Whole-Cell PVA Cryogel-Immobilized Microbial Consortium LE-C1 for Xanthan Depolymerization
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening, Isolation, and Identification of Xanthan-Degrading Microorganisms
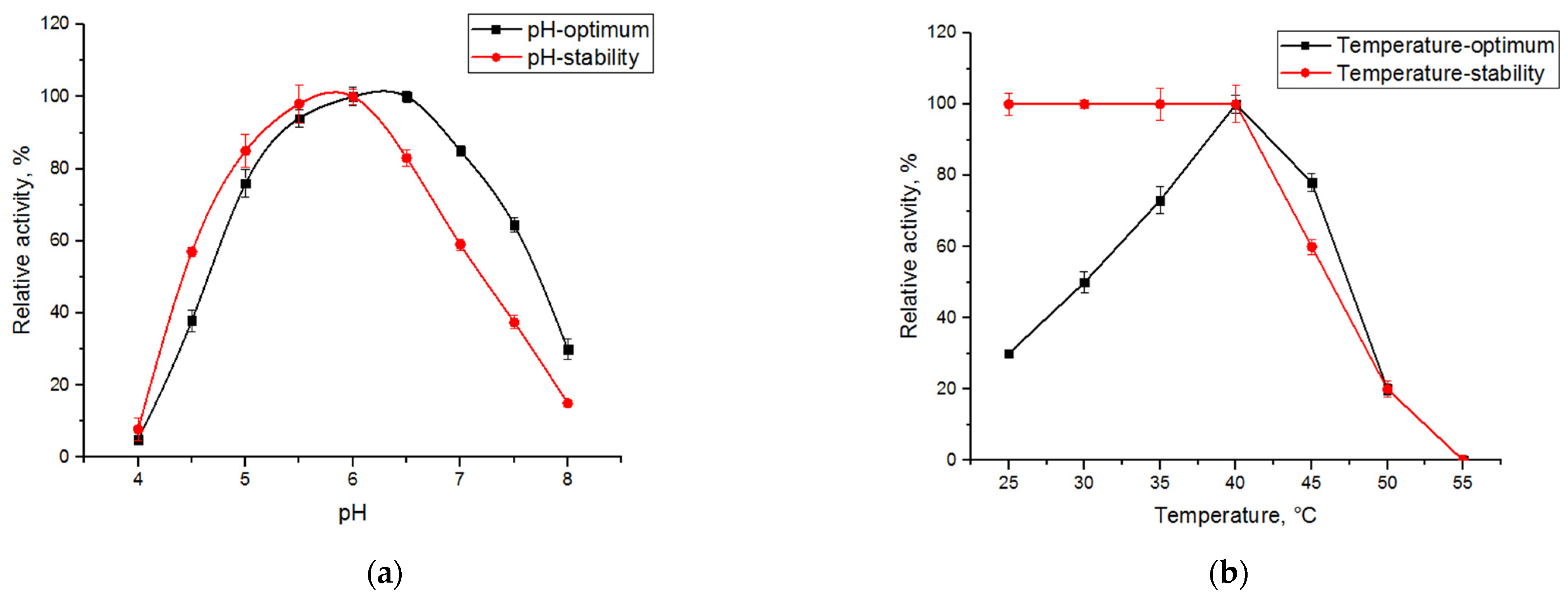
2.2. Characterization of Xanthan-Degrading Activity
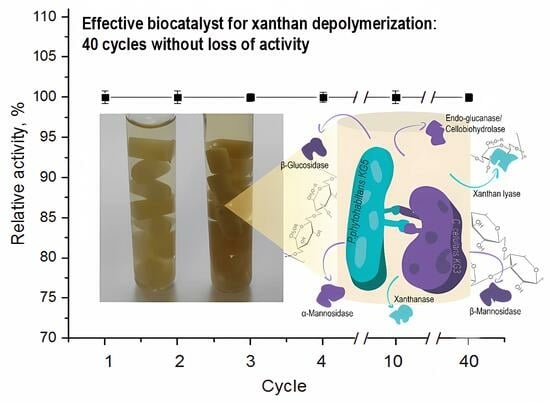
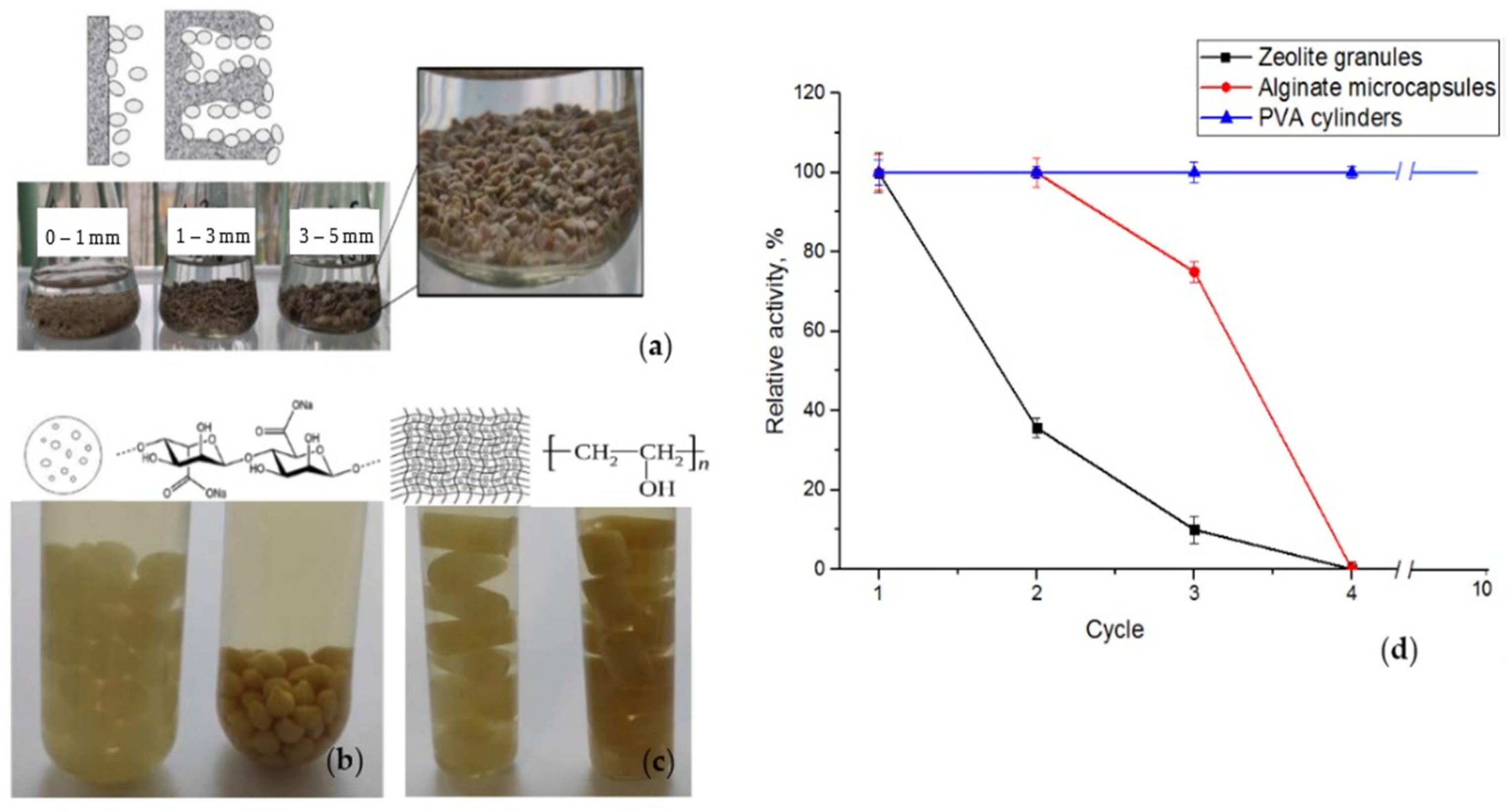
2.3. Immobilization of the LE-C1 Consortium
3. Materials and Methods
3.1. Chemicals
3.2. Screening for a Xanthan-Degrading Microorganisms
3.3. Genomic Identification
3.4. Analytic Methods
3.5. Enzyme Assays
3.6. Immobilization of the LE-C1 Consortium
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Becker, A.; Katzen, F.; Pühler, A.; Ielpi, L. Xanthan Gum Biosynthesis and Application: A Biochemical/Genetic Perspective. Appl. Microbiol. Biotechnol. 1998, 50, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Furtado, I.F.S.P.C.; Sydney, E.B.; Rodrigues, S.A.; Sydney, A.C.N. Xanthan Gum: Applications, Challenges, and Advantages of This Asset of Biotechnological Origin. Biotechnol. Res. Innov. 2022, 6, e202204. [Google Scholar] [CrossRef]
- More, T.T.; Yadav, J.S.S.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Extracellular Polymeric Substances of Bacteria and Their Potential Environmental Applications. J. Environ. Manag. 2014, 144, 1–25. [Google Scholar] [CrossRef]
- Wu, M.; Qu, J.; Tian, X.; Zhao, X.; Shen, Y.; Shi, Z.; Chen, P.; Li, G.; Ma, T. Tailor-Made Polysaccharides Containing Uniformly Distributed Repeating Units Based on the Xanthan Gum Skeleton. Int. J. Biol. Macromol. 2019, 131, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Bhat, I.M.; Wani, S.M.; Mir, S.A.; Masoodi, F.A. Advances in Xanthan Gum Production, Modifications and Its Applications. Biocatal. Agric. Biotechnol. 2022, 42, 102328. [Google Scholar] [CrossRef]
- Carvalho, L.T.; Vieira, T.A.; Zhao, Y.; Celli, A.; Medeiros, S.F.; Lacerda, T.M. Recent Advances in the Production of Biomedical Systems Based on Polyhydroxyalkanoates and Exopolysaccharides. Int. J. Biol. Macromol. 2021, 183, 1514–1539. [Google Scholar] [CrossRef]
- BeMiller, J.N. Carbohydrate Chemistry for Food Scientists, 3rd ed.; Elsevier Inc.: Philadelphia, PA, USA; Woodhead Publishing: Cambridge, UK; AACC International Press: St. Paul, MN, USA, 2018; Volume 11, pp. 261–269. [Google Scholar]
- Kool, M.M.; Gruppen, H.; Sworn, G.; Schols, H.A. The Influence of the Six Constituent Xanthan Repeating Units on the Order–Disorder Transition of Xanthan. Carbohydr. Polym. 2014, 104, 94–100. [Google Scholar] [CrossRef]
- Riaz, T.; Iqbal, M.W.; Jiang, B.; Chen, J. A Review of the Enzymatic, Physical, and Chemical Modification Techniques of Xanthan Gum. Int. J. Biol. Macromol. 2021, 186, 472–489. [Google Scholar] [CrossRef]
- Palaniraj, A.; Jayaraman, V. Production, Recovery and Applications of Xanthan Gum by Xanthomonas campestris. J. Food Eng. 2011, 106, 1–12. [Google Scholar] [CrossRef]
- Nsengiyumva, E.M.; Alexandridis, P. Xanthan Gum in Aqueous Solutions: Fundamentals and Applications. Int. J. Biol. Macromol. 2022, 216, 583–604. [Google Scholar] [CrossRef] [PubMed]
- Bagnol, R.; Grijpma, D.; Eglin, D.; Moriarty, T.F. The Production and Application of Bacterial Exopolysaccharides as Biomaterials for Bone Regeneration. Carbohydr. Polym. 2022, 291, 119550. [Google Scholar] [CrossRef]
- Han, G.; Chen, Q.; Liu, F.; Cui, Z.; Shao, H.; Liu, F.; Ma, A.; Liao, J.; Guo, B.; Guo, Y. Low Molecular Weight Xanthan Gum for Treating Osteoarthritis. Carbohydr. Polym. 2017, 164, 386–395. [Google Scholar] [CrossRef]
- Chen, Q.; Shao, X.; Ling, P.; Liu, F.; Shao, H.; Ma, A.; Wu, J.; Zhang, W.; Liu, F.; Han, G. Low Molecular Weight Xanthan Gum Suppresses Oxidative Stress-Induced Apoptosis in Rabbit Chondrocytes. Carbohydr. Polym. 2017, 169, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Xie, J.-L.; Liu, K.-Y.; Shan, J.-L.; Sun, Y.-G.; Ying, W.-G. Xanthan Gum Protects Temporomandibular Chondrocytes from IL-1β through Pin1/NF-κB Signaling Pathway. Mol. Med. Rep. 2020, 22, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, J.; Zhang, F.; Dou, X.; Ma, A.; Zhang, X.; Shao, H.; Zhao, S.; Ling, P.; Liu, F. Lower Range of Molecular Weight of Xanthan Gum Inhibits Apoptosis of Chondrocytes through MAPK Signaling Pathways. Int. J. Biol. Macromol. 2019, 130, 79–87. [Google Scholar] [CrossRef]
- Shao, X.; Chen, Q.; Dou, X.; Chen, L.; Wu, J.; Zhang, W.; Shao, H.; Ling, P.; Liu, F.; Wang, F. Lower Range of Molecular Weight of Xanthan Gum Inhibits Cartilage Matrix Destruction via Intrinsic Bax-Mitochondria Cytochrome c-Caspase Pathway. Carbohydr. Polym. 2018, 198, 354–363. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, Z.; Leng, K.; Li, B.; Feng, C.; Huo, X. Can Supramolecular Polymers Become Another Material Choice for Polymer Flooding to Enhance Oil Recovery? Polymers 2022, 14, 4405. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.E.; Smidsroed, O.; Elgsaeter, A.; Stokke, B.T. Depolymerization of Double-Stranded Xanthan by Acid Hydrolysis: Characterization of Partially Degraded Double Strands and Single-Stranded Oligomers Released from the Ordered Structures. Macromolecules 1993, 26, 6111–6120. [Google Scholar] [CrossRef]
- Cheetham, N.W.H.; Mashimba, E.N.M. Characterisation of Some Enzymic Hydrolysis Products of Xanthan. Carbohydr. Polym. 1991, 15, 195–206. [Google Scholar] [CrossRef]
- Christensen, B.E.; Smidsrød, O. Dependence of the Content of Unsubstituted (Cellulosic) Regions in Prehydrolysed Xanthans on the Rate of Hydrolysis by Trichoderma reesei Endoglucanase. Int. J. Biol. Macromol. 1996, 18, 93–99. [Google Scholar] [CrossRef]
- Yang, F.; Li, H.; Sun, J.; Guo, X.; Zhang, X.; Tao, M.; Chen, X.; Li, X. Novel Endotype Xanthanase from Xanthan-Degrading Microbacterium sp. Strain XT11. Appl. Environ. Microbiol. 2019, 85, e01800-18. [Google Scholar] [CrossRef] [PubMed]
- Nankai, H.; Hashimoto, W.; Miki, H.; Kawai, S.; Murata, K. Microbial System for Polysaccharide Depolymerization: Enzymatic Route for Xanthan Depolymerization by Bacillus sp. Strain GL1. Appl. Environ. Microbiol. 1999, 65, 2520–2526. [Google Scholar] [CrossRef]
- Ashraf, S.; Soudi, M.R.; Amoozegar, M.A.; Moshtaghi Nikou, M.; Spröer, C. Paenibacillus xanthanilyticus sp. nov., a Xanthan-Degrading Bacterium Isolated from Soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 76–80. [Google Scholar] [CrossRef]
- Muchová, M.; Růžička, J.; Julinová, M.; Doležalová, M.; Houser, J.; Koutný, M.; Buňková, L. Xanthan and Gellan Degradation by Bacteria of Activated Sludge. Water Sci. Technol. 2009, 60, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.; Soudi, M.R.; Ghadam, P. Production of Xanthanases by Paenibacillus spp.: Complete Xanthan Degradation and Possible Applications. Iran. J. Biotechnol. 2017, 15, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Cadmus, M.C.; Jackson, L.K.; Burton, K.A.; Plattner, R.D.; Slodki, M.E. Biodegradation of Xanthan Gum by Bacillus sp. Appl. Environ. Microbiol. 1982, 44, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Ruijssenaars, H.J.; de Bont, J.A.; Hartmans, S. A Pyruvated Mannose-Specific Xanthan Lyase Involved in Xanthan Degradation by Paenibacillus alginolyticus XL-1. Appl. Environ. Microbiol. 1999, 65, 2446–2452. [Google Scholar] [CrossRef]
- Ruijssenaars, H.J.; Hartmans, S.; Verdoes, J.C. A Novel Gene Encoding Xanthan Lyase of Paenibacillus alginolyticus Strain XL-1. Appl. Environ. Microbiol. 2000, 66, 3945–3950. [Google Scholar] [CrossRef]
- Liu, H.; Huang, C.; Dong, W.; Du, Y.; Bai, X.; Li, X. Biodegradation of Xanthan by Newly Isolated Cellulomonas Sp. LX, Releasing Elicitor-Active Xantho-Oligosaccharides-Induced Phytoalexin Synthesis in Soybean Cotyledons. Process Biochem. 2005, 40, 3701–3706. [Google Scholar] [CrossRef]
- Qian, F.; An, L.; Wang, M.; Li, C.; Li, X. Isolation and Characterization of a Xanthan-Degrading Microbacterium sp. Strain XT11 from Garden Soil. J. Appl. Microbiol. 2007, 102, 1362–1371. [Google Scholar] [CrossRef]
- Hu, X.; Wang, K.; Yu, M.; He, P.; Qiao, H.; Zhang, H.; Wang, Z. Characterization and Antioxidant Activity of a Low-Molecular-Weight Xanthan Gum. Biomolecules 2019, 9, 730. [Google Scholar] [CrossRef]
- Kool, M.M.; Gruppen, H.; Sworn, G.; Schols, H.A. Comparison of Xanthans by the Relative Abundance of Its Six Constituent Repeating Units. Carbohydr. Polym. 2013, 98, 914–921. [Google Scholar] [CrossRef] [PubMed]
- Cadmus, M.C.; Slodki, M.E.; Nicholson, J.J. High-Temperature, Salt-Tolerant Xanthanase. J. Ind. Microbiol. 1989, 4, 127–133. [Google Scholar] [CrossRef]
- Hou, C.T.; Barnabe, N.; Greaney, K. Biodegradation of Xanthan by Salt-Tolerant Aerobic Microorganisms. J. Ind. Microbiol. 1986, 1, 31–37. [Google Scholar] [CrossRef]
- Martins, S.C.S.; Martins, C.M.; Fiúza, L.M.C.G.; Santaella, S.T. Immobilization of Microbial Cells: A Promising Tool for Treatment of Toxic Pollutants in Industrial Wastewater. Afr. J. Biotechnol. 2013, 12, 4412–4418. [Google Scholar] [CrossRef]
- Yang, P.Y.; Cai, T.; Wang, M.-L. Immobilized Mixed Microbial Cells for Wastewater Treatment. Biol. Wastes 1988, 23, 295–312. [Google Scholar] [CrossRef]
- Abdul Manaf, S.A.; Mohamad Fuzi, S.F.Z.; Abdul Manas, N.H.; Md Illias, R.; Low, K.O.; Hegde, G.; Che Man, R.; Wan Azelee, N.I.; Matias-Peralta, H.M. Emergence of Nanomaterials as Potential Immobilization Supports for Whole Cell Biocatalysts and Cell Toxicity Effects. Biotechnol. Appl. Biochem. 2021, 68, 1128–1138. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, J.; Massey, I.Y.; Peng, T.; Yang, F. Immobilization of Microbes for Biodegradation of Microcystins: A Mini Review. Toxins 2022, 14, 573. [Google Scholar] [CrossRef] [PubMed]
- Polakovič, M.; Švitel, J.; Bučko, M.; Filip, J.; Neděla, V.; Ansorge-Schumacher, M.B.; Gemeiner, P. Progress in Biocatalysis with Immobilized Viable Whole Cells: Systems Development, Reaction Engineering and Applications. Biotechnol. Lett. 2017, 39, 667–683. [Google Scholar] [CrossRef]
- Mühlmann, M.; Kunze, M.; Ribeiro, J.; Geinitz, B.; Lehmann, C.; Schwaneberg, U.; Commandeur, U.; Büchs, J. Cellulolytic RoboLector—Towards an Automated High-Throughput Screening Platform for Recombinant Cellulase Expression. J. Biol. Eng. 2017, 11, 1. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jensen, P.F.; Kadziola, A.; Comamala, G.; Segura, D.R.; Anderson, L.; Poulsen, J.-C.N.; Rasmussen, K.K.; Agarwal, S.; Sainathan, R.K.; Monrad, R.N.; et al. Structure and Dynamics of a Promiscuous Xanthan Lyase from Paenibacillus nanensis and the Design of Variants with Increased Stability and Activity. Cell Chem. Biol. 2019, 26, 191–202.e6. [Google Scholar] [CrossRef] [PubMed]
- Dasman; Kajiyama, S.; Okazawa, A.; Fukusaki, E.; Kobayashi, A. Purification and Properties of an Enzyme Capable of Degrading the Polysaccharide of the Cyanobacterium, Nostoc commune. Z. Naturforschung C J. Biosci. 2002, 57, 1042–1046. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Cheng, L.; Meng, Y.; Li, S.; Zhao, X.; Du, Y.; Yin, H. Cellulosimicrobium cellulans Strain E4-5 Enzymatic Hydrolysis of Curdlan for Production of (1→3)-Linked β-d-Glucan Oligosaccharides. Carbohydr. Polym. 2015, 134, 740–744. [Google Scholar] [CrossRef]
- Dou, T.-Y.; Chen, J.; Hao, Y.-F.; Qi, X. Xylanosomes Produced by Cellulosimicrobium cellulans F16 Were Diverse in Size, but Resembled in Subunit Composition. Arch. Microbiol. 2019, 201, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Vu, N.T.-H.; Quach, T.N.; Dao, X.T.-T.; Le, H.T.; Le, C.P.; Nguyen, L.T.; Le, L.T.; Ngo, C.C.; Hoang, H.; Chu, H.H.; et al. A Genomic Perspective on the Potential of Termite-Associated Cellulosimicrobium cellulans MP1 as Producer of Plant Biomass-Acting Enzymes and Exopolysaccharides. PeerJ 2021, 9, e11839. [Google Scholar] [CrossRef]
- Kool, M.M.; Schols, H.A.; Delahaije, R.J.B.M.; Sworn, G.; Wierenga, P.A.; Gruppen, H. The Influence of the Primary and Secondary Xanthan Structure on the Enzymatic Hydrolysis of the Xanthan Backbone. Carbohydr. Polym. 2013, 97, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, S.R.; Shockley, K.R.; Conners, S.B.; Scott, K.L.; Wolfinger, R.D.; Kelly, R.M. Carbohydrate-Induced Differential Gene Expression Patterns in the Hyperthermophilic Bacterium Thermotoga maritima. J. Biol. Chem. 2003, 278, 7540–7552. [Google Scholar] [CrossRef]
- Emami Moghaddam, S.A.; Harun, R.; Mokhtar, M.N.; Zakaria, R. Potential of Zeolite and Algae in Biomass Immobilization. BioMed Res. Int. 2018, 2018, 6563196. [Google Scholar] [CrossRef]
- Wang, S.; Peng, Y. Natural Zeolites as Effective Adsorbents in Water and Wastewater Treatment. Chem. Eng. J. 2010, 156, 11–24. [Google Scholar] [CrossRef]
- Mery, C.; Guerrero, L.; Alonso-Gutiérrez, J.; Figueroa, M.; Lema, J.M.; Montalvo, S.; Borja, R. Evaluation of Natural Zeolite as Microorganism Support Medium in Nitrifying Batch Reactors: Influence of Zeolite Particle Size. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2012, 47, 420–427. [Google Scholar] [CrossRef]
- Kubota, M.; Nakabayashi, T.; Matsumoto, Y.; Shiomi, T.; Yamada, Y.; Ino, K.; Yamanokuchi, H.; Matsui, M.; Tsunoda, T.; Mizukami, F.; et al. Selective Adsorption of Bacterial Cells onto Zeolites. Colloids Surf. B Biointerfaces 2008, 64, 88–97. [Google Scholar] [CrossRef]
- Smidsrod, O.; Skjåk-Braek, G. Alginate as Immobilization Matrix for Cells. Trends Biotechnol. 1990, 8, 71–78. [Google Scholar] [CrossRef]
- Lozinsky, V.I. Polymeric Cryogels as a New Family of Macroporous and Supermacroporous Materials for Biotechnological Purposes. Russ. Chem. Bull. 2008, 57, 1015–1032. [Google Scholar] [CrossRef]
- Berillo, D.; Al-Jwaid, A.; Caplin, J. Polymeric Materials Used for Immobilisation of Bacteria for the Bioremediation of Contaminants in Water. Polymers 2021, 13, 1073. [Google Scholar] [CrossRef] [PubMed]
- Razavi, M.; Qiao, Y.; Thakor, A.S. Three-Dimensional Cryogels for Biomedical Applications. J. Biomed. Mater. Res. A 2019, 107, 2736–2755. [Google Scholar] [CrossRef]
- Plieva, F.M.; Galaev, I.Y.; Noppe, W.; Mattiasson, B. Cryogel Applications in Microbiology. Trends Microbiol. 2008, 16, 543–551. [Google Scholar] [CrossRef]
- Ibatullin, F.; Selivanov, S.; Shavva, A. A General Procedure for Conversion of S-Glycosyl Isothiourea Derivatives into Thioglycosides, Thiooligosaccharides and Glycosyl Thioesters. Synthesis 2001, 2001, 0419–0422. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.; Palmer, J.D. Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit RRNA Sequence Analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Rudi, K.; Skulberg, O.M.; Larsen, F.; Jakobsen, K.S. Strain Characterization and Classification of Oxyphotobacteria in Clone Cultures on the Basis of 16S RRNA Sequences from the Variable Regions V6, V7, and V8. Appl. Environ. Microbiol. 1997, 63, 2593–2599. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hashimoto, W.; Miki, H.; Tsuchiya, N.; Nankai, H.; Murata, K. Xanthan Lyase of Bacillus sp. Strain GL1 Liberates Pyruvylated Mannose from Xanthan Side Chains. Appl. Environ. Microbiol. 1998, 64, 3765–3768. [Google Scholar] [CrossRef]
- Gür, S.D.; İdil, N.; Aksöz, N. Optimization of Enzyme Co-Immobilization with Sodium Alginate and Glutaraldehyde-Activated Chitosan Beads. Appl. Biochem. Biotechnol. 2018, 184, 538–552. [Google Scholar] [CrossRef]
- Stepanov, N.; Efremenko, E. “Deceived” Concentrated Immobilized Cells as Biocatalyst for Intensive Bacterial Cellulose Production from Various Sources. Catalysts 2018, 8, 33. [Google Scholar] [CrossRef]


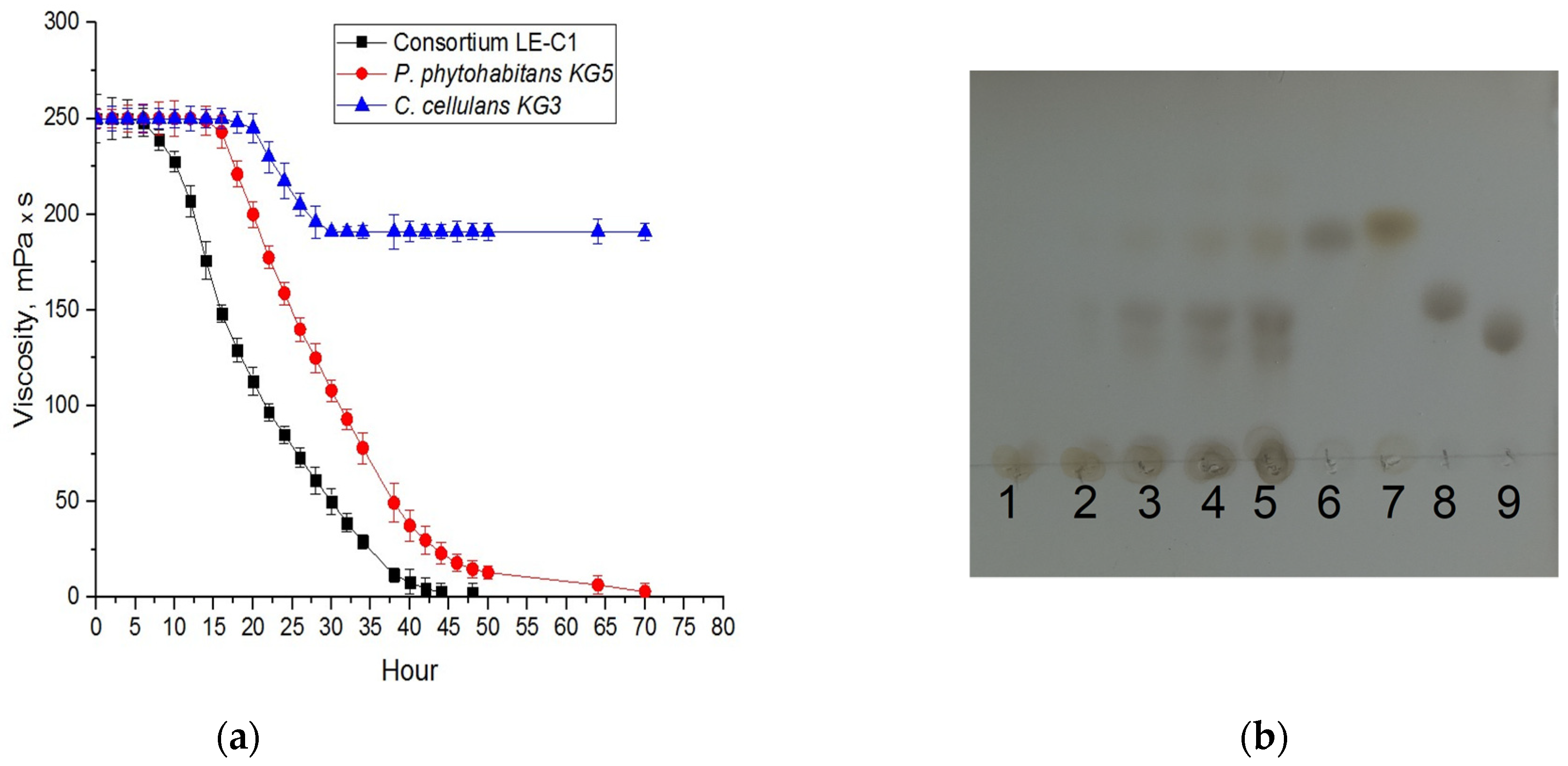
| Microorganism(s) | Specific Activity * | |||||
|---|---|---|---|---|---|---|
| Xanthanase, U/g | Xanthan Lyase, U/g | β-Glucosidase, U/g | Endo-Glucanase/Cellobiohydrolase, U/g | α-Mannosidase, U/g | β-Mannosidase, U/g | |
| LE-C1 | 19.6 ± 0.6 | 2200 ± 70 | 3.4 ± 0.1 | 4.0 ± 0.1 | 68.0 ± 2.0 | 0.40 ± 0.01 |
| P. phytohabitans KG5 | 19.3 ± 0.6 | 1660 ± 50 | n.d. | n.d. | n.d. | n.d. |
| C. cellulans KG3 | n.d. | n.d. | 80.0 ± 2.4 | 30.0 ± 0.9 | 130.0 ± 3.9 | 70.0 ± 2.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhurishkina, E.V.; Eneyskaya, E.V.; Shvetsova, S.V.; Yurchenko, L.V.; Bobrov, K.S.; Kulminskaya, A.A. Whole-Cell PVA Cryogel-Immobilized Microbial Consortium LE-C1 for Xanthan Depolymerization. Catalysts 2023, 13, 1249. https://doi.org/10.3390/catal13091249
Zhurishkina EV, Eneyskaya EV, Shvetsova SV, Yurchenko LV, Bobrov KS, Kulminskaya AA. Whole-Cell PVA Cryogel-Immobilized Microbial Consortium LE-C1 for Xanthan Depolymerization. Catalysts. 2023; 13(9):1249. https://doi.org/10.3390/catal13091249
Chicago/Turabian StyleZhurishkina, Elena V., Elena V. Eneyskaya, Svetlana V. Shvetsova, Lyudmila V. Yurchenko, Kirill S. Bobrov, and Anna A. Kulminskaya. 2023. "Whole-Cell PVA Cryogel-Immobilized Microbial Consortium LE-C1 for Xanthan Depolymerization" Catalysts 13, no. 9: 1249. https://doi.org/10.3390/catal13091249
APA StyleZhurishkina, E. V., Eneyskaya, E. V., Shvetsova, S. V., Yurchenko, L. V., Bobrov, K. S., & Kulminskaya, A. A. (2023). Whole-Cell PVA Cryogel-Immobilized Microbial Consortium LE-C1 for Xanthan Depolymerization. Catalysts, 13(9), 1249. https://doi.org/10.3390/catal13091249