scholar] [CrossRef]
- Freakley, S.J.; Kochius, S.; van Marwijk, J.; Fenner, C.; Lewis, R.J.; Baldenius, K.; Marais, S.S.; Opperman, D.J.; Harrison, S.T.L.; Alcalde, M.; et al. A Chemo-Enzymatic Oxidation Cascade to Activate C–H Bonds with in Situ Generated H2O2. Nat. Commun. 2019, 10, 4178. [Google Scholar] [CrossRef] [Green Version]
- Horst, A.E.W.; Bormann, S.; Meyer, J.; Steinhagen, M.; Ludwig, R.; Drews, A.; Ansorge-Schumacher, M.; Holtmann, D. Electro-Enzymatic Hydroxylation of Ethylbenzene by the Evolved Unspecific Peroxygenase of Agrocybe aegerita. J. Mol. Catal. B Enzym. 2016, 133, S137–S142. [Google Scholar] [CrossRef]
- Churakova, E.; Kluge, M.; Ullrich, R.; Arends, I.; Hofrichter, M.; Hollmann, F. Specific Photobiocatalytic Oxyfunctionalization Reactions. Angew. Chem. Int. Ed. 2011, 50, 10716–10719. [Google Scholar] [CrossRef]
- Sundaramoorthy, M.; Terner, J.; Poulos, T.L. The Crystal Structure of Chloroperoxidase: A Heme Peroxidase-Cytochrome P450 Functional Hybrid. Structure 1995, 3, 1367–1378. [Google Scholar] [CrossRef] [Green Version]
- Piontek, K.; Strittmatter, E.; Ullrich, R.; Gröbe, G.; Pecyna, M.J.; Kluge, M.; Scheibner, K.; Hofrichter, M.; Plattner, D.A. Structural Basis of Substrate Conversion in a New Aromatic Peroxygenase: Cytochrome P450 Functionality with Benefits. J. Biol. Chem. 2013, 288, 34767–34776. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Escudero, M.; Molina-Espeja, P.; Gomez De Santos, P.; Hofrichter, M.; Sanz-Aparicio, J.; Alcalde, M. Structural Insights into the Substrate Promiscuity of a Laboratory-Evolved Peroxygenase. ACS Chem. Biol. 2018, 13, 3259–3268. [Google Scholar] [CrossRef]
- Linde, D.; Santillana, E.; Fernández-Fueyo, E.; González-Benjumea, A.; Carro, J.; Gutiérrez, A.; Martínez, A.T.; Romero, A. Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements. Antioxidants 2022, 11, 891. [Google Scholar] [CrossRef]
- Conesa, A.; Van De Velde, F.; Van Rantwijk, F.; Sheldon, R.A.; Van Den Hondel, C.A.M.J.J.; Punt, P.J. Expression of the Caldariomyces fumago Chloroperoxidase in Aspergillus niger and Characterization of the Recombinant Enzyme. J. Biol. Chem. 2001, 276, 17635–17640. [Google Scholar] [CrossRef]
- Babot, E.D.; del Río, J.C.; Kalum, L.; Martínez, A.T.; Gutiérrez, A. Oxyfunctionalization of Aliphatic Compounds by a Recombinant Peroxygenase from Coprinopsis cinerea. Biotechnol. Bioeng. 2013, 110, 2323–2332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Espeja, P.; Ma, S.; Mate, D.M.; Ludwig, R.; Alcalde, M. Tandem-Yeast Expression System for Engineering and Producing Unspecific Peroxygenase. Enzyme Microb. Technol. 2015, 73–74, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Püllmann, P.; Knorrscheidt, A.; Münch, J.; Palme, P.R.; Hoehenwarter, W.; Marillonnet, S.; Alcalde, M.; Westermann, B.; Weissenborn, M.J. A Modular Two Yeast Species Secretion System for the Production and Preparative Application of Unspecific Peroxygenases. Commun. Biol. 2021, 4, 562. [Google Scholar] [CrossRef] [PubMed]
- Bormann, S.; Kellner, H.; Hermes, J.; Herzog, R.; Ullrich, R.; Liers, C.; Ulber, R.; Hofrichter, M.; Holtmann, D. Broadening the Biocatalytic Toolbox—Screening and Expression of New Unspecific Peroxygenases. Antioxidants 2022, 11, 223. [Google Scholar] [CrossRef] [PubMed]
- Kimani, V.W. New Secretory Peroxidases and Peroxygenases from Saprotrophic Fungi of Kenyan Forests. Ph.D. Thesis, Technical University Dresden, Dresden, Germany, 2019. [Google Scholar]
- Kiebist, J.; Schmidtke, K.; Zimmermann, J.; Kellner, H.; Jehmlich, N.; Ullrich, R.; Zänder, D.; Hofrichter, M.; Scheibner, K. A Peroxygenase from Chaetomium globosum Catalyzes the Selective Oxygenation of Testosterone. ChemBioChem 2017, 18, 563–569. [Google Scholar] [CrossRef] [Green Version]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Kiebist, J.; Scheibner, K.; Ullrich, R. Peroxide-Mediated Oxygenation of Organic Compounds by Fungal Peroxygenases. Antioxidants 2022, 11, 163. [Google Scholar] [CrossRef]
- Kuipers, R.K.; Joosten, H.-J.; van Berkel, W.J.H.; Leferink, N.G.H.; Rooijen, E.; Ittmann, E.; van Zimmeren, F.; Jochens, H.; Bornscheuer, U.; Vriend, G.; et al. 3DM: Systematic Analysis of Heterogeneous Superfamily Data to Discover Protein Functionalities. Proteins 2010, 78, 2101–2113. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [Green Version]
- Abad, S.; Nahalka, J.; Bergler, G.; Arnold, S.A.; Speight, R.; Fotheringham, I.; Nidetzky, B.; Glieder, A. Stepwise Engineering of a Pichia Pastoris D-Amino Acid Oxidase Whole Cell Catalyst. Microb. Cell Fact. 2010, 9, 24. [Google Scholar] [CrossRef] [Green Version]
- Vogl, T.; Fischer, J.E.; Hyden, P.; Wasmayer, R.; Sturmberger, L.; Glieder, A. Orthologous Promoters from Related Methylotrophic Yeasts Surpass Expression of Endogenous Promoters of Pichia pastoris. AMB Express 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Poraj-Kobielska, M.; Kinne, M.; Ullrich, R.; Scheibner, K.; Hofrichter, M. A Spectrophotometric Assay for the Detection of Fungal Peroxygenases. Anal. Biochem. 2012, 421, 327–329. [Google Scholar] [CrossRef]
- Pütter, J. Peroxidases. In Methods of Enzymatic Analysis, 2nd ed.; Bergmeyer, H.U., Ed.; Academic Press: San Diego, CA, USA, 1974; pp. 685–690. ISBN 978-0-12-091302-2. [Google Scholar]
- Breslmayr, E.; Hanžek, M.; Hanrahan, A.; Leitner, C.; Kittl, R.; Šantek, B.; Oostenbrink, C.; Ludwig, R. A Fast and Sensitive Activity Assay for Lytic Polysaccharide Monooxygenase. Biotechnol. Biofuels 2018, 11, 1–13. [Google Scholar] [CrossRef]
- Joshi, H.J.; Gupta, R. Eukaryotic Glycosylation: Online Methods for Site Prediction on Protein Sequences. In Glycoinformatics; Lütteke, T., Frank, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 127–137. ISBN 978-1-4939-2343-4. [Google Scholar]
- Gupta, R.; Brunak, S. Prediction of Glycosylation Across the Human Proteome and the Correlation to Protein Function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar] [CrossRef] [Green Version]
- Faiza, M.; Huang, S.; Lan, D.; Wang, Y. New Insights on Unspecific Peroxygenases: Superfamily Reclassification and Evolution. BMC Evol. Biol. 2019, 19, 76. [Google Scholar] [CrossRef] [Green Version]
- Faiza, M.; Lan, D.; Huang, S.; Wang, Y. UPObase: An Online Database of Unspecific Peroxygenases. Database 2019, 2019, baz122. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Liang, H.; Zhao, Z.; Wu, B.; Lan, D.; Hollmann, F.; Wang, Y. A Novel Unspecific Peroxygenase from Galatian Marginata for Biocatalytic Oxyfunctionalization Reactions. Mol. Catal. 2022, 531, 112707. [Google Scholar] [CrossRef]
- Püllmann, P.; Weissenborn, M.J. Improving the Heterologous Production of Fungal Peroxygenases through an Episomal Pichia pastoris Promoter and Signal Peptide Shuffling System. ACS Synth. Biol. 2021, 10, 1360–1372. [Google Scholar] [CrossRef]
- Tonin, F.; Tieves, F.; Willot, S.; van Troost, A.; van Oosten, R.; Breestraat, S.; van Pelt, S.; Alcalde, M.; Hollmann, F. Pilot-Scale Production of Peroxygenase from Agrocybe Aegerita. Org. Proc. Res. Dev. 2021, 25, 1414–1418. [Google Scholar] [CrossRef]
- Patricia, G.d.S.; Dat, H.M.; Jan, K.; Harald, K.; René, U.; Katrin, S.; Martin, H.; Christiane, L.; Miguel, A. Functional Expression of Two Unusual Acidic Peroxygenases from Candolleomyces aberdarensis in Yeasts by Adopting Evolved Secretion Mutations. Appl. Environ. Microbiol. 2021, 87, e00878-21. [Google Scholar] [CrossRef]
- Ruth, C.; Buchetics, M.; Vidimce, V.; Kotz, D.; Naschberger, S.; Mattanovich, D.; Pichler, H.; Gasser, B. Pichia pastoris Aft1—A Novel Transcription Factor, Enhancing Recombinant Protein Secretion. Microb. Cell Fact. 2014, 13, 120. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R Language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic Analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Villalobos, A.; Ness, J.E.; Gustafsson, C.; Minshull, J.; Govindarajan, S. Gene Designer: A Synthetic Biology Tool for Constructing Artificial DNA Segments. BMC Bioinform. 2006, 7, 285. [Google Scholar] [CrossRef]
- Brodskiĭ, L.I.; Ivanov, V.V.; Kalaĭdzidis, I.L.; Leontovich, A.M.; Nikolaev, V.K.; Feranchuk, S.I.; Drachev, V.A. GeneBee-NET: An Internet Based Server for Biopolymer Structure Analysis. Biokhimiia 1995, 60, 1221–1230. [Google Scholar]




| Motif Number | 3DM Number | Number of Proteins | Percentage | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 23 | 40 | 66 | 73 | 114 | 117 | 118 | 188 | 196 | |||
| 1 | A | P | N | V | L | G | K | F | L | F | 51 | 2.01% |
| 2 | A | P | N | I | I | R | I | L | G | S | 31 | 1.22% |
| 3 | P | L | S | M | F | T | V | F | F | F | 29 | 1.15% |
| 4 | A | P | N | I | L | R | I | L | G | S | 21 | 0.83% |
| 5 | A | P | N | L | L | K | V | I | Q | L | 20 | 0.79% |
| 6 | A | P | N | M | V | R | T | V | I | F | 20 | 0.79% |
| 7 | A | P | N | M | F | K | R | F | A | F | 18 | 0.71% |
| 8 | A | P | N | I | T | E | T | F | P | F | 16 | 0.63% |
| 9 | A | P | N | F | T | K | V | I | T | V | 16 | 0.63% |
| 10 | K | P | N | M | A | H | P | I | V | L | 15 | 0.59% |
| Motif Number | 3DM Number | Number of Proteins | Percentage | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | 34 | 40 | 44 | 120 | 125 | 218 | 223 | 230 | |||
| 1 | W | M | N | L | H | S | Y | E | G | 50 | 58.14% |
| 2 | W | M | N | L | H | S | F | E | G | 4 | 4.65% |
| 3 | W | F | N | L | H | S | Y | E | G | 2 | 2.33% |
| 4 | W | M | N | L | H | S | Y | E | G | 2 | 2.33% |
| Organism of Origin | Protein Accession No. | UPO Family | Shortcut | Systematic Name | Criteria |
|---|---|---|---|---|---|
| Galerina marginata CBS 339.88 | KDR77412.1 | II | UPO42 | GmaUPO-II | M4F & PLSTV |
| Leucoagaricus sp. SymC. cos | KXN81289.1 | II | UPO43 | - | M4F & PLSTV |
| Psilocybe cyanescens | PPQ78776.1 | II | UPO44 | - | M4F & PLSTV |
| Candolleomyces aberdarensis | RXW15716.1 | II | UPO46 | PabUPO-III | PLSVT |
| Coprinopsis marcescibilis | TFK24496.1 | II | UPO47 | CmaUPO-I | PLSHT |
| Coprinopsis marcescibilis | TFK18510.1 | II | UPO48 | Putative CmaUPO-II | PLSNS |
| Leucoagaricus sp. SymC. cos | KXN91485.1 | II | UPO50 | LspUPO | PPSTP |
| Coprinopsis marcescibilis | TFK18228.1 | II | UPO51 | Putative CmaUPO-III | PLSNT |
| Gymnopilus dilepis | PPQ67339.1 | II | UPO53 | GdiUPO | M4F & PLSTV |
| Crucibulum laeve | TFK34946.1 | II | UPO54 | - | M4F & PLSTV |
| Crucibulum laeve | TFK34139.1 | II | UPO55 | - | M4F & PLSTV |
| Coprinellus micaceus | TEB20562.1 | II | UPO56 | CmiUPO | M4F & PLSTV |
| Coprinellus micaceus | TEB37025.1 | II | UPO57 | M4F & PLSTV | |
| Galerina marginata CBS 339.88 | KDR72033.1 | II | UPO58 | - | M4F & PLSTV |
| Penicillium steckii | OQE16359.1 | I | UPO59 | - | WMNLHSYEG |
| Aspergillus novoparasiticus | KAB8223135.1 | I | UPO60 | AnoUPO | WMNLHSYEG and M34, R114, H115, N116, I117, L118 |
| Gymnopus luxurians FD-317 M1 | KIK53163.1 | I | UPO63 | - | G30, G34 |
| Dendrothele bispora CBS 962.96 | THV03356.1 | I | UPO64 | DbiUPO | G30, G34 |
| Fusarium pseudograminearum | KAF0645823.1 | I | UPO65 | - | G30, G34 |
| Monosporascus sp. mg162 | RYP51541.1 | I | UPO66 | - | WMNLHSYE and M34, R114, H115, N116, I117, L118 |
| Aspergillus brasiliensis CBS 101740 | OJJ67899.1 | I | UPO67 | AbrUPO | M34, R114, H115, N116, I117, L118 |
| Aspergillus homomorphus CBS 101889 | XP_025551759.1 | I | UPO68 | - | WMNLHSYE and M34, R114, H115, N116, I117, L118 |
| Aspergillus pseudotamarii | KAE8141564.1 | I | UPO70 | ApsUPO | WMNLHSYEG |
| Aspergillus candidus | XP_024669203.1 | I | UPO71 | - | WMNLHSYEG |
| Aspergillus bombycis | XP_022384340.1 | I | UPO74 | AboUPO | WMNLHSYEG |
| Stachybotrys chartarum IBT 40292 | KFA56383.1 | I | UPO75 | SchUPO | WMNLHSYEG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebner, K.; Pfeifenberger, L.J.; Rinnofner, C.; Schusterbauer, V.; Glieder, A.; Winkler, M. Discovery and Heterologous Expression of Unspecific Peroxygenases. Catalysts 2023, 13, 206. https://doi.org/10.3390/catal13010206
Ebner K, Pfeifenberger LJ, Rinnofner C, Schusterbauer V, Glieder A, Winkler M. Discovery and Heterologous Expression of Unspecific Peroxygenases. Catalysts. 2023; 13(1):206. https://doi.org/10.3390/catal13010206
Chicago/Turabian StyleEbner, Katharina, Lukas J. Pfeifenberger, Claudia Rinnofner, Veronika Schusterbauer, Anton Glieder, and Margit Winkler. 2023. "Discovery and Heterologous Expression of Unspecific Peroxygenases" Catalysts 13, no. 1: 206. https://doi.org/10.3390/catal13010206
APA StyleEbner, K., Pfeifenberger, L. J., Rinnofner, C., Schusterbauer, V., Glieder, A., & Winkler, M. (2023). Discovery and Heterologous Expression of Unspecific Peroxygenases. Catalysts, 13(1), 206. https://doi.org/10.3390/catal13010206