Thermostable CaCO3-Immobilized Bacillus subtilis Lipase for Sustainable Biodiesel Production from Waste Cooking Oil
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening and Molecular Identification of the Lipase-Producing Strain
2.2. Growth and Lipase Activity Characteristics of Bacillus subtilis
2.3. Optimization of B. subtilis Lipase Production
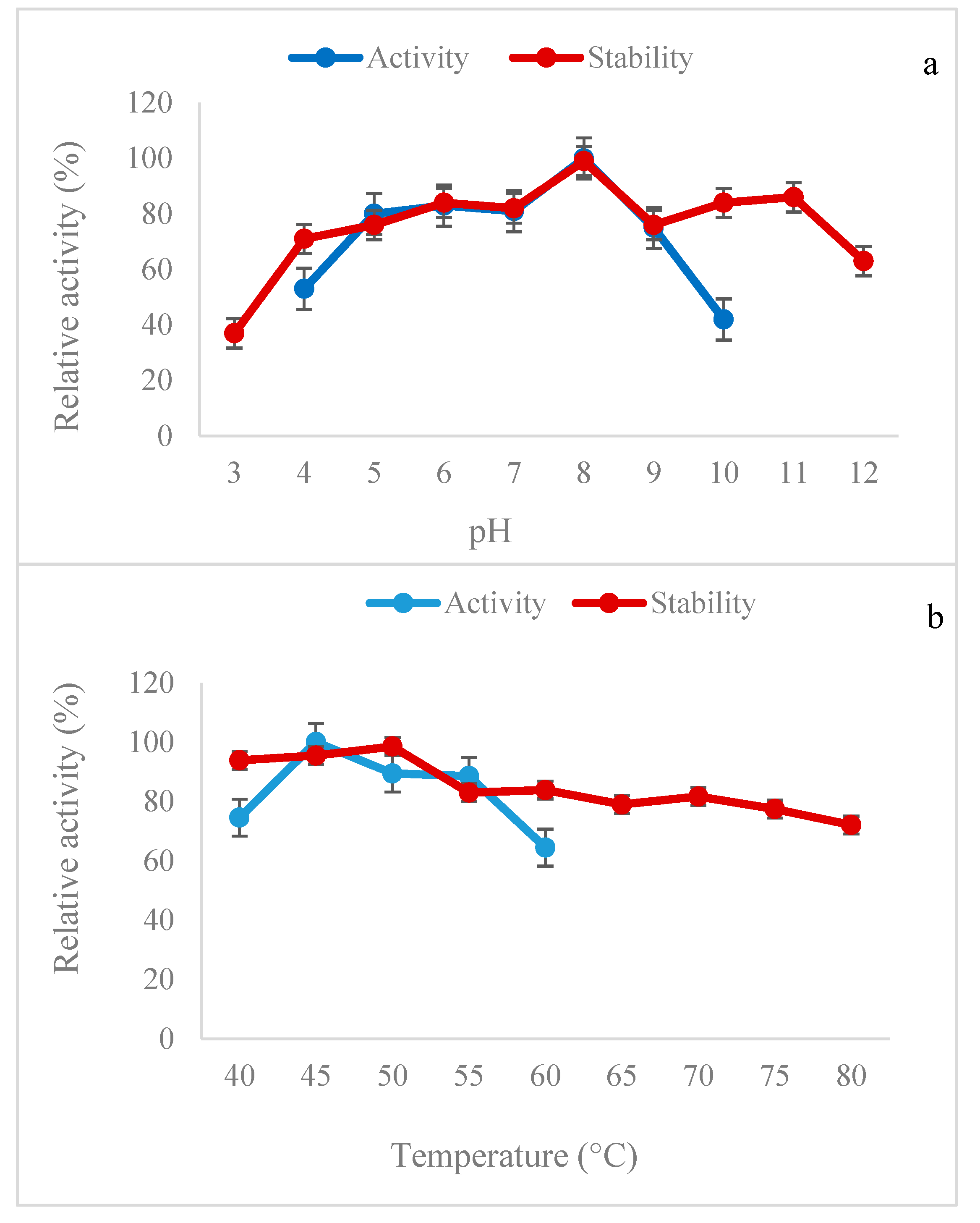
2.4. Effects of pH and Temperature on B. subtilis Lipase Activity and Stability
2.5. Effects of Incubation Time and Substrate Concentration on Lipase Activity
2.6. Effects of Metal Ions on Lipase Activity
2.7. Effects of Enzyme Inhibitors and Surfactants on the Stability of the Lipase
2.8. Effects of Organic Solvents on the Stability of Lipase
2.9. Optimization of the Transesterification Reaction with the Immobilized Lipase
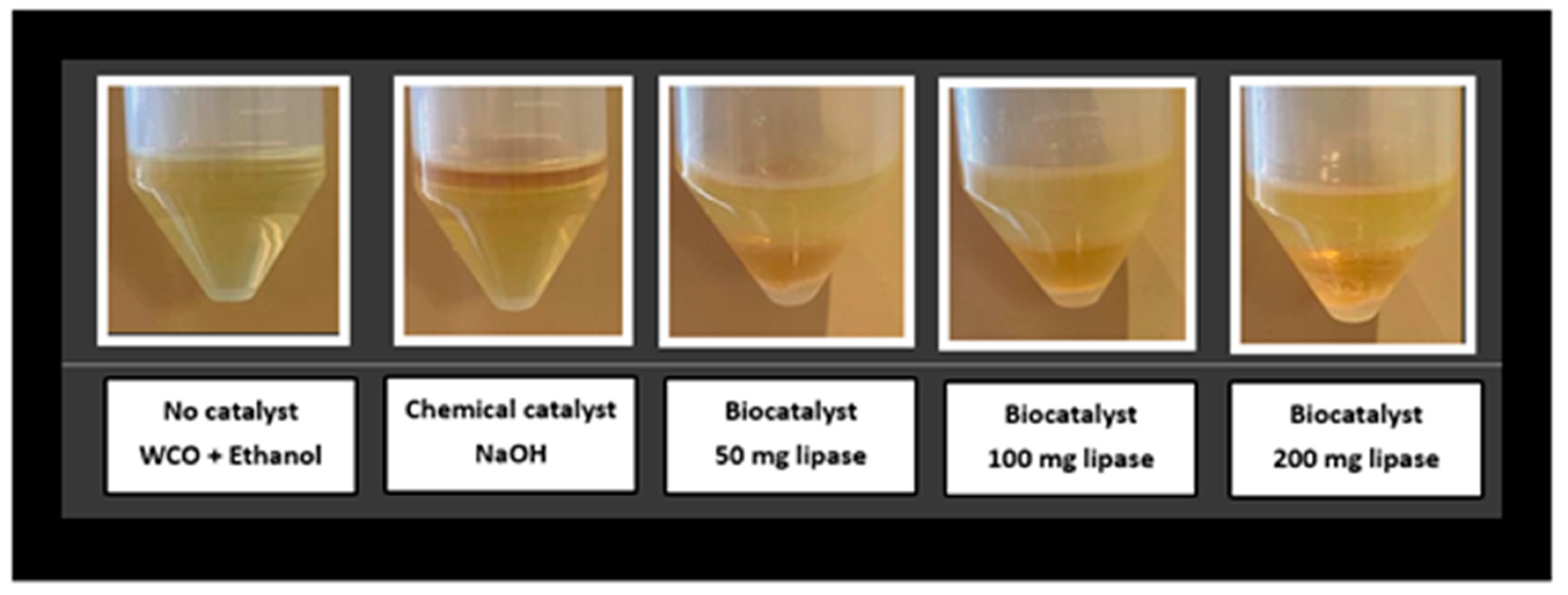
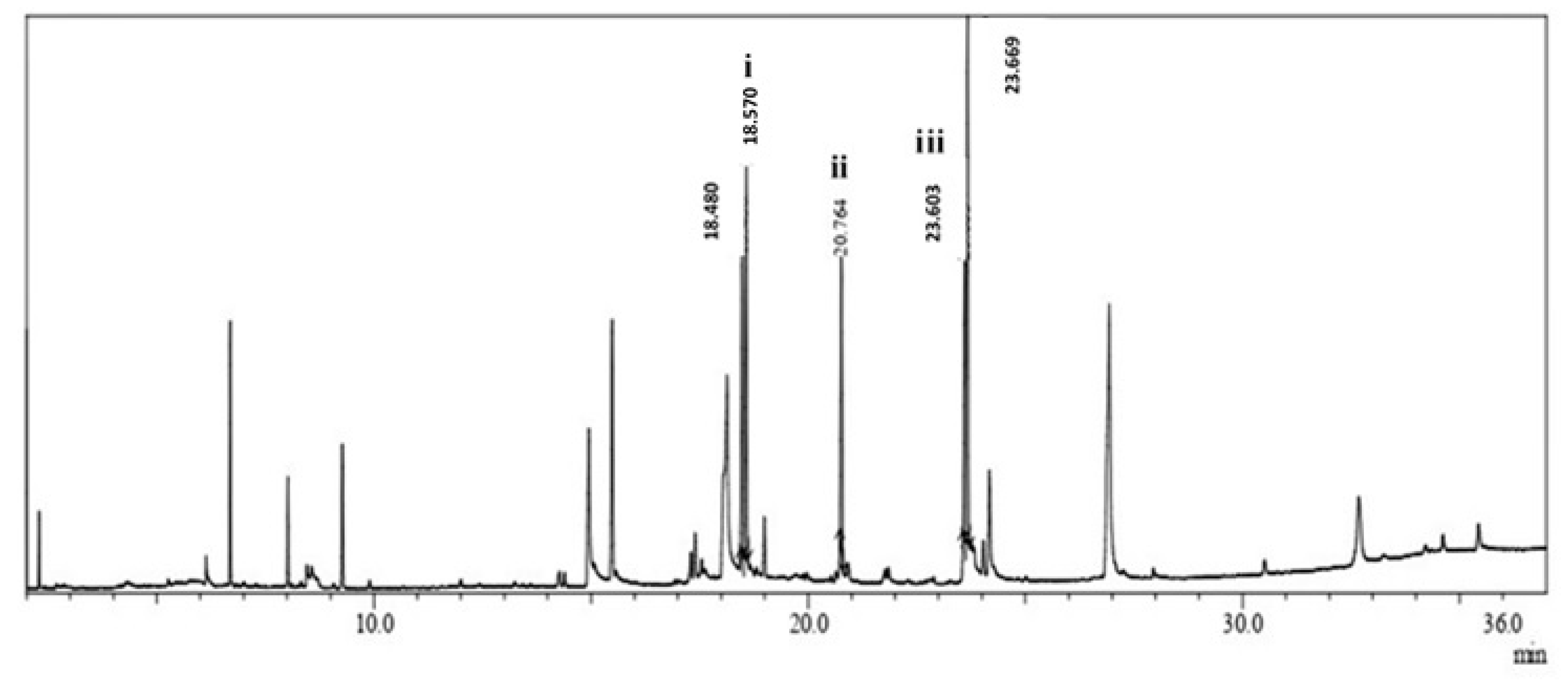
2.10. Transesterification Reaction
2.11. Reusability of the Immobilized Lipase
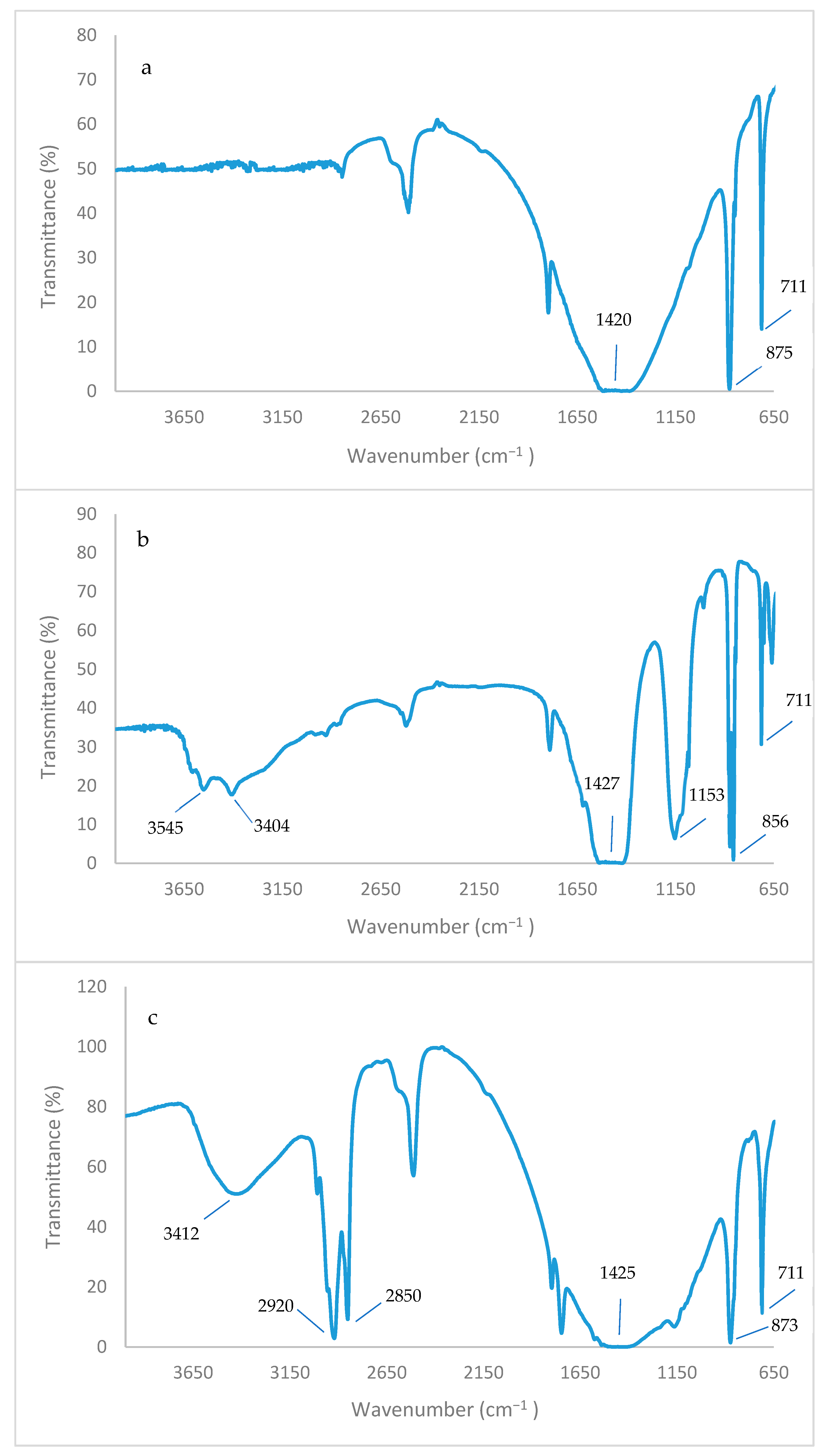
2.12. FTIR Analysis
3. Material and Methods
3.1. Chemicals
3.2. Screening of Lipase-Producing Bacteria
3.3. Identification of the Selected Strain
3.4. Growth of Lipase-Producing Bacteria
3.5. Lipase Activity Measurement
3.6. Media Optimization for Bacillus subtilis Lipase Production
3.7. Effects of pH and Temperature on Lipase Activity and Stability
3.8. Effects of Incubation Time and Substrate Concentration on Lipase Activity
3.9. Effects of Metal Ions, Enzyme Inhibitors, and Surfactants on Bacillus subtilis Lipase Activity
3.10. Effects of Organic Solvents on the Stability of Lipase
3.11. Immobilization of Bacillus subtilis Lipase
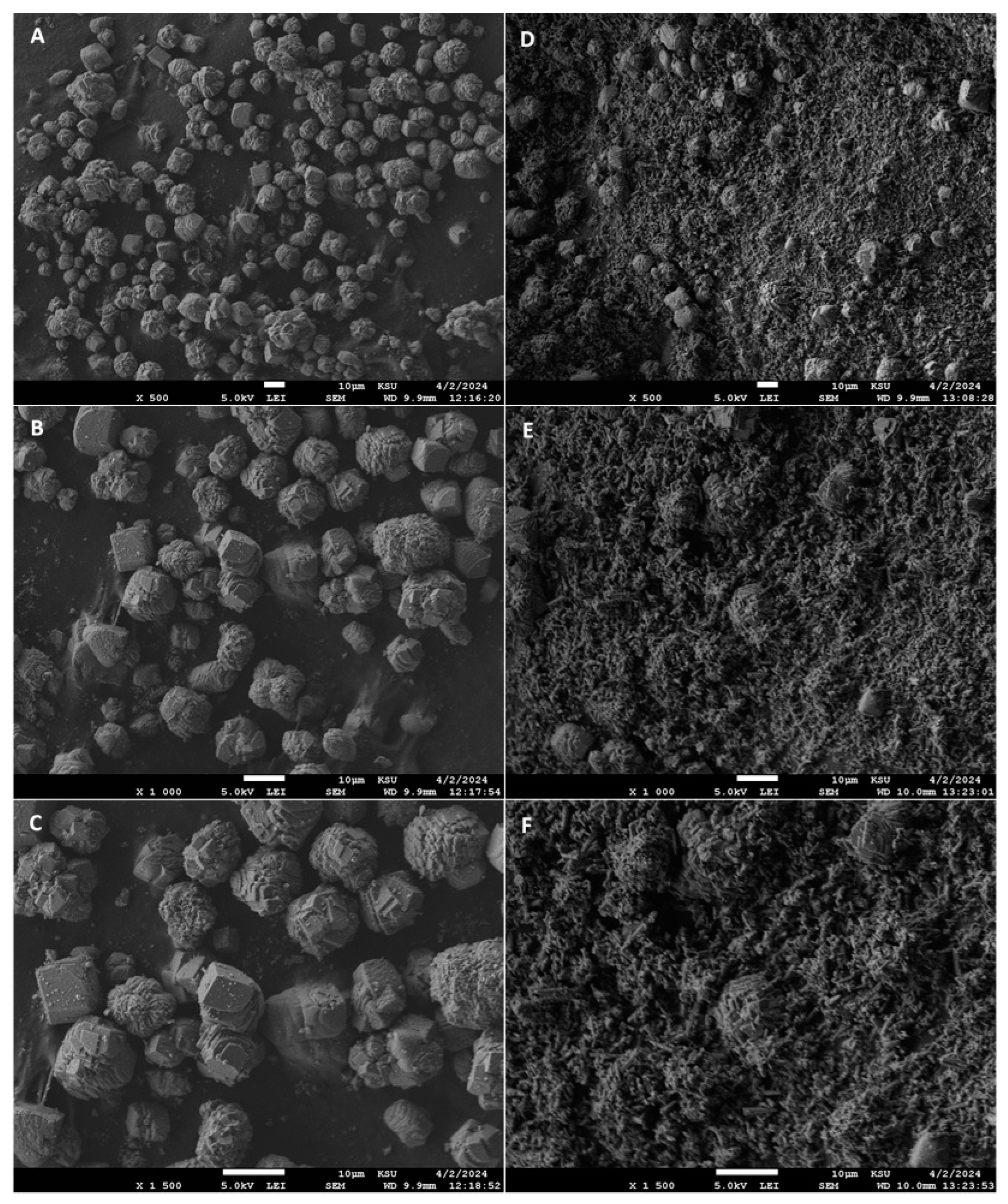
3.12. FT-IR and FE-SEM Analyses
3.13. Optimization of the Biodiesel Synthesis Process from Waste Cooking Oil
3.14. Fatty Acid Ethyl Ester Analysis
3.15. Reusability of the Immobilized Lipase
3.16. Statistical Analyses
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Souza, A.B.; Ochoa, A.A.V.; Costa, J.Â.P.d.; Leite, G.d.N.P.; Silva, H.C.N.; Tómas, A.C.C.; Barbosa, D.C.; Michima, P.S.A. A Review of Tropical Organic Materials for Biodiesel as a Substitute Energy Source in Internal Combustion Engines: A Viable Solution? Energies 2023, 16, 3736. [Google Scholar] [CrossRef]
- Deshmukh, S.; Kumar, R.; Bala, K. Microalgae biodiesel: A review on oil extraction, fatty acid composition, properties and effect on engine performance and emissions. Fuel Process. Technol. 2019, 191, 232–247. [Google Scholar] [CrossRef]
- Muniru, O.; Ezeanyanaso, C.; Fagbemigun, T.; Emmanuel, A.; Oyewole, A.; Okunola, O.; Asieba, G.; Shifatu, A.; Igwe, C.; Elemo, G. Valorization of Biodiesel Production: Focus on Crude Glycerine Refining/Purification. J. Sci. Res. Rep. 2016, 11, 1–8. [Google Scholar] [CrossRef]
- Brahma, S.; Nath, B.; Basumatary, B.; Das, B.; Saikia, P.; Patir, K.; Basumatary, S. Biodiesel production from mixed oils: A sustainable approach towards industrial biofuel production. Chem. Eng. J. Adv. 2022, 10, 100284. [Google Scholar] [CrossRef]
- Thangaraj, B.; Solomon, P.R.; Muniyandi, B.; Ranganathan, S.; Lin, L. Catalysis in biodiesel production—A review. Clean Energy 2018, 3, 2–23. [Google Scholar] [CrossRef]
- Smith, M.R.; Khera, E.; Wen, F. Engineering Novel and Improved Biocatalysts by Cell Surface Display. Ind. Eng. Chem. Res. 2015, 54, 4021–4032. [Google Scholar] [CrossRef]
- Horchani, H.; Bouaziz, A.; Gargouri, Y.; Sayari, A. Immobilized Staphylococcus xylosus lipase-catalyzed synthesis of ricinoleic acid esters. J. Mol. Catal. B-Enzym. 2012, 75, 35–42. [Google Scholar] [CrossRef]
- Bouaziz, A.; Horchani, H.; Ben Salem, N.; Chaari, A.; Chaabouni, M.; Gargouri, Y.; Sayari, A. Enzymatic propyl gallate synthesis in solvent-free system: Optimization by response surface methodology. J. Mol. Catal. B-Enzym. 2020, 67, 242–250. [Google Scholar] [CrossRef]
- Patchimpet, J.; Simpson, B.K.; Sangkharak, K.; Klomklao, S. Optimization of process variables for the production of biodiesel by transesterification of used cooking oil using lipase from Nile tilapia viscera. Renew. Energy 2020, 153, 861–869. [Google Scholar] [CrossRef]
- Ugur, A.; Sarac, N.; Boran, R.; Ayaz, B.; Ceylan, O.; Okmen, G. New Lipase for Biodiesel Production: Partial Purification and Characterization of LipSB 25-4. ISRN Biochem. 2014, 2014, 289749. [Google Scholar] [CrossRef]
- Boas, R.N.V.; Ceron, A.A.; Bento, H.B.; de Castro, H.F. Application of an immobilized Rhizopus oryzae lipase to batch and continuous ester synthesis with a mixture of a lauric acid and fusel oil. Biomass Bioenerg. 2018, 119, 61–68. [Google Scholar] [CrossRef]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic synthesis for industrial applications. Angew. Chem. Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef] [PubMed]
- Kinner, A.; Nerke, P.; Siedentop, R.; Steinmetz, T.; Classen, T.; Rosenthal, K.; Lütz, S. Recent advances in biocatalysis for drug synthesis. Biomedicines 2022, 10, 964. [Google Scholar] [CrossRef] [PubMed]
- Horchani, H.; Chaabouni, M.; Gargouri, Y.; Sayari, A. Solvent-free lipase-catalyzed synthesis of long-chain starch esters using microwave heating. Optimization by response surface methodology. Carbohydr. Polym. 2010, 79, 466–474. [Google Scholar] [CrossRef]
- Mahfoudhi, A.; Ben Mabrouk, S.; Hadrich, B.; Mhadhbi, M.; Abderrazak, H.; Alghamdi, O.A.; Fendri, A.; Sayari, A. Efficient green enzymatic synthesis of lipophilic piperic acid esters by immobilized Rhizopus oryzae lipase: Optimization and antioxydant activities. Catal. Lett. 2024, 1–19. [Google Scholar] [CrossRef]
- Maheshwari, P.; Haider, M.B.; Yusuf, M.; Klemeš, J.J.; Bokhari, A.; Beg, M.; Al-Othman, A.; Kumar, R.; Jaiswal, A.K. A review on latest trends in cleaner biodiesel production: Role of feedstock, production methods, and catalysts. J. Clean. Prod. 2022, 355, 131588. [Google Scholar] [CrossRef]
- Moazeni, F.; Chen, Y.-C.; Zhang, G. Enzymatic transesterification for biodiesel production from used cooking oil, a review. J. Clean. Prod. 2019, 216, 117–128. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories 2020, 19, 169. [Google Scholar] [CrossRef] [PubMed]
- Horchani, H.; Ben Salem, N.; Zarai, Z.; Sayari, A.; Gargouri, A.; Chaâbouni, M. Enzymatic synthesis of eugenol benzoate by immobilized Staphylococcus aureus lipase: Optimization using response surface methodology and determination of antioxidant activity. Bioresour. Technol. 2010, 101, 2809–2817. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Liu, K.; Liu, H.; Jiang, Y.; Wang, R.; Wang, W.; Wang, T. A Valuable Product of Microbial Cell Factories: Microbial Lipase. Front. Microbiol. 2021, 12, 743377. [Google Scholar] [CrossRef]
- Mahfoudhi, A.; Benmabrouk, S.; Fendri, A.; Sayari, A. Fungal lipases as biocatalysts: A promising platform in several industrial biotechnology applications. Biotechnol. Bioeng. 2022, 119, 3370–3392. [Google Scholar] [CrossRef]
- Nguyen, L.N.; Dao, T.T.; Živković, T.; Fehrholz, M.; Schäfer, W.; Salomon, S. Enzymatic properties and expression patterns of five extracellular lipases of Fusarium graminearum in vitro. Enzym. Microb. Technol. 2010, 46, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, G.; Ahmad, E.; Ahmad, A.; Khan, R.H. Structural features, temperature adaptation and industrial applications of microbial lipases from psychrophilic, mesophilic and thermophilic origins. Int. J. Biol. Macromol. 2023, 225, 822–839. [Google Scholar] [CrossRef]
- Mazhar, H.; Abbas, N.; Hussain, Z.; Sohail, A.; Ali, S. Extracellular lipase production from Bacillus subtilis using agro-industrial waste and fruit peels. Punjab Univ. J. Zool. 2016, 31, 261–267. [Google Scholar]
- Ayaz, B.; Ugur, A.; Boran, R. Purification and characterization of organic solvent-tolerant lipase from Streptomyces sp. OC119-7 for biodiesel production. Biocatal. Agric. Biotechnol. 2015, 4, 103–108. [Google Scholar] [CrossRef]
- Toldrá-Reig, F.; Mora, L.; Toldrá, F. Developments in the Use of Lipase Transesterification for Biodiesel Production from Animal Fat Waste. Appl. Sci. 2020, 10, 5085. [Google Scholar] [CrossRef]
- Emmanouilidou, E.; Lazaridou, A.; Mitkidou, S.; Kokkinos, N.C. A comparative study on biodiesel production from edible and non-edible biomasses. J. Mol. Struct. 2024, 1306, 137870. [Google Scholar] [CrossRef]
- Pullen, J.; Saeed, K. An overview of biodiesel oxidation stability. Renew. Sustain. Energy Rev. 2012, 16, 5924–5950. [Google Scholar] [CrossRef]
- Kokkinos, N.C.; Emmanouilidou, E.; Sharma, S.K. Waste-To-Biofuel Production for the Transportation Sector. In Intelligent Transportation System and Advanced Technology. Energy, Environment, and Sustainability; Upadhyay, R.K., Sharma, S.K., Kumar, V., Eds.; Springer: Singapore, 2024. [Google Scholar] [CrossRef]
- Hannane, F.; Elmossaoui, H.; Nguyen, T.V.; Petit, P.; Aillerie, M.; Charles, J.P. Forecasting the PV Panel Operating Conditions Using the Design of Experiments Method. Energy Procedia 2013, 36, 479–487. [Google Scholar] [CrossRef]
- Draper, N.R. Introduction to Box and Wilson (1951) on the Experimental Attainment of Optimum Conditions. In Breakthroughs in Statistics: Methodology and Distribution; Kotz, S., Johnson, N.L., Eds.; Springer: New York, NY, USA, 1992; pp. 267–269. [Google Scholar]
- Nassiri Mahallati, M. Chapter 9—Advances in Modeling Saffron Growth and Development at Different Scales; Saffron Koocheki, A., Khajeh-Hosseini, M., Eds.; Woodhead Publishing: Sawston, UK, 2020; pp. 139–167. [Google Scholar]
- Abdella Akhter, K.; Karim, I.; Aziz, B.; Bibi, A.; Khan, J.; Akhtar, T. Optimization and characterization of alkaliphilic lipase from a novel Bacillus cereus NC7401 strain isolated from diesel fuel polluted soil. PLoS ONE 2022, 17, e0273368. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, M.; Zeng, Z.; Yu, P.; Gong, D.; Deng, S. Production, purification and biochemical characterisation of a novel lipase from a newly identified lipolytic bacterium Staphylococcus caprae NCU S6. J. Enzym. Inhib. Med. Chem. 2021, 36, 248–256. [Google Scholar] [CrossRef]
- Mazhar, H.; Abbas, N.; Ali, S.; Sohail, A.; Hussain, Z.; Ali, S.S. Optimized production of lipase from Bacillus subtilis PCSIRNL-39. Afr. J. Biotechnol. 2017, 16, 1106–1115. [Google Scholar]
- Jigajinni, S.K.; Meti, B.S. Immobilization optimization and characterization of immobilized lipase from Lysinibacillus macroides FS1 for biodiesel production. Int. J. Curr. Microbiol. App. Sci 2021, 10, 232–245. [Google Scholar]
- Al-Haidari, A.; Alsaadawi, I.; Khudhair, S.H. Determination the Optimum Conditions of the Activity and Stability of Lipase Extracted from Sunflower Germinated Seeds. Iraqi J. Sci. 2021, 62, 431–440. [Google Scholar] [CrossRef]
- Abdella, B.; Youssif, A.M.; Sabry, S.A.; Ghozlan, H.A. Production, purification, and characterization of cold-active lipase from the psychrotroph Pseudomonas sp. A6. Braz. J. Microbiol. 2023, 54, 1623–1633. [Google Scholar] [CrossRef]
- Park, J.-Y.; Ha, J.; Choi, Y.; Chang, P.-S.; Park, K.-M. Optimization of spectrophotometric and fluorometric assays using alternative substrates for the high-throughput screening of lipase activity. J. Chem. 2021, 2021, 3688124. [Google Scholar] [CrossRef]
- Martínez, R.; Bernal, C.; Álvarez, R.; Concha, C.; Araya, F.; Cabrera, R.; Dhoke, G.V.; Davari, M.D. Deletion and Randomization of Structurally Variable Regions in B. subtilis Lipase A (BSLA) Alter Its Stability and Hydrolytic Performance Against Long Chain Fatty Acid Esters. Int. J. Mol. Sci. 2020, 21, 1990. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, A.; Tani, T.; Tominaga, Y. Purification and characterization of a novel thermostable lipase from Bacillus sp. J. Biochem. 1991, 109, 211–216. [Google Scholar] [PubMed]
- Hasanah, N.; Laila, A.; Satria, H.; Juliasih, N.L.G.R.; Husna, Q.N. Characterization of Organic Solvent Tolerance Lipase from Compost Indigenous Bacteria. Atl. Highlights Chem. Pharm. Sci. 2023, 20–29. [Google Scholar] [CrossRef]
- Ghori, M.; Iqbal, M.; Hameed, A. Characterization of a novel lipase from Bacillus sp. isolated from tannery wastes. Braz. J. Microbiol. 2011, 42, 22–29. [Google Scholar] [CrossRef]
- Bharathiraja, D.B. Optimization of biodiesel production from waste cooking oil using pure lipase and Rhizopus oryzae through response surface methodology. Asian J. Microbiol. Biotech. Environ. Sci. 2013, 16, 63–70. [Google Scholar]
- Binhayeeding, N.; Klomklao, S.; Prasertsan, P.; Sangkharak, K. Improvement of biodiesel production using waste cooking oil and applying single and mixed immobilised lipases on polyhydroxyalkanoate. Renew. Energy 2020, 162, 1819–1827. [Google Scholar] [CrossRef]
- Tran, D.-T.; Chen, C.-L.; Chang, J.-S. Effect of solvents and oil content on direct transesterification of wet oil-bearing microalgal biomass of Chlorella vulgaris ESP-31 for biodiesel synthesis using immobilized lipase as the biocatalyst. Bioresour. Technol. 2013, 135, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Ben Bacha, A.; Alonazi, M.; Alharbi, M.G.; Horchani, H.; Ben Abdelmalek, I. Biodiesel Production by Single and Mixed Immobilized Lipases Using Waste Cooking Oil. Molecules 2022, 27, 8736. [Google Scholar] [CrossRef]
- Kasirajan, R. Biodiesel production by two step process from an energy source of Chrysophyllum albidum oil using homogeneous catalyst. S. Afr. J. Chem. Eng. 2021, 37, 161–166. [Google Scholar] [CrossRef]
- Shi, S.; Valle-Rodríguez, J.O.; Khoomrung, S.; Siewers, V.; Nielsen, J. Functional expression and characterization of five wax ester synthases in Saccharomyces cerevisiae and their utility for biodiesel production. Biotechnol. Biofuels 2012, 5, 7. [Google Scholar] [CrossRef]
- Moreira, K.; de Oliveira, A.L.; Júnior, L.; Monteiro, R.; Rocha, T.; Menezes, F.; Dutra, L.; Denardin, J.; Michea, S.; Freire, R.; et al. Lipase From Rhizomucor miehei Immobilized on Magnetic Nanoparticles: Performance in Fatty Acid Ethyl Ester (FAEE) Optimized Production by the Taguchi Method. Front. Bioeng. Biotechnol. 2020, 8, 693. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Martín, E.; Otero, C. Different enzyme requirements for the synthesis of biodiesel: Novozym® 435 and Lipozyme® TL IM. Bioresour. Technol. 2008, 99, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Marín-Suárez, M.; Méndez-Mateos, D.; Guadix, A.; Guadix, E.M. Reuse of immobilized lipases in the transesterification of waste fish oil for the production of biodiesel. Renew. Energy 2019, 140, 1–8. [Google Scholar] [CrossRef]
- Długosz, O.; Lis, K.; Banach, M. Synthesis and antimicrobial properties of CaCO3-nAg and nAg-CaCO3 nanocomposites. Nanotechnology 2020, 32, 025715. [Google Scholar] [CrossRef]
- Demir, S.; Gök, S.B.; Kahraman, M.V. α-Amylase immobilization on functionalized nano CaCO3 by covalent attachment. Starch–Stärke 2011, 64, 3–9. [Google Scholar] [CrossRef]
- Ilesanmi, O.I.; Adekunle, A.E.; Omolaiye, J.A.; Olorode, E.M.; Ogunkanmi, A.L. Isolation, optimization and molecular characterization of lipase producing bacteria from contaminated soil. Sci. Afr. 2020, 8, e00279. [Google Scholar] [CrossRef]
- Buonaurio, R.; Stravato, V.M.; Cappelli, C. Brown spot caused by Sphingomonas sp. on yellow Spanish melon fruits in Spain. Plant Pathol. 2001, 50, 397–401. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Singh, J.; Bharti, R.k.; Thakur, I.S. Isolation, Purification and Characterization of Lipase from Microbacterium sp. and its Application in Biodiesel Production. Energy Procedia 2014, 54, 518–529. [Google Scholar] [CrossRef]
- Mahnashi, M.H.; Hombalimath, V.S.; Shaikh, I.A.; Muddapur, U.M.; Desai, S.V.; Achappa, S.; El-Sherbiny, M.; Ghoneim, M.M.; Jefri, O.A.; Alshahrani, M.M.; et al. Production of Extracellular Lipase by Bacillus halotolerans from Oil-Contaminated Soil in a Pilot-Scale Submerged Bioreactor. Processes 2022, 10, 1548. [Google Scholar] [CrossRef]
- Haq, A.; Adeel, S.; Khan, A.; Rana, Q.U.A.; Khan, M.A.N.; Rafiq, M.; Ishfaq, M.; Khan, S.; Shah, A.A.; Hasan, F.; et al. Screening of Lipase-Producing Bacteria and Optimization of Lipase-Mediated Biodiesel Production from Jatropha curcas Seed Oil Using Whole Cell Approach. BioEnergy Res. 2020, 13, 1280–1296. [Google Scholar] [CrossRef]


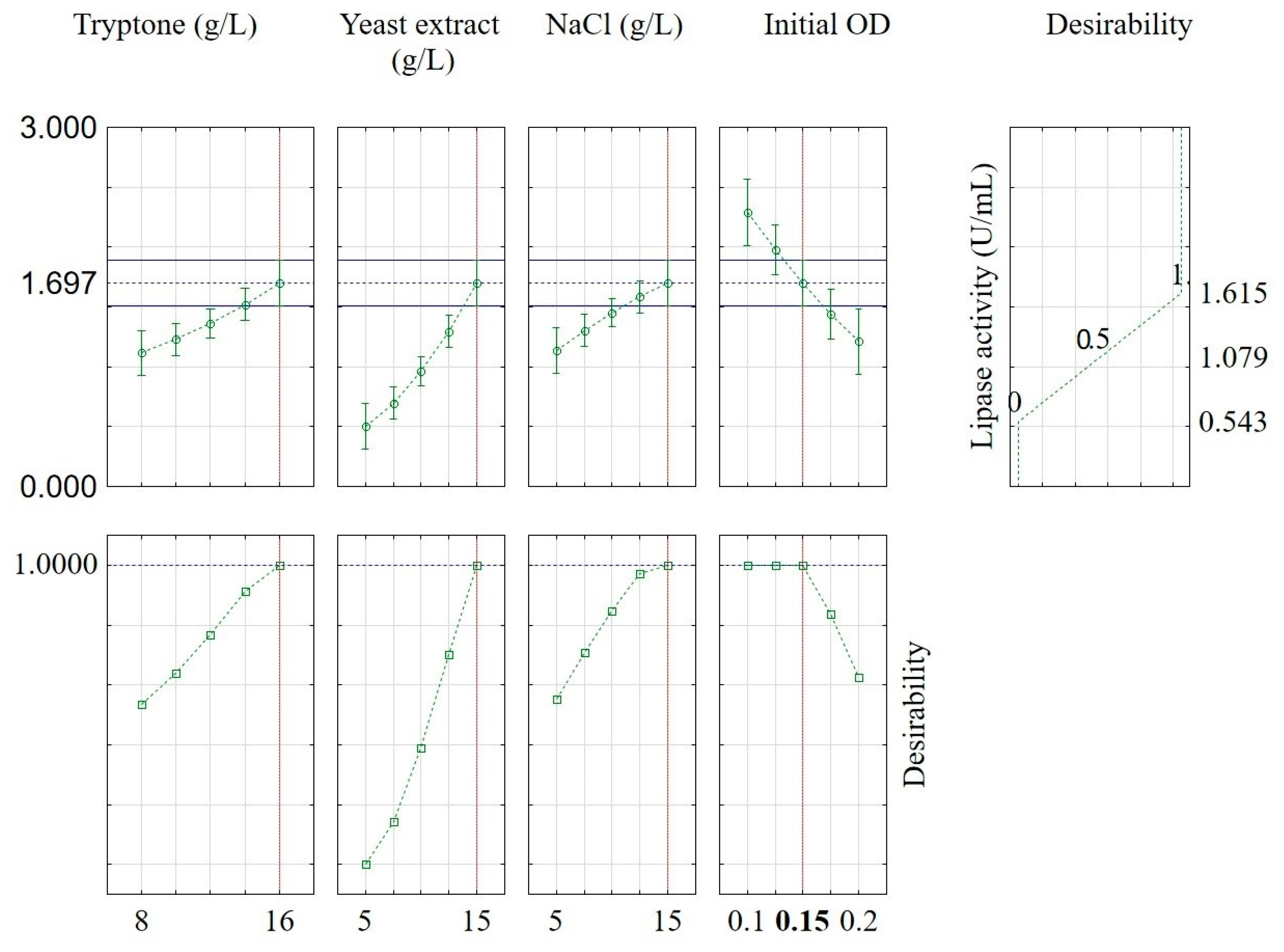

| Variable | Low Level (−1) | Central Level (0) | High Level (+1) |
|---|---|---|---|
| Tryptone (g/L) | 8 | 12 | 16 |
| Yeast extract (g/L) | 5 | 10 | 15 |
| NaCl (g/L) | 5 | 10 | 15 |
| Initial OD | 0.10 | 0.15 | 0.20 |
| Run | Tryptone (g/L) | Yeast Extract (g/L) | NaCl (g/L) | Initial OD | Lipase Activity (U/mL) |
|---|---|---|---|---|---|
| 1 | 8 | 5 | 10 | 0.15 | 0.549 ± 0.006 |
| 2 | 16 | 5 | 10 | 0.15 | 0.691 ± 0.006 |
| 3 | 8 | 15 | 10 | 0.15 | 0.767 ± 0.018 |
| 4 | 16 | 15 | 10 | 0.15 | 1.526 ± 0.018 |
| 5 | 12 | 10 | 5 | 0.10 | 0.752 ± 0.029 |
| 6 | 12 | 10 | 15 | 0.10 | 0.695 ± 0.005 |
| 7 | 12 | 10 | 5 | 0.20 | 0.650 ± 0.003 |
| 8 | 12 | 10 | 15 | 0.20 | 0.781± 0.029 |
| 9 | 8 | 10 | 10 | 0.10 | 0.722 ± 0.010 |
| 10 | 16 | 10 | 10 | 0.10 | 1.587 ± 0.047 |
| 11 | 8 | 10 | 10 | 0.20 | 0.735 ± 0.006 |
| 12 | 16 | 10 | 10 | 0.20 | 0.623 ± 0.006 |
| 13 | 12 | 5 | 5 | 0.15 | 0.775 ± 0.050 |
| 14 | 12 | 15 | 5 | 0.15 | 0.721 ± 0.007 |
| 15 | 12 | 5 | 15 | 0.15 | 0.704 ± 0.001 |
| 16 | 12 | 15 | 15 | 0.15 | 1.508 ± 0.005 |
| 17 | 8 | 10 | 5 | 0.15 | 0.699 ± 0.003 |
| 18 | 16 | 10 | 5 | 0.15 | 0.725 ± 0.016 |
| 19 | 8 | 10 | 15 | 0.15 | 0.731 ± 0.005 |
| 20 | 16 | 10 | 15 | 0.15 | 0.741 ± 0.003 |
| 21 | 12 | 5 | 10 | 0.10 | 0.598 ± 0.007 |
| 22 | 12 | 15 | 10 | 0.10 | 1.497 ± 0.006 |
| 23 | 12 | 5 | 10 | 0.20 | 0.758 ± 0.024 |
| 24 | 12 | 15 | 10 | 0.20 | 0.796 ± 0.005 |
| 25 | 12 | 10 | 10 | 0.15 | 0.748 ± 0.031 |
| Factor | Coeff. | Std. Err. | t (60) | p | −95% Cnf. Limt | +95% Cnf. Limt |
|---|---|---|---|---|---|---|
| Mean/Interc. | 0.880 | 0.034 | 25.898 | <0.001 | 0.812 | 0.948 |
| (1) Tryptone (g/L) (L) | 0.141 | 0.023 | 6.171 | <0.001 | 0.095 | 0.186 |
| Tryptone (g/L) (Q) | −0.021 | 0.024 | −0.873 | 0.386 | −0.068 | 0.027 |
| (2) Yeast extract (g/L) (L) | 0.228 | 0.023 | 10.016 | <0.001 | 0.183 | 0.274 |
| Yeast extract (g/L) (Q) | −0.070 | 0.024 | −2.983 | 0.004 | −0.117 | −0.023 |
| (3) NaCl (g/L) (L) | 0.070 | 0.023 | 3.062 | 0.003 | 0.024 | 0.115 |
| NaCl (g/L) (Q) | 0.018 | 0.024 | 0.760 | 0.450 | −0.029 | 0.065 |
| (4) Initial OD (L) | −0.126 | 0.023 | −5.508 | <0.001 | −0.171 | −0.080 |
| Initial OD (Q) | −0.027 | 0.024 | −1.135 | 0.261 | −0.074 | 0.020 |
| 1 L by 2 L | 0.154 | 0.039 | 3.910 | <0.001 | 0.075 | 0.233 |
| 1 L by 3 L | −0.004 | 0.039 | −0.097 | 0.923 | −0.083 | 0.075 |
| 1 L by 4 L | −0.244 | 0.039 | −6.183 | <0.001 | −0.323 | −0.165 |
| 2 L by 3 L | 0.215 | 0.039 | 5.434 | <0.001 | 0.136 | 0.294 |
| 2 L by 4 L | −0.216 | 0.039 | −5.459 | <0.001 | −0.295 | −0.137 |
| 3 L by 4 L | 0.047 | 0.039 | 1.186 | 0.240 | −0.032 | 0.126 |
| Factor | SS | df | MS | F | p |
|---|---|---|---|---|---|
| (1) Tryptone (g/L) (L) | 0.713 | 1 | 0.713 | 38.079 | <0.001 |
| Tryptone (g/L) (Q) | 0.014 | 1 | 0.014 | 0.762 | 0.386 |
| (2) Yeast extract (g/L) (L) | 1.877 | 1 | 1.877 | 100.317 | <0.001 |
| Yeast extract (g/L) (Q) | 0.166 | 1 | 0.166 | 8.896 | 0.004 |
| (3) NaCl (g/L) (L) | 0.175 | 1 | 0.175 | 9.374 | 0.003 |
| NaCl (g/L) (Q) | 0.011 | 1 | 0.011 | 0.578 | 0.450 |
| (4) Initial OD (L) | 0.568 | 1 | 0.568 | 30.338 | <0.001 |
| Initial OD (Q) | 0.024 | 1 | 0.024 | 1.289 | 0.261 |
| 1 L by 2 L | 0.286 | 1 | 0.286 | 15.290 | <0.001 |
| 1 L by 3 L | 0.0002 | 1 | 0.0002 | 0.009 | 0.923 |
| 1 L by 4 L | 0.715 | 1 | 0.715 | 38.228 | <0.001 |
| 2 L by 3 L | 0.553 | 1 | 0.553 | 29.526 | <0.001 |
| 2 L by 4 L | 0.558 | 1 | 0.558 | 29.802 | <0.001 |
| 3 L by 4 L | 0.026 | 1 | 0.026 | 1.406 | 0.240 |
| Error | 1.123 | 60 | 0.019 | ||
| Total | 6.997 | 74 |
| Metal Ions | Relative Activity (%) | Inhibitors and Surfactants | Relative Activity (%) |
|---|---|---|---|
| 5 mM | 0.1% | ||
| Control | 100.0 | Control | 100.0 |
| NaCl2 | 77.2 | EDTA | 32.4 |
| CaCl2 | 68.6 | DTT | 11.5 |
| NiCl2 | 72.3 | BME | 37.4 |
| KCl | 10.0 | SDS | 14.2 |
| MnCl2 | 67.5 | H2O2 | 59.2 |
| CoCl2 | 13.3 | Tween 20 | 17.0 |
| MgSO4 | 87.2 | Tween 80 | 16.4 |
| FeSO4 | 5.1 |
| Solvent | Concentration (%) | Relative Activity (%) |
|---|---|---|
| Control | 0 | 100.0 |
| Isopropanol | 0.1 | 86.7 |
| 0.3 | 75.7 | |
| 0.5 | 23.4 | |
| Ethanol | 0.1 | 90.2 |
| 0.3 | 63.2 | |
| 0.5 | 20.0 | |
| Acetone | 0.1 | 39.1 |
| 0.3 | 35.9 | |
| 0.5 | 7.0 | |
| Chloroform | 0.1 | 59.5 |
| 0.3 | 60.5 | |
| 0.5 | 11.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshehri, W.A.; Alghamdi, N.H.; Khalel, A.F.; Almalki, M.H.; Hadrich, B.; Sayari, A. Thermostable CaCO3-Immobilized Bacillus subtilis Lipase for Sustainable Biodiesel Production from Waste Cooking Oil. Catalysts 2024, 14, 253. https://doi.org/10.3390/catal14040253
Alshehri WA, Alghamdi NH, Khalel AF, Almalki MH, Hadrich B, Sayari A. Thermostable CaCO3-Immobilized Bacillus subtilis Lipase for Sustainable Biodiesel Production from Waste Cooking Oil. Catalysts. 2024; 14(4):253. https://doi.org/10.3390/catal14040253
Chicago/Turabian StyleAlshehri, Wafa A., Nouf H. Alghamdi, Ashjan F. Khalel, Meshal H. Almalki, Bilel Hadrich, and Adel Sayari. 2024. "Thermostable CaCO3-Immobilized Bacillus subtilis Lipase for Sustainable Biodiesel Production from Waste Cooking Oil" Catalysts 14, no. 4: 253. https://doi.org/10.3390/catal14040253









