The Immobilization and Stabilization of Trypsin from the Porcine Pancreas on Chitosan and Its Catalytic Performance in Protein Hydrolysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Influence of the Solution pH on Trypsin Immobilization on the Different Supports
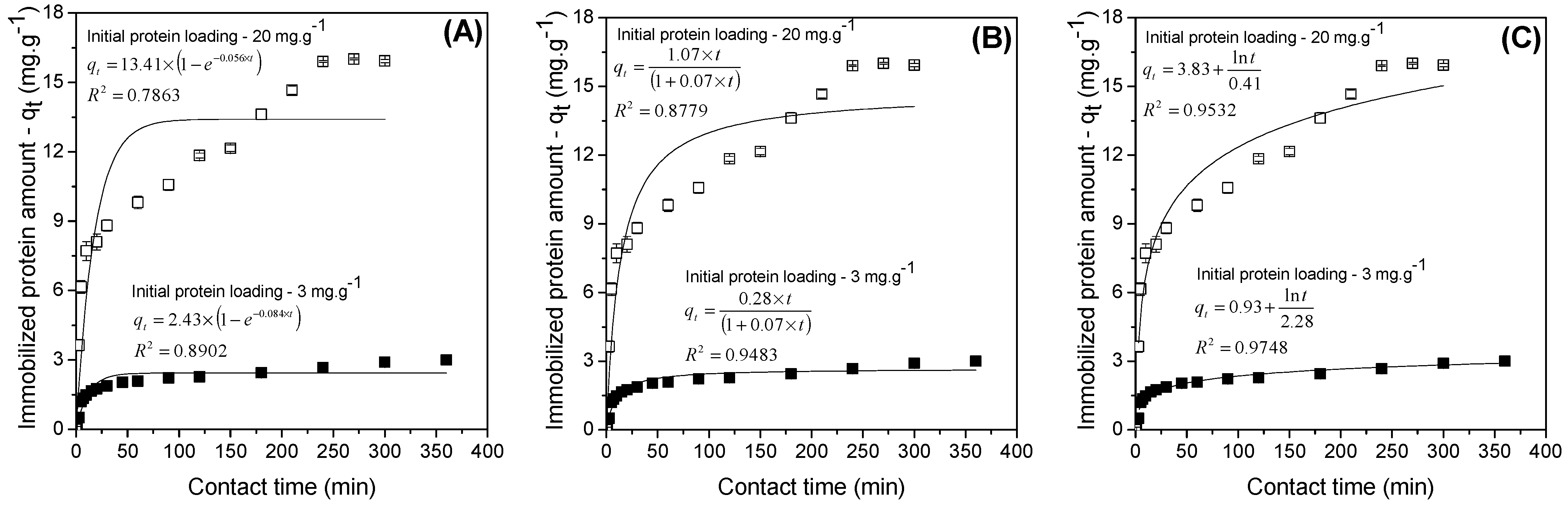
2.2. Influence of Time on the Immobilization Process: Kinetic Adsorption Studies
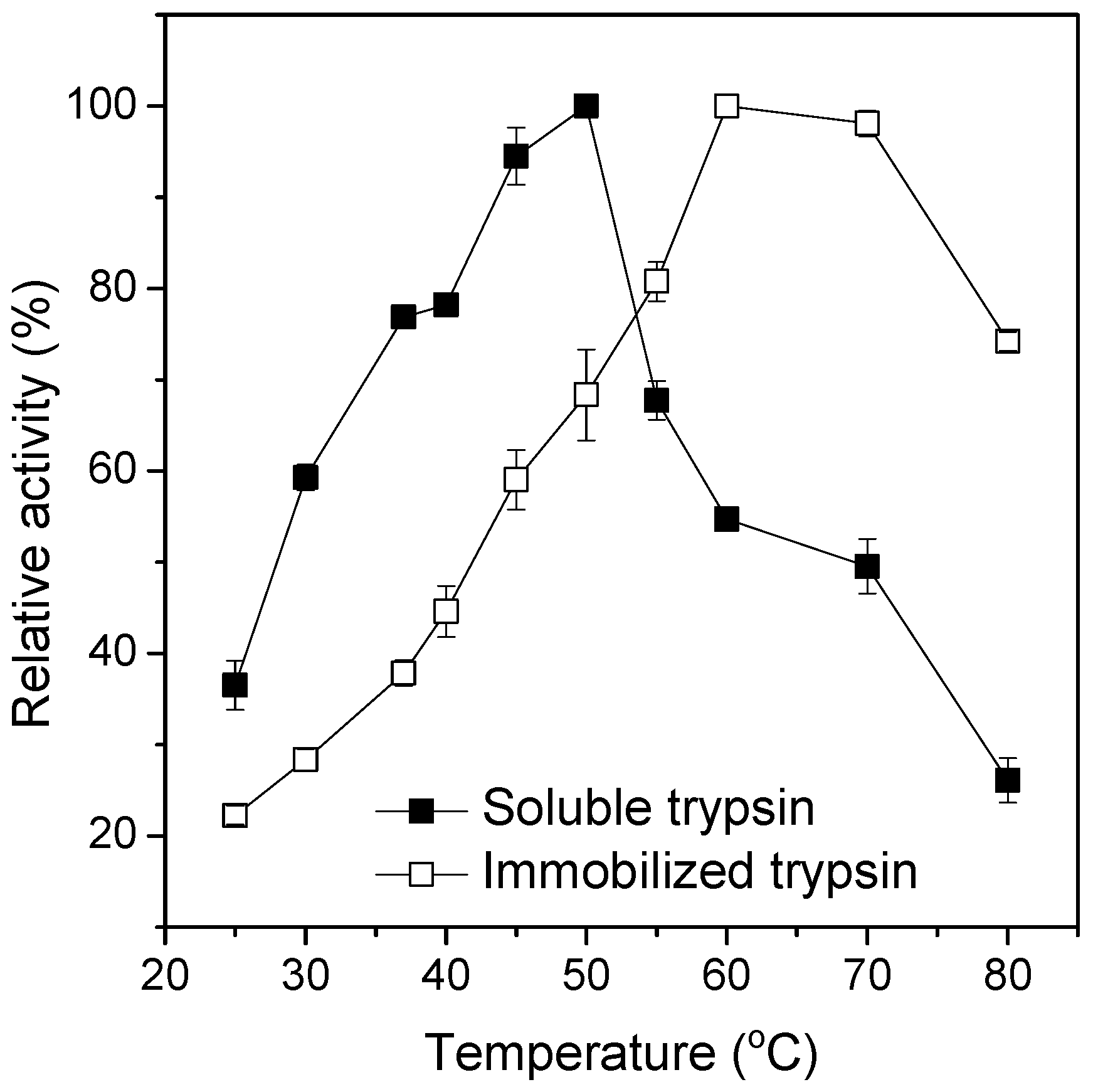
2.3. Influence of Temperature on Soluble and Immobilized Trypsin Activity
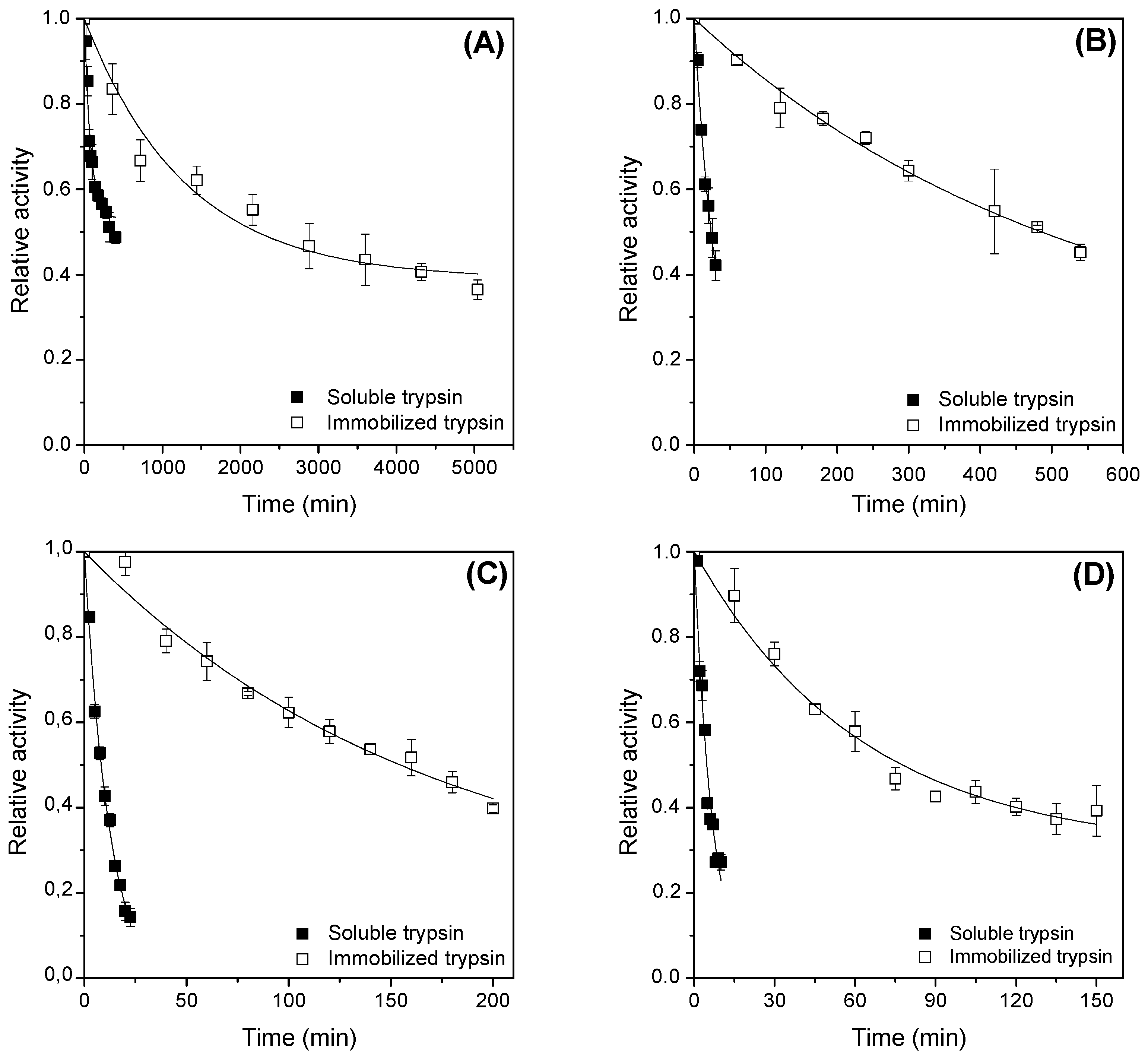
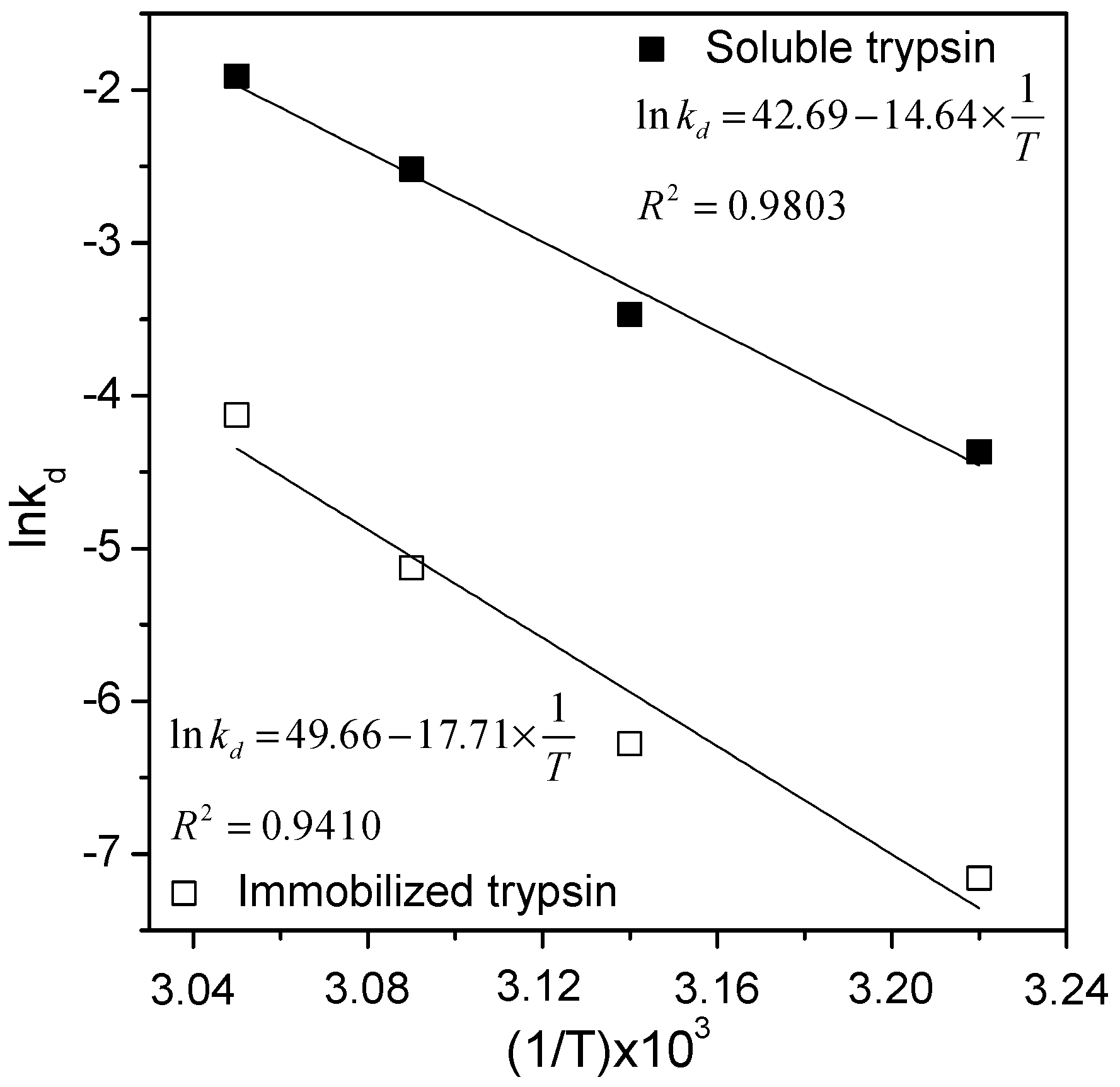
2.4. Thermal Stability Tests and Estimating Kinetic and Thermodynamic Parameters
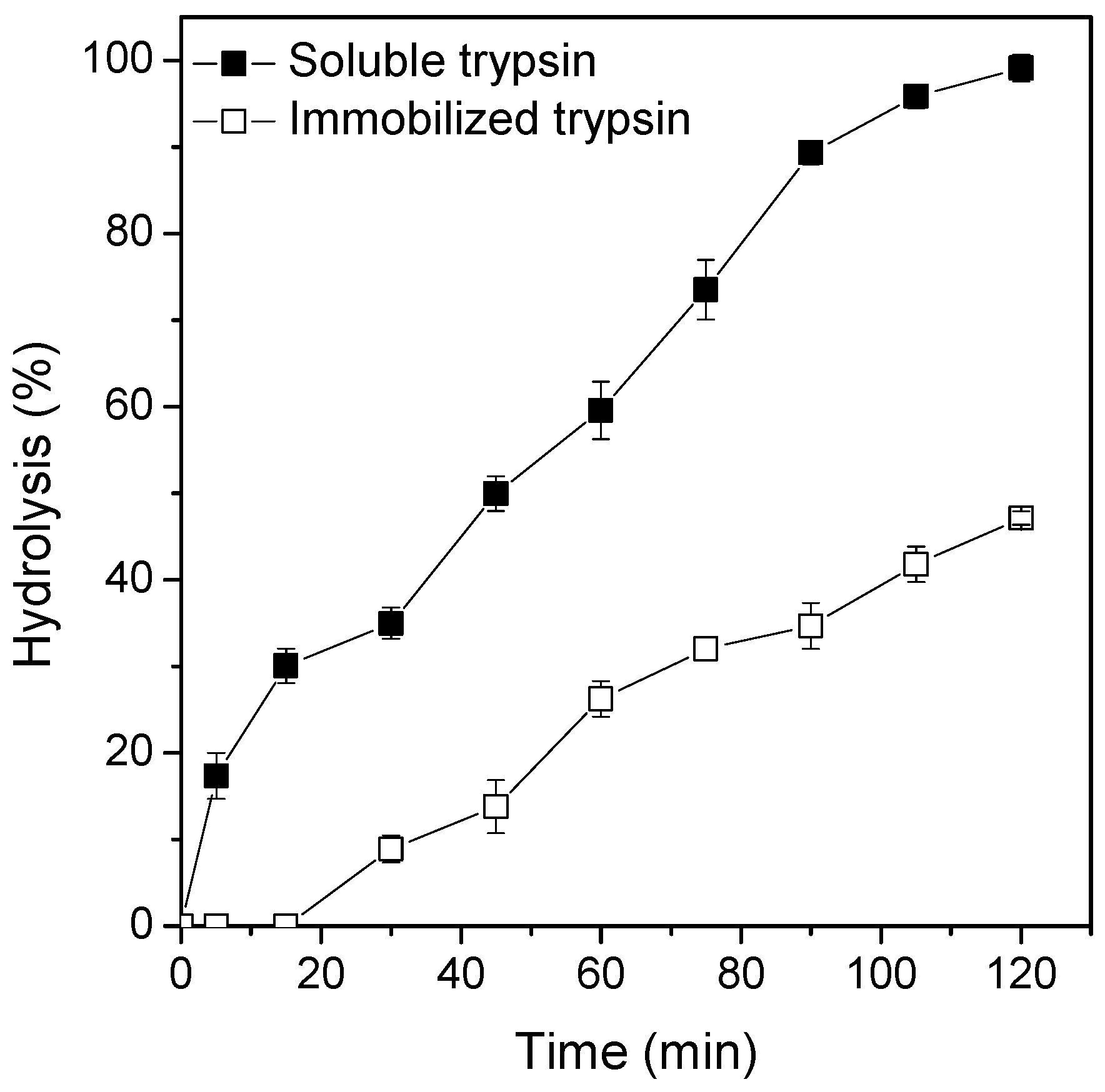
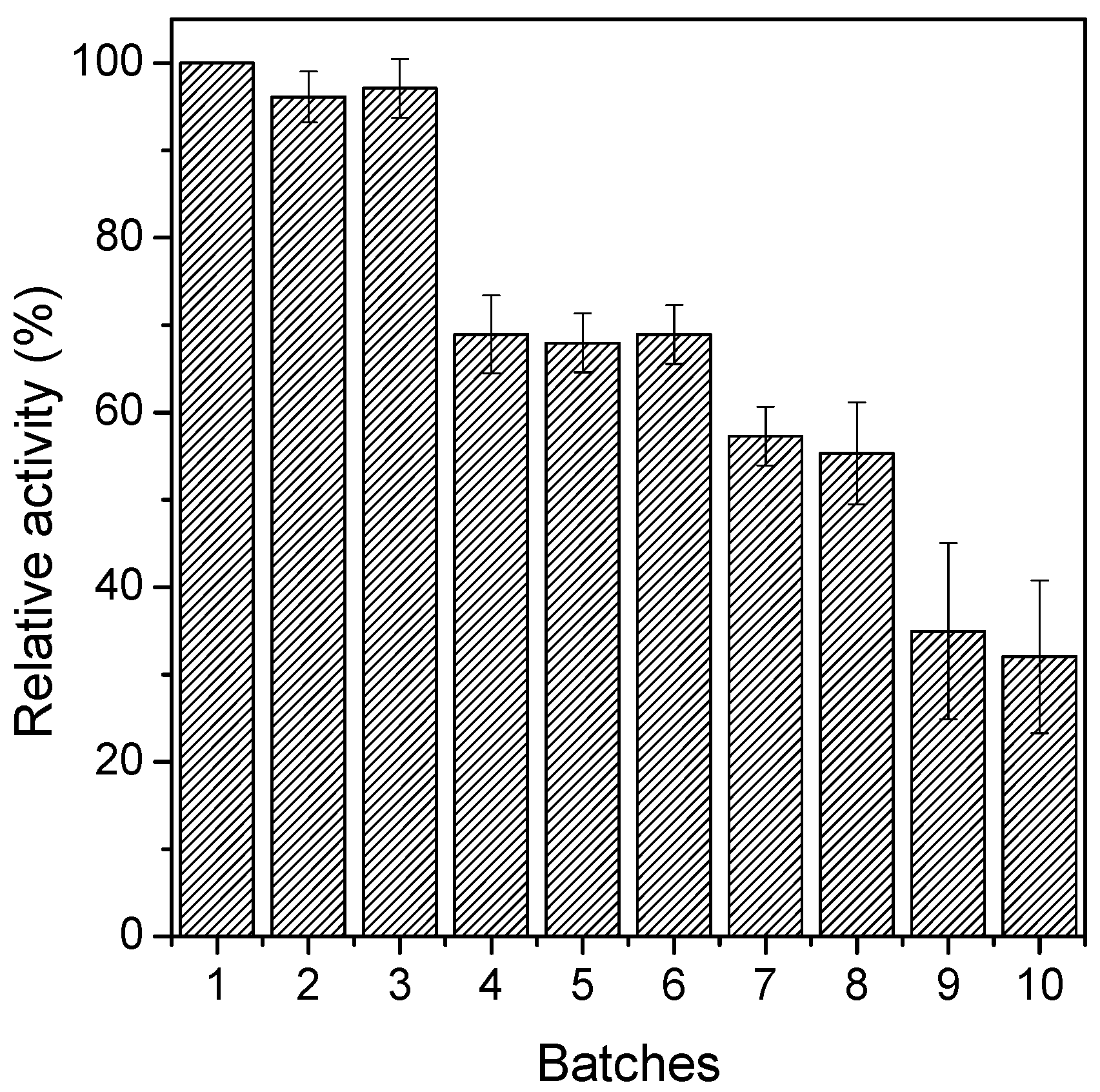
2.5. Evaluating the Catalytic Performance of Soluble and Immobilized Trypsin in the BSA Hydrolysis Reaction and Operations Stability Tests
3. Materials and Methods
3.1. Materials
3.2. Preparing the Heterofunctional Supports
3.3. Immobilization of Trypsin on Chit–GA–Gly Hydrogel
3.4. Influence of pH on the Immobilization Process
3.5. Influence of Immobilization Time: Kinetic Adsorption Studies
3.6. Influence of Temperature on the Activity of Soluble and Immobilized Trypsin
3.7. Thermal Stability: Estimating the Kinetic and Thermodynamic Parameters
3.8. BSA Hydrolysis Catalyzed by Soluble and Immobilized Trypsin and Operational Stability Tests
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naveed, M.; Nadeem, F.; Mehmood, T.; Bilal, M.; Anwar, Z.; Amjad, F. Protease—A Versatile and Ecofriendly Biocatalyst with Multi-Industrial Applications: An Updated Review. Catal. Lett. 2021, 151, 307–323. [Google Scholar] [CrossRef]
- Deng, Y.; Gruppen, H.; Wierenga, P.A. Comparison of Protein Hydrolysis Catalyzed by Bovine, Porcine, and Human Trypsins. J. Agric. Food Chem. 2018, 66, 4219–4232. [Google Scholar] [CrossRef] [PubMed]
- Perera, E.; Rodríguez-Viera, L.; Perdomo-Morales, R.; Montero-Alejo, V.; Moyano, F.J.; Martínez-Rodríguez, G.; Mancera, J.M. Trypsin Isozymes in the Lobster Panulirus argus (Latreille, 1804): From Molecules to Physiology. J. Comp. Physiol. B 2015, 185, 17–35. [Google Scholar] [CrossRef]
- Kaur, J.; Singh, P.K. Trypsin Detection Strategies: A Review. Crit. Rev. Anal. Chem. 2022, 52, 949–967. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, J.M.; Schumbrutzki, C.; Wortelkamp, S.; Sickmann, A.; Zahedi, R.P. Systematic and Quantitative Comparison of Digest Efficiency and Specificity Reveals the Impact of Trypsin Quality on MS-Based Proteomics. J. Proteom. 2012, 75, 1454–1462. [Google Scholar] [CrossRef]
- Bougatef, A. Trypsins from Fish Processing Waste: Characteristics and Biotechnological Applications—Comprehensive Review. J. Clean. Prod. 2013, 57, 257–265. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Chen, X.M.; Kitts, D.D.; Li-Chan, E.C.Y. Investigation into the Bioavailability of Milk Protein-Derived Peptides with Dipeptidyl-Peptidase IV Inhibitory Activity Using Caco-2 Cell Monolayers. Food Funct. 2017, 8, 701–709. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Fu, X.; Li, S.; Wei, J. A Novel Antioxidant and ACE Inhibitory Peptide from Rice Bran Protein: Biochemical Characterization and Molecular Docking Study. LWT 2017, 75, 93–99. [Google Scholar] [CrossRef]
- Nath, A.; Eren, B.A.; Csighy, A.; Pásztorné-Huszár, K.; Kiskó, G.; Abrankó, L.; Tóth, A.; Szerdahelyi, E.; Kovács, Z.; Koris, A.; et al. Production of Liquid Milk Protein Concentrate with Antioxidant Capacity, Angiotensin Converting Enzyme Inhibitory Activity, Antibacterial Activity, and Hypoallergenic Property by Membrane Filtration and Enzymatic Modification of Proteins. Processes 2020, 8, 871. [Google Scholar] [CrossRef]
- Tavano, O.L.; Berenguer-Murcia, A.; Secundo, F.; Fernandez-Lafuente, R. Biotechnological Applications of Proteases in Food Technology. Compr. Rev. Food Sci. Food Saf. 2018, 17, 412–436. [Google Scholar] [CrossRef]
- Basso, A.; Serban, S. Industrial Applications of Immobilized Enzymes—A Review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Woodley, J.M.; Fernandez-Lafuente, R. Is Enzyme Immobilization a Mature Discipline? Some Critical Considerations to Capitalize on the Benefits of Immobilization. Chem. Soc. Rev. 2022, 51, 6251–6290. [Google Scholar] [CrossRef] [PubMed]
- Maghraby, Y.R.; El-Shabasy, R.M.; Ibrahim, A.H.; Azzazy, H.M.E.S. Enzyme Immobilization Technologies and Industrial Applications. ACS Omega 2023, 8, 5184–5196. [Google Scholar] [CrossRef] [PubMed]
- Bernal, C.; Rodríguez, K.; Martínez, R. Integrating Enzyme Immobilization and Protein Engineering: An Alternative Path for the Development of Novel and Improved Industrial Biocatalysts. Biotechnol. Adv. 2018, 36, 1470–1480. [Google Scholar] [CrossRef]
- Cavalcante, F.T.T.; Cavalcante, A.L.G.; de Sousa, I.G.; Neto, F.S.; dos Santos, J.C.S. Current Status and Future Perspectives of Supports and Protocols for Enzyme Immobilization. Catalysts 2021, 11, 1222. [Google Scholar] [CrossRef]
- Bashir, N.; Sood, M.; Bandral, J.D. Enzyme Immobilization and Its Applications in Food Processing: A Review. Int. J. Chem. Stud. 2020, 8, 254–261. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszynska, D. Immobilization as a Strategy for Improving Enzyme Properties-Application to Oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Carballares, D.; Morellon-Sterling, R.; Fernandez-Lafuente, R. Stabilization of Enzymes via Immobilization: Multipoint Covalent Attachment and Other Stabilization Strategies. Biotechnol. Adv. 2021, 52, 107821. [Google Scholar] [CrossRef]
- Zdarta, J.; Meyer, A.S.; Jesionowski, T.; Pinelo, M. A General Overview of Support Materials for Enzyme Immobilization: Characteristics, Properties, Practical Utility. Catalysts 2018, 8, 92. [Google Scholar] [CrossRef]
- Chang, Q.; Zhu, L.; Jiang, G.; Tang, H. Sensitive Fluorescent Probes for Determination of Hydrogen Peroxide and Glucose Based on Enzyme-Immobilized Magnetite/Silica Nanoparticles. Anal. Bioanal. Chem. 2009, 395, 2377–2385. [Google Scholar] [CrossRef]
- Aybastier, Ö.; Demir, C. Optimization of Immobilization Conditions of Thermomyces lanuginosus Lipase on Styrene-Divinylbenzene Copolymer Using Response Surface Methodology. J. Mol. Catal. B Enzym. 2010, 63, 170–178. [Google Scholar] [CrossRef]
- Tonini, D.; Astrup, T. Life-Cycle Assessment of a Waste Refinery Process for Enzymatic Treatment of Municipal Solid Waste. Waste Manag. 2012, 32, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.R.; Fidantsef, A.L. Directed Evolution of Industrial Enzymes: An Update. Curr. Opin. Biotechnol. 2003, 14, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Massolini, G.; Calleri, E. Immobilized Trypsin Systems Coupled On-Line to Separation Methods: Recent Developments and Analytical Applications. J. Sep. Sci. 2005, 28, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Sasai, Y.; Kanno, H.; Doi, N.; Yamauchi, Y.; Kuzuya, M.; Kondo, S.I. Synthesis and Characterization of Highly Stabilized Polymer-Trypsin Conjugates with Autolysis Resistance. Catalysts 2017, 7, 4. [Google Scholar] [CrossRef]
- Transue, T.R.; Krahn, J.M.; Gabel, S.A.; DeRose, E.F.; London, R.E. X-Ray and NMR Characterization of Covalent Complexes of Trypsin, Borate, and Alcohols. Biochemistry 2004, 43, 2829–2839. [Google Scholar] [CrossRef]
- Carneiro, L.A.B.C.; Costa-Silva, T.A.; Souza, C.R.F.; Bachmann, L.; Oliveira, W.P.; Said, S. Immobilization of Lipases Produced by the Endophytic Fungus Cercospora kikuchii on Chitosan Microparticles. Braz. Arch. Biol. Technol. 2014, 57, 578–586. [Google Scholar] [CrossRef]
- Urrutia, P.; Bernal, C.; Wilson, L.; Illanes, A. Use of Chitosan Heterofunctionality for Enzyme Immobilization: β-Galactosidase Immobilization for Galacto-Oligosaccharide Synthesis. Int. J. Biol. Macromol. 2018, 116, 182–193. [Google Scholar] [CrossRef]
- Nunes, Y.L.; de Menezes, F.L.; de Sousa, I.G.; Cavalcante, A.L.G.; Cavalcante, F.T.T.; da Silva Moreira, K.; de Oliveira, A.L.B.; Mota, G.F.; da Silva Souza, J.E.; de Aguiar Falcão, I.R.; et al. Chemical and Physical Chitosan Modification for Designing Enzymatic Industrial Biocatalysts: How to Choose the Best Strategy? Int. J. Biol. Macromol. 2021, 181, 1124–1170. [Google Scholar] [CrossRef]
- Okura, N.S.; Sabi, G.J.; Crivellenti, M.C.; Gomes, R.A.B.; Fernandez-Lafuente, R.; Mendes, A.A. Improved Immobilization of Lipase from Thermomyces lanuginosus on a New Chitosan-Based Heterofunctional Support: Mixed Ion Exchange plus Hydrophobic Interactions. Int. J. Biol. Macromol. 2020, 163, 550–561. [Google Scholar] [CrossRef]
- Diviesti, K.; Holz, R.C. Catalytic Biomaterials for Atrazine Degradation. Catalysts 2023, 13, 140. [Google Scholar] [CrossRef]
- Kou, S.; Peters, L.M.; Mucalo, M.R. Chitosan: A Review of Sources and Preparation Methods. Int. J. Biol. Macromol. 2021, 169, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Aranaz, I.; Mengíbar, M.; Harris, R.; Paños, I.; Miralles, B.; Acosta, N.; Galed, G.; Heras, Á. Functional Characterization of Chitin and Chitosan; Bentham Science Publishers: Sharjah, United Arab Emirates, 2009; Volume 3. [Google Scholar]
- Ribeiro, E.S.; de Farias, B.S.; Sant’Anna Cadaval Junior, T.R.; de Almeida Pinto, L.A.; Diaz, P.S. Chitosan–Based Nanofibers for Enzyme Immobilization. Int. J. Biol. Macromol. 2021, 183, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Soozanipour, A.; Taheri-Kafrani, A.; Barkhori, M.; Nasrollahzadeh, M. Preparation of a Stable and Robust Nanobiocatalyst by Efficiently Immobilizing of Pectinase onto Cyanuric Chloride-Functionalized Chitosan Grafted Magnetic Nanoparticles. J. Colloid. Interface Sci. 2019, 536, 261–270. [Google Scholar] [CrossRef]
- Wahba, M.I. Porous Chitosan Beads of Superior Mechanical Properties for the Covalent Immobilization of Enzymes. Int. J. Biol. Macromol. 2017, 105, 894–904. [Google Scholar] [CrossRef]
- Pinheiro, B.B.; Rios, N.S.; Rodríguez Aguado, E.; Fernandez-Lafuente, R.; Freire, T.M.; Fechine, P.B.A.; dos Santos, J.C.S.; Gonçalves, L.R.B. Chitosan Activated with Divinyl Sulfone: A New Heterofunctional Support for Enzyme Immobilization. Application in the Immobilization of Lipase B from Candida antarctica. Int. J. Biol. Macromol. 2019, 130, 798–809. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in Bio-Catalysts Design: A Useful Crosslinker and a Versatile Tool in Enzyme Immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Monteiro, O.A.C.; Airoldi, C. Some Studies of Crosslinking Chitosan-Glutaraldehyde Interaction in a Homogeneous System; Elsevier: Amsterdam, The Netherlands, 1999; Volume 26. [Google Scholar]
- Elias, N.; Chandren, S.; Razak, F.I.A.; Jamalis, J.; Widodo, N.; Wahab, R.A. Characterization, Optimization and Stability Studies on Candida rugosa Lipase Supported on Nanocellulose Reinforced Chitosan Prepared from Oil Palm Biomass. Int. J. Biol. Macromol. 2018, 114, 306–316. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, Á.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional Supports in Enzyme Immobilization: From Traditional Immobilization Protocols to Opportunities in Tuning Enzyme Properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, S. Immobilization of Trypsin from Porcine Pancreas onto Chitosan Nonwoven by Covalent Bonding. Polymers 2019, 11, 1462. [Google Scholar] [CrossRef]
- Shu, M.; Shen, W.; Wang, X.; Wang, F.; Ma, L.; Zhai, C. Expression, Activation and Characterization of Porcine Trypsin in Pichia pastoris GS115. Protein Expr. Purif. 2015, 114, 149–155. [Google Scholar] [CrossRef]
- Bujacz, A. Structures of Bovine, Equine and Leporine Serum Albumin. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- Zdarta, J.; Antecka, K.; Jędrzak, A.; Synoradzki, K.; Łuczak, M.; Jesionowski, T. Biopolymers Conjugated with Magnetite as Support Materials for Trypsin Immobilization and Protein Digestion. Colloids Surf. B Biointerfaces 2018, 169, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Atacan, K.; Çakiroǧlu, B.; Özacar, M. Improvement of the Stability and Activity of Immobilized Trypsin on Modified Fe3O4 Magnetic Nanoparticles for Hydrolysis of Bovine Serum Albumin and Its Application in the Bovine Milk. Food Chem. 2016, 212, 460–468. [Google Scholar] [CrossRef]
- Buck, F.F.; Vithayathil, A.J.; Sord, F.F. On the Mechanism of Enzyme Action. LXXIII. Studies on Trypsins from Beef, Sheep and Pig Pancreas. Arch. Biochem. Biophys. 1962, 97, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Soto, I.D.; Escobar, S.; Mesa, M. Study of the Physicochemical Interactions between Thermomyces lanuginosus Lipase and Silica-Based Supports and Their Correlation with the Biochemical Activity of the Biocatalysts. Mater. Sci. Eng. C 2017, 79, 525–532. [Google Scholar] [CrossRef]
- Tan, K.L.; Hameed, B.H. Insight into the Adsorption Kinetics Models for the Removal of Contaminants from Aqueous Solutions. J. Taiwan Inst. Chem. Eng. 2017, 74, 25–48. [Google Scholar] [CrossRef]
- Miranda, J.S.; Silva, N.C.A.; Bassi, J.J.; Corradini, M.C.C.; Lage, F.A.P.; Hirata, D.B.; Mendes, A.A. Immobilization of Thermomyces lanuginosus Lipase on Mesoporous Poly-Hydroxybutyrate Particles and Application in Alkyl Esters Synthesis: Isotherm, Thermodynamic and Mass Transfer Studies. Chem. Eng. J. 2014, 251, 392–403. [Google Scholar] [CrossRef]
- Manrich, A.; Galvão, C.M.A.; Jesus, C.D.F.; Giordano, R.C.; Giordano, R.L.C. Immobilization of Trypsin on Chitosan Gels: Use of Different Activation Protocols and Comparison with Other Supports. Int. J. Biol. Macromol. 2008, 43, 54–61. [Google Scholar] [CrossRef]
- Mendes, A.A.; de Castro, H.F.; Rodrigues, D.d.S.; Adriano, W.S.; Tardioli, P.W.; Mammarella, E.J.; Giordano, R.d.C.; Giordano, R.d.L.C. Multipoint Covalent Immobilization of Lipase on Chitosan Hybrid Hydrogels: Influence of the Polyelectrolyte Complex Type and Chemical Modification on the Catalytic Properties of the Biocatalysts. J. Ind. Microbiol. Biotechnol. 2011, 38, 1055–1066. [Google Scholar] [CrossRef]
- Bezerra, R.M.; Neto, D.M.A.; Galvão, W.S.; Rios, N.S.; Carvalho, A.C.L.d.M.; Correa, M.A.; Bohn, F.; Fernandez-Lafuente, R.; Fechine, P.B.A.; de Mattos, M.C.; et al. Design of a Lipase-Nano Particle Biocatalysts and Its Use in the Kinetic Resolution of Medicament Precursors. Biochem. Eng. J. 2017, 125, 104–115. [Google Scholar] [CrossRef]
- Bezerra, R.M.; Monteiro, R.R.C.; Neto, D.M.A.; da Silva, F.F.M.; de Paula, R.C.M.; de Lemos, T.L.G.; Fechine, P.B.A.; Correa, M.A.; Bohn, F.; Gonçalves, L.R.B.; et al. A New Heterofunctional Support for Enzyme Immobilization: PEI Functionalized Fe3O4 MNPs Activated with Divinyl Sulfone. Application in the Immobilization of Lipase from Thermomyces lanuginosus. Enzym. Microb. Technol. 2020, 138, 109560. [Google Scholar] [CrossRef] [PubMed]
- Faria, L.L.; Morales, S.A.V.; Prado, J.P.Z.; Dias, G.d.S.; de Almeida, A.F.; Xavier, M.d.C.A.; da Silva, E.S.; Maiorano, A.E.; Perna, R.F. Biochemical Characterization of Extracellular Fructosyltransferase from Aspergillus oryzae IPT-301 Immobilized on Silica Gel for the Production of Fructooligosaccharides. Biotechnol. Lett. 2021, 43, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Nagar, S.; Bhushan, I.; Kumar, L.; Parshad, R.; Gupta, V.K. Covalent Immobilization of Organic Solvent Tolerant Lipase on Aluminum Oxide Pellets and Its Potential Application in Esterification Reaction. J. Mol. Catal. B Enzym. 2013, 87, 51–61. [Google Scholar] [CrossRef]
- Tran, D.T.; Chang, J.S. Kinetics of Enzymatic Transesterification and Thermal Deactivation Using Immobilized Burkholderia Lipase as Catalyst. Bioprocess. Biosyst. Eng. 2014, 37, 481–491. [Google Scholar] [CrossRef]
- Vieille, C.; Zeikus, J.G. Thermozymes: Identifying Molecular Determinants of Protein Structural and Functional Stability. Trends Biotechnol. 1996, 14, 183–190. [Google Scholar] [CrossRef]
- Saqib, A.A.N.; Siddiqui, K.S. How to Calculate Thermostability of Enzymes Using a Simple Approach. Biochem. Mol. Biol. Educ. 2018, 46, 398–402. [Google Scholar] [CrossRef]
- Gohel, S.D.; Singh, S.P. Characteristics and Thermodynamics of a Thermostable Protease from a Salt-Tolerant Alkaliphilic Actinomycete. Int. J. Biol. Macromol. 2013, 56, 20–27. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; Castañeda-Valbuena, D.; Morellon-Sterling, R.; Tavano, O.; Berenguer-Murcia, Á.; Vela-Gutiérrez, G.; Rather, I.A.; Fernandez-Lafuente, R. Bioactive Peptides from Fisheries Residues: A Review of Use of Papain in Proteolysis Reactions. Int. J. Biol. Macromol. 2021, 184, 415–428. [Google Scholar] [CrossRef]
- Morellon-Sterling, R.; Tavano, O.; Bolivar, J.M.; Berenguer-Murcia, Á.; Vela-Gutiérrez, G.; Sabir, J.S.M.; Tacias-Pascacio, V.G.; Fernandez-Lafuente, R. A Review on the Immobilization of Pepsin: A Lys-Poor Enzyme That Is Unstable at Alkaline pH Values. Int. J. Biol. Macromol. 2022, 210, 682–702. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; Morellon-Sterling, R.; Castañeda-Valbuena, D.; Berenguer-Murcia, Á.; Kamli, M.R.; Tavano, O.; Fernandez-Lafuente, R. Immobilization of Papain: A Review. Int. J. Biol. Macromol. 2021, 188, 94–113. [Google Scholar] [PubMed]
- Manoel, E.A.; Ribeiro, M.F.P.; Dos Santos, J.C.S.; Coelho, M.A.Z.; Simas, A.B.C.; Fernandez-Lafuente, R.; Freire, D.M.G. Accurel MP 1000 as a Support for the Immobilization of Lipase from Burkholderia cepacia: Application to the Kinetic Resolution of Myo-Inositol Derivatives. Process Biochem. 2015, 50, 1557–1564. [Google Scholar] [CrossRef]
- Mageed, H.A.; Ezz, N.A.; Radwan, R. Bio-Inspired Trypsin-Chitosan Cross-Linked Enzyme Aggregates: A Versatile Approach for Stabilization through Carrier-Free Immobilization. Biotechnologia 2019, 100, 301–309. [Google Scholar] [CrossRef]
- Aslani, E.; Abri, A.; Pazhang, M. Immobilization of Trypsin onto Fe3O4@SiO2 –NH2 and Study of Its Activity and Stability. Colloids Surf. B Biointerfaces 2018, 170, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Poonsin, T.; Simpson, B.K.; Visessanguan, W.; Yoshida, A.; Klomklao, S. Optimal Immobilization of Trypsin from the Spleen of Albacore Tuna (Thunnus alalunga) and Its Characterization. Int. J. Biol. Macromol. 2020, 143, 462–471. [Google Scholar] [CrossRef]
- Alves, M.D.; Aracri, F.M.; Cren, É.C.; Mendes, A.A. Isotherm, Kinetic, Mechanism and Thermodynamic Studies of Adsorption of a Microbial Lipase on a Mesoporous and Hydrophobic Resin. Chem. Eng. J. 2017, 311, 1–12. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lage, F.A.P.; Bassi, J.J.; Corradini, M.C.C.; Todero, L.M.; Luiz, J.H.H.; Mendes, A.A. Preparation of a Biocatalyst via Physical Adsorption of Lipase from Thermomyces lanuginosus on Hydrophobic Support to Catalyze Biolubricant Synthesis by Esterification Reaction in a Solvent-Free System. Enzym. Microb. Technol. 2016, 84, 56–67. [Google Scholar] [CrossRef]
- Miguez, J.P.; Sousa, A.E.A.; Tavano, O.L. Increased Trypsin Resilience in Aqueous-Acetonitrile Environment When Immobilized on Glyoxyl-Agarose May Improve Its Applicability. Biocatal. Biotransform. 2023, 1–8. [Google Scholar] [CrossRef]
- Sadana, A.; Henley, J.P. Single-Step Unimolecular Non-First-Order Enzyme Deactivation Kinetics. Biotechnol. Bioeng. 1987, 30, 717–723. [Google Scholar] [CrossRef]
- Pessato, T.B.; Tavano, O.L. Hydrolysis of Casein and β-Lactoglobulin by Immobilized Papain after Pre-Treatment with Immobilized Trypsin. Acta Aliment. 2015, 44, 570–577. [Google Scholar] [CrossRef]
- Church, F.C.; Swaisgood, H.E.; Porter, D.H.; Catignani, G.L. Spectrophotometric Assay Using O-Phthaldialdehyde for Determination of Proteolysis in Milk and Isolated Milk Proteins. J. Dairy. Sci. 1983, 66, 1219–1227. [Google Scholar] [CrossRef]

| Support | pH | IP a (mg·g−1) | IY b (%) | HA c (nmol·min−1·g−1) | SA d (nmol·min−1·mg−1) |
|---|---|---|---|---|---|
| Chit–GA–Gly | 4.0 | 6.8 ± 0.2 | 30.7 ± 0.5 | 18.1 ± 2.0 | 2.7 ± 0.3 |
| 5.0 | 5.4 ± 0.3 | 25.6 ± 1.1 | 29.7 ± 0.8 | 4.4 ± 0.1 | |
| 6.0 | 6.8 ± 0.1 | 32.4 ± 0.5 | 45.5 ± 0.9 | 6.7 ± 0.1 | |
| 7.0 | 9.1 ± 0.3 | 44.5 ± 1.1 | 115.5 ± 3.7 | 17.3 ± 0.5 | |
| 8.0 | 11.7 ± 0 | 56.2 ± 0.2 | 163.3 ± 2.4 | 23.8± 0.4 | |
| 9.0 | 16.2 ± 0.2 | 81.0 ± 0.2 | 224.8 ± 1.6 | 33.1 ± 0.2 | |
| 10.0 | 15.1 ± 0.1 | 71.6 ± 0.2 | 117.2 ± 5.9 | 26.1 ± 0.9 | |
| Chit | 8.0 | 2.1 ± 0.4 | 10.5 ± 1.8 | 57.6 ± 5.7 | 26.9 ± 2.7 |
| 9.0 | 3.8 ± 0.3 | 19.8 ± 1.4 | 84.3 ± 2.6 | 21.9 ± 0.7 | |
| Chit–GA | 7.0 | 20.0 | 100.0 | 161.5 ± 1.1 | 8.1 ± 0.1 |
| 8.0 | 20.0 | 100.0 | 139.3 ± 2.2 | 7.1 ± 0.2 | |
| 9.0 | 19.1 ± 0.2 | 98.6 ± 0.8 | 129.1 ± 2.1 | 6.7 ± 0.1 |
| Parameter | Biocatalyst | Temperature (°C) | |||
|---|---|---|---|---|---|
| 37 | 45 | 50 | 55 | ||
| R2 | S a | 0.97521 | 0.98830 | 0.99503 | 0.96078 |
| I b | 0.96722 | 0.98837 | 0.97837 | 0.98275 | |
| kd (min−1) | S a | 0.01262 | 0.03123 | 0.07999 | 0.14721 |
| I b | 0.00077 | 0.00188 | 0.00593 | 0.01600 | |
| t1/2 (min) | S a | 340 | 23 | 8 | 4.5 |
| I b | 2100 | 485 | 164 | 72 | |
| SF | S a | 1 | 1 | 1 | 1 |
| I b | 6.2 | 21.1 | 20.5 | 16.0 | |
| Ed (kJ·mol−1) | S a | 121.7 | |||
| I b | 147.2 | ||||
| ΔH (kJ·mol−1) | S a | 119.1 | 119.0 | 119.0 | 118.9 |
| I b | 144.6 | 144.5 | 144.5 | 144.4 | |
| ΔG (kJ·mol−1) | S a | 85.9 | 85.8 | 84.7 | 84.4 |
| I b | 93.1 | 93.3 | 91.7 | 90.4 | |
| ΔS (J·mol−1·K−1) | S a | 106.9 | 107.1 | 110.6 | 111.5 |
| I b | 166.0 | 165.4 | 170.4 | 174.2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miguez, J.P.; Fernandez-Lafuente, R.; Tavano, O.L.; Mendes, A.A. The Immobilization and Stabilization of Trypsin from the Porcine Pancreas on Chitosan and Its Catalytic Performance in Protein Hydrolysis. Catalysts 2023, 13, 1344. https://doi.org/10.3390/catal13101344
Miguez JP, Fernandez-Lafuente R, Tavano OL, Mendes AA. The Immobilization and Stabilization of Trypsin from the Porcine Pancreas on Chitosan and Its Catalytic Performance in Protein Hydrolysis. Catalysts. 2023; 13(10):1344. https://doi.org/10.3390/catal13101344
Chicago/Turabian StyleMiguez, João Pedro, Roberto Fernandez-Lafuente, Olga Luisa Tavano, and Adriano Aguiar Mendes. 2023. "The Immobilization and Stabilization of Trypsin from the Porcine Pancreas on Chitosan and Its Catalytic Performance in Protein Hydrolysis" Catalysts 13, no. 10: 1344. https://doi.org/10.3390/catal13101344
APA StyleMiguez, J. P., Fernandez-Lafuente, R., Tavano, O. L., & Mendes, A. A. (2023). The Immobilization and Stabilization of Trypsin from the Porcine Pancreas on Chitosan and Its Catalytic Performance in Protein Hydrolysis. Catalysts, 13(10), 1344. https://doi.org/10.3390/catal13101344








