Production of Thermostable T1 Lipase Using Agroindustrial Waste Medium Formulation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Analysis of Agroindustrial Wastes in Relation to Carbon and Nitrogen Sources
2.2. Effect of Molasses Concentration
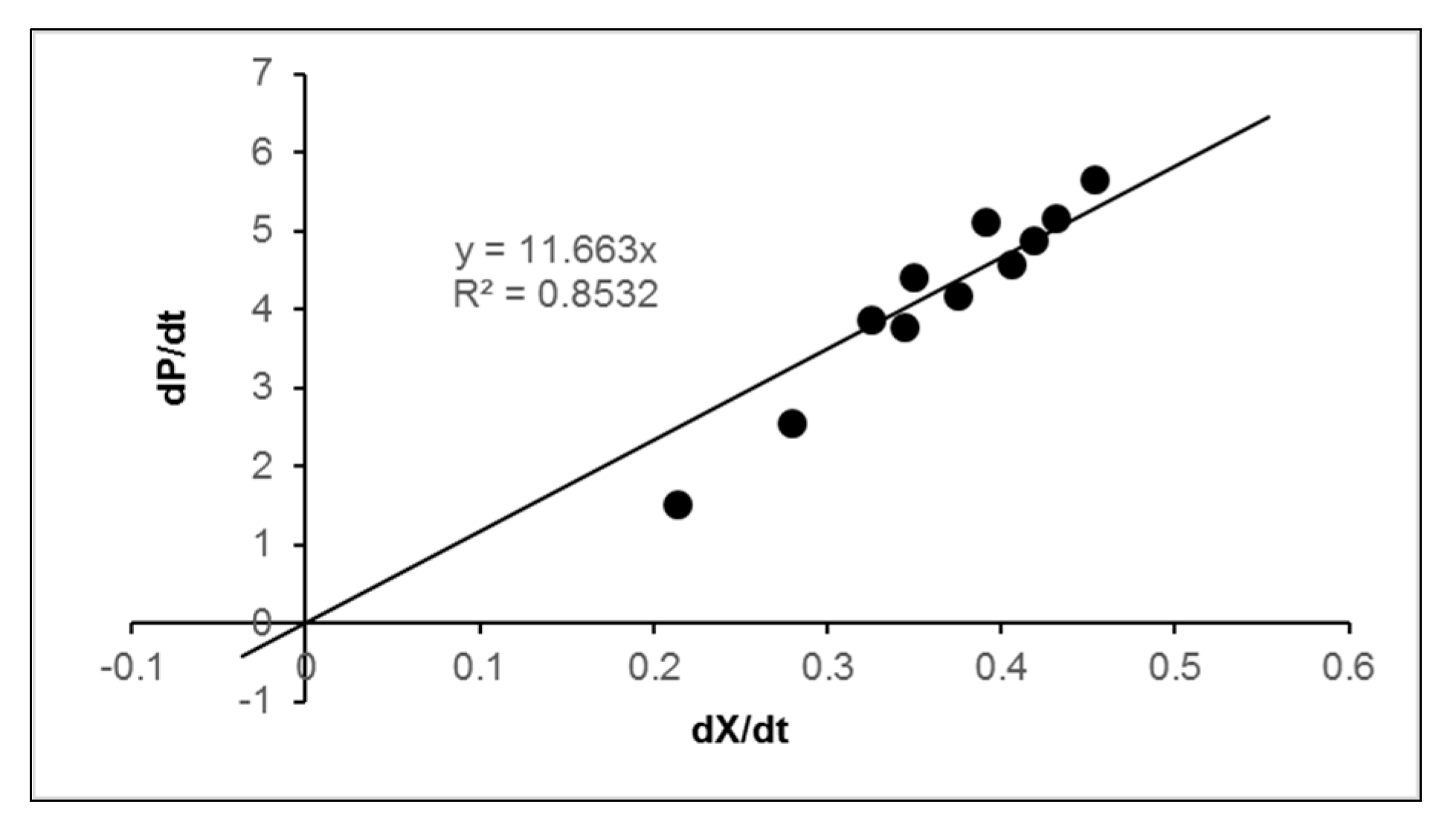
2.3. Effect of Fish Waste Concentration
2.4. Model Fitting and Analysis of Variance (ANOVA)
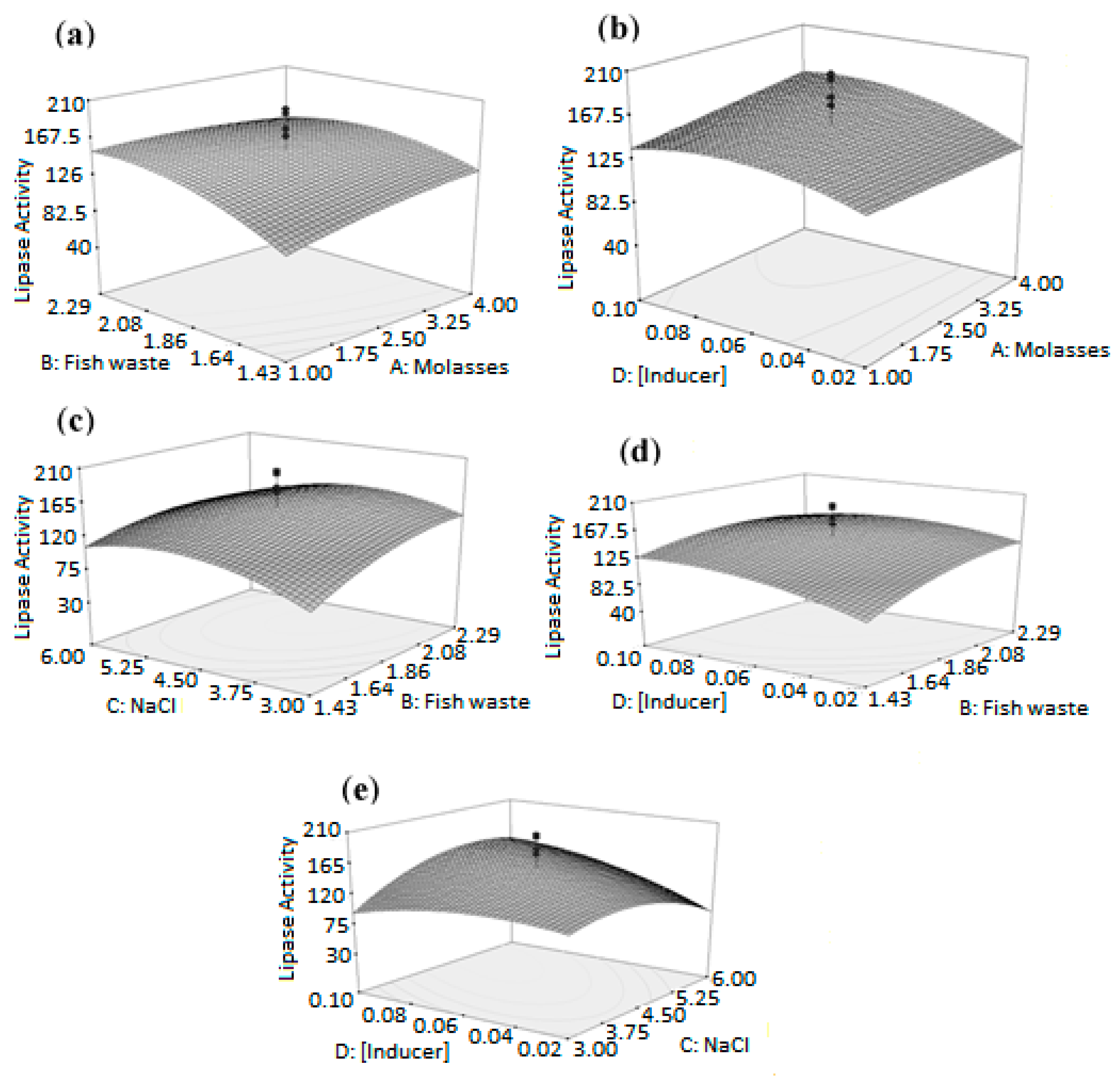
2.5. Effect of Parameters on T1 Lipase Production
2.6. Optimum Conditions
3. Materials and Methods
3.1. Bacterial Strain, Media Compound Preparation and Materials
3.2. Media Preparation and Cultivation
3.3. Determination of Cell Growth
3.4. Determination of Carbohydrate by Anthrone Analysis
3.5. Determination of Nitrogen Content by Kjeldahl Analysis
3.6. Lipase Assay
3.7. Acetate Concentration Detection
3.8. Experimental Design and Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| t | time |
| Xm | maximum cell concentration |
| Pm | maximum product formed |
| Sm | maximum substrate consumed |
| A | acetate concentration |
| μm | maximum specific growth rate |
| YX/S | cell yield per substrate |
| YP/S | lipase yield per substrate |
| YP/X | lipase yield per cell |
| qP | specific product formation rate |
| qS | specific substrate consumption rate |
References
- Singh, A.K.; Mukhopadhyay, M. Overview of fungal lipase: A review. Appl. Biochem. Biotechnol. 2012, 166, 486–520. [Google Scholar] [CrossRef] [PubMed]
- Andualema, B.; Gessesse, A. Microbial lipases and their industrial applications: Review. Biotechnology 2012, 11, 100–118. [Google Scholar] [CrossRef]
- Smaniotto, A.; Skovronski, A.; Rigo, E.; Tsai, S.M.; Durrer, A.; Foltran, L.L.; Paroul, N.; Di Luccio, M.; Oliveira, J.V.; de Oliveira, D.; et al. Concentration, characterization and application of lipases from Sporidiobolus pararoseus strain. Braz. J. Microbiol. 2014, 45, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.R.; Teixeira, J.A.; Oliveira, R. Low-cost fermentative medium for biosurfactant production by probiotic bacteria. Biochem. Eng. J. 2006, 32, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, L.; Isar, J.; Meghwanshi, G.K.; Saxena, R.K. A cost effective fermentative production of succinic acid from cane molasses and corn steep liquor by Escherichia coli. J. Appl. Microbiol. 2006, 100, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Ben Rebah, F.; Miled, N. Fish processing wastes for microbial enzyme production: A review. 3 Biotech. 2013, 3, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Kacem, M.; Sellami, M.; Kammoun, W.; Frikha, F.; Miled, N.; Ben, R.F. Seasonal variations in proximate and fatty acid composition of viscera of Sardinella aurita, Sarpa salpa, and Sepia officinalis from Tunisia. J. Aquat. Food Prod. Technol. 2011, 20, 233–246. [Google Scholar] [CrossRef]
- Haddar, A.; Fakhfakh-Zouari, N.; Hmidet, N.; Frikha, F.; Nasri, M.; Kamoun, A.S. Low-cost fermentation medium for alkaline protease production by Bacillus mojavensis A21 using hulled grain of wheat and sardinella peptone. J. Biosci. Bioeng. 2010, 110, 288–294. [Google Scholar] [CrossRef] [PubMed]
- El-Boulifi, N.; Ashari, S.E.; Serrano, M.; Aracil, J.; Martínez, M. Solvent-free lipase-catalyzed synthesis of a novel hydroxyl-fatty acid derivative of kojic acid. Enzyme Microb. Technol. 2014, 55, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Ashari, S.E.; Mohamad, R.; Ariff, A.; Basri, M.; Salleh, A.B. Optimization of enzymatic synthesis of palm-based kojic acid ester using response surface methodology. J. Oleo. Sci. 2009, 58, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kang, S.K.; Choi, Y.J. A low-cost Lactobacillus salivarius L29 growth medium containing molasses and corn steep liquor allows the attainment of high levels of cell mass and lactic acid production. Afr. J. Biotechnol. 2013, 12, 2013–2018. [Google Scholar]
- Michailides, M.K.; Tekerlekopoulou, A.G.; Akratos, C.S.; Coles, S.; Pavlou, S.; Vayenas, D.V. Molasses as an efficient low-cost carbon source for biological Cr(VI) removal. J. Hazard. Mater. 2015, 281, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Mohd, Y.N.H.; Fukuzaki, M.; Maeda, T.; Miyazaki, T.; Hakiman, C.M.; Ariffin, H.; Wood, T.K. Biohydrogen production from oil palm frond juice and sewage sludge by a metabolically engineered Escherichia coli strain. Int. J. Hydrogen Energy 2013, 38, 10277–10283. [Google Scholar] [CrossRef]
- Vital, M.; Stucki, D.; Egli, T.; Hammes, F. Evaluating the growth potential of pathogenic bacteria in water. Appl. Environ. Microbiol. 2010, 76, 6477–6484. [Google Scholar] [CrossRef] [PubMed]
- Sekar, N.; Veetil, S.; Neerathilingam, M. Tender coconut water an economical growth medium for the production of recombinant proteins in Escherichia coli. BMC Biotechnol. 2013, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Luli, G.; Strohl, W. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Envir. Microbiol. 1990, 6, 1004–1011. [Google Scholar]
- Lee, L.P.; Karbul, H.M.; Citartan, M.; Gopinath, S.C.B.; Lakshmipriya, T.; Tang, T.H. Lipase-secreting Bacillus species in an oil-contaminated habitat: Promising strains to alleviate oil pollution. Biomed Res. Int. 2015, 1–9. [Google Scholar] [CrossRef]
- Hamzaoui, A.H.; Jamoussi, B.; M’nif, A. Lithium recovery from highly concentrated solutions: Response surface methodology (RSM) process parameters optimization. Hydrometallurgy 2008, 90, 1–7. [Google Scholar] [CrossRef]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.P.; Zheng, P.; Sun, Z.H.; Ni, Y.; Dong, J.J.; Zhu, L.L. Economical succinic acid production from cane molasses by Actinobacillus succinogenes. Bioresour. Technol. 2008, 99, 1736–1742. [Google Scholar] [CrossRef] [PubMed]
- Abdulkarim, S.M.; Fatimah, A.B.; Anderson, J.G. Effect of salt concentrations on the growth of heat-stressed and unstressed Escherichia coli. J. Food Agric. Environ. 2009, 7, 51–54. [Google Scholar]
- Malakar, P.; Venkatesh, K.V. Effect of substrate and IPTG concentrations on the burden to growth of Escherichia coli on glycerol due to the expression of Lac proteins. Appl. Microbiol. Biotechnol. 2012, 93, 2543–2549. [Google Scholar] [CrossRef] [PubMed]
- Boonchaidung, T.; Papone, T. Effect of carbon and nitrogen sources on lipase production by isolated lipase-producing soil yeast. J. Life Sci. Tech. 2013, 1, 176–179. [Google Scholar] [CrossRef]
- Ramani, K.; Chockalingam, E.; Sekaran, G. Production of a novel extracellular acidic lipase from Pseudomonas gessardii using slaughterhouse waste as a substrate. J. Ind. Microbiol. 2010, 37, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Kanmani, P.; Karthik, S.; Aravind, J.; Kumaresan, K. The use of response surface methodology as a statistical tool for media optimization in lipase production from the dairy effluent isolate Fusarium solani. ISRN Biotechnol. 2013, 2013, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Velez, A.; Horta, A.; da Silva, A.; Iemma, M.; Giordano, R.; Zangirolami, T. Enhanced production of recombinant thermo-stable lipase in Escherichia coli at high induction temperature. Protein Expr. Purif. 2013, 90, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Leow, T.C.; Rahman, R.N.Z.R.A.; Basri, M.; Salleh, A.B. A thermoalkaliphilic lipase of Geobacillus sp. T1. Extremophiles 2007, 11, 527–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, R.N.Z.R.A.; Leow, T.C.; Basri, M.; Salleh, A.B. Secretory expression of thermostable T1 lipase through bacteriocin release protein. Protein Expr. Purif. 2005, 40, 411–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shasaltaneh, M.D.; Fooladi, J.; Moosavi-Nejad, S.Z. L-tryptophan production by Escherichia coli in the presence of Iranian cane molasses. J. Paramed. Sci. 2010, 1. [Google Scholar] [CrossRef]
- Norziah, M.H.; Al-Hassan, A.; Khairulnizam, A.B.; Mordi, M.N.; Norita, M. Characterization of fish gelatin from surimi processing wastes: Thermal analysis and effect of transglutaminase on gel properties. Food Hydrocoll. 2009, 23, 1610–1616. [Google Scholar] [CrossRef]
- Ghaley, A.E.M.; Brooks, M.; Ramakrishnan, V.V.; Budge, S.M.; Dave, D. Fish processing wastes as a potential source of proteins, amino acids and oils: A critical review. J. Microb. Biochem. Technol. 2013, 5, 107–129. [Google Scholar] [CrossRef]
- Behera, S.; Ghanty, S.; Ahmad, F.; Santra, S.; Banerjee, S. UV-visible spectrophotometric method development and validation of assay of paracetamol tablet formulation. J. Anal. Bioanal. Tech. 2012, 3. [Google Scholar] [CrossRef]
- Dey, S.S.; Dora, K.C. Effect of sodium lactate as cryostabilizer on physico-chemical attributes of croaker (Johnius gangeticus) muscle protein. J. Food Sci. Technol. 2010, 47, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Loewus, F.A. Improvement in anthrone method for determination of carbohydrates. Anal. Chem. 1952, 24, 219. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Rhee, J.S. A simple and rapid colorimetric method for determination of free fatty acids for lipase assay. J. Am. Oil Chem. Soc. 1986, 63, 89–92. [Google Scholar] [CrossRef]
- Li, S.; Kim, Y.; Phunysho, S.; Chekli, L.; Shon, H.K.; Leiknes, T.; Ghaffour, N. Methane production in an anaerobic osmotic membrane bioreactor using forward osmosis: Effect of reverse salt flux. Bioresour. Technol. 2017, 239, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Liu, K.J.; Lou, Y.H.; Shieh, C.J. Optimisation of kojic acid monolaurate synthesis with lipase PS from Pseudomonas cepacia. J. Sci. Food Agric. 2002, 82, 601–605. [Google Scholar] [CrossRef]
- Chaibakhsh, N.; Abdul, R.M.B.; Abd-Aziz, S.; Basri, M.; Salleh, A.B.; Rahman, R.N.Z.R.A. Optimized lipase-catalyzed synthesis of adipate ester in a solvent-free system. J. Ind. Microbiol. Biotechnol. 2009, 36, 1149–1155. [Google Scholar] [CrossRef] [PubMed]


| Sample | Glucose (g/L) | Nitrogen (%) |
|---|---|---|
| POME | - | 7.11 ± 3.402 |
| Fruit bunch | - | 1.32 ± 4.460 |
| Fish waste | 0.08 ± 0.074 | 19.08 ± 3.585 |
| Influent | - | 2.35 ± 2.131 |
| Effluent | - | 0.17 ± 4.895 |
| Fish hydrolysate | - | 9.59 ± 1.222 |
| 5th grade molasses | 19.80 ± 0.021 | - |
| Mature coconut water | 0.19 ± 0.065 | - |
| Kinetic Parameters | Molasses (g/L) | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| t (h) | 12 | 12 | 12 | 12 | 12 |
| Xm (g/L) | 3.90 | 4.69 | 4.18 | 3.35 | 2.56 |
| Pm (U/mL) | 46.53 | 61.56 | 53.11 | 30.78 | 18.14 |
| Sm (g/L) | 15.30 | 16.90 | 16.6 | 14.8 | 10.00 |
| A (g/L) | 0.43 | 0.75 | 0.55 | 0.41 | 0.24 |
| µmax (h−1) | 0.46 | 0.47 | 0.45 | 0.38 | 0.21 |
| Yx/s (gcells/gsugar) | 0.254 | 0.278 | 0.252 | 0.226 | 0.256 |
| Yp/s (U/gsugar) | 3.041 | 3.643 | 3.199 | 2.079 | 1.814 |
| Yp/x (U/gcells) | 11.931 | 13.126 | 12.706 | 9.188 | 7.086 |
| qp (U/g/h) | 6.373 | 8.185 | 7.790 | 5.473 | 3.755 |
| qs (g/g/h) | 1.122 | 1.242 | 1.114 | 1.001 | 0.878 |
| Kinetic Parameters | Fish Waste (g/L) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1.91 | 2.29 | 2.67 | 3.05 | 3.43 | 3.82 | 4.20 | 4.58 | |
| t (h) | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 |
| Xm (g/L) | 5.17 | 5.44 | 5.02 | 4.87 | 4.50 | 4.13 | 4.00 | 3.97 |
| Pm (U/mL) | 62.13 | 67.99 | 58.64 | 54.98 | 50.21 | 45.3 | 31.70 | 30.22 |
| Sm (g/L) | 16.36 | 16.97 | 16.00 | 15.35 | 15.10 | 14.3 | 12.10 | 12.00 |
| Am (g/L) | 0.43 | 0.40 | 0.55 | 0.73 | 0.75 | 0.81 | 0.92 | 0.95 |
| µmax (h−1) | 0.427 | 0.431 | 0.412 | 0.400 | 0.387 | 0.355 | 0.285 | 0.280 |
| Yx/s (gcells/gsugar) | 0.316 | 0.321 | 0.3014 | 0.317 | 0.298 | 0.286 | 0.331 | 0.331 |
| Yp/s (U/gsugar) | 3.798 | 4.006 | 3.665 | 3.582 | 3.325 | 3.168 | 2.620 | 3.299 |
| Yp/x (U/gcells) | 12.017 | 12.498 | 11.681 | 11.289 | 11.158 | 10.968 | 7.925 | 7.612 |
| qp (U/g/h) | 2.532 | 2.669 | 2.500 | 2.465 | 2.445 | 2.331 | 2.277 | 2.230 |
| qs (g/g/h) | 3.260 | 3.3373 | 3.195 | 3.184 | 2.965 | 2.743 | 2.333 | 2.300 |
| Standard Order | Molasses, A (g/L) | Fish Waste, B (g/L) | NaCl C, (g/L) | [IPTG], D (mM) | Lipase Activity (U/mL) | |
|---|---|---|---|---|---|---|
| Actual | Predicted | |||||
| 1 | 1.00 (−1) | 1.43 (−1) | 3.00 (−1) | 0.10 (+1) | 51.32 | 43.83 |
| 2 | 4.00 (+1) | 1.43 (−1) | 3.00 (−1) | 0.02 (–1) | 23.249 | 25.546 |
| 3 | 1.00 (−1) | 2.29 (+1) | 3.00 (−1) | 0.02 (–1) | 125.49 | 136.764 |
| 4 | 4.00 (+1) | 2.29 (+1) | 3.00 (−1) | 0.10 (+1) | 101.96 | 103.49 |
| 5 | 1.00 (−1) | 1.43 (−1) | 6.00 (+1) | 0.02 (–1) | 66.67 | 64.44 |
| 6 | 4.00 (+1) | 1.43 (−1) | 6.00 (+1) | 0.10 (+1) | 88.782 | 90.71 |
| 7 | 1.00 (−1) | 2.29 (+1) | 6.00 (+1) | 0.10 (+1) | 147.62 | 146.92 |
| 8 | 4.00 (+1) | 2.29 (+1) | 6.00 (+1) | 0.02 (−1) | 93.48 | 93.48 |
| 9 | 2.50 (0) | 1.86 (0) | 4.50 (0) | 0.06 (0) | 175.59 | 175.00 |
| 10 | 2.50 (0) | 1.86 (0) | 4.50 (0) | 0.06 (0) | 178.02 | 175.00 |
| 11 | 1.00 (−1) | 1.43 (−1) | 3.00 (−1) | 0.02 (−1) | 71.05 | 79.17 |
| 12 | 4.00 (+1) | 1.43 (−1) | 3.00 (−1) | 0.10 (+1) | 133.89 | 144.03 |
| 13 | 1.00 (−1) | 2.29 (+1) | 3.00 (−1) | 0.10 (+1) | 159.38 | 153.02 |
| 14 | 4.00 (+1) | 2.29 (+1) | 3.00 (−1) | 0.02 (–1) | 151.98 | 153.59 |
| 15 | 1.00 (−1) | 1.43 (−1) | 6.00 (+1) | 0.10 (+1) | 198.319 | 182.345 |
| 16 | 4.00 (+1) | 1.43 (0) | 6.00 (+1) | 0.02 (−1) | 80.55 | 89.22 |
| 17 | 1.00 (−1) | 2.29 (+1) | 6.00 (+1) | 0.02 (−1) | 135.56 | 127.03 |
| 18 | 4.00 (+1) | 2.29 (+1) | 6.00 (+1) | 0.10 (+1) | 186.56 | 180.74 |
| 19 | 2.50 (0) | 1.86 (0) | 4.50 (0) | 0.06 (0) | 195.19 | 193.83 |
| 20 | 2.50 (0) | 1.86 (0) | 4.50 (0) | 0.06 (0) | 200.28 | 193.83 |
| 21 | 0.40 (−2) | 1.86 (0) | 4.50 (0) | 0.06 (0) | 57.58 | 77.07 |
| 22 | 4.60 (+2) | 1.86 (0) | 4.50 (0) | 0.06 (0) | 101.40 | 91.74 |
| 23 | 2.50 (0) | 1.26 (−2) | 4.50 (0) | 0.06 (0) | 43.14 | 31.51 |
| 24 | 2.50 (0) | 2.46 (+2) | 4.50 (0) | 0.06 (0) | 70.59 | 78.94 |
| 25 | 2.50 (0) | 1.86 (0) | 2.40 (0) | 0.06 (0) | 30.81 | 26.09 |
| 26 | 2.50 (0) | 1.86 (0) | 6.60 (+1) | 0.06 (0) | 36.41 | 37.86 |
| 27 | 2.50 (0) | 1.86 (0) | 4.50 (0) | 0.00 (−2) | 45.94 | 35.79 |
| 28 | 2.50 (0) | 1.86 (0) | 4.50 (0) | 0.12 (+2) | 72.14 | 72.14 |
| 29 | 2.50 (0) | 1.86 (0) | 4.50 (0) | 0.06 (0) | 144.58 | 151.22 |
| 30 | 2.50 (0) | 1.86 (0) | 4.50 (0) | 0.06 (0) | 156.21 | 151.22 |
| Source | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 35,117.19 | 13 | 2701.32 | 15.03 | 0.0003 |
| Molasses, A | 357.25 | 1 | 357.25 | 1.99 | 0.1963 |
| Fish waste, B | 3714.89 | 1 | 3714.99 | 20.66 | 0.0019 |
| NaCl, C | 180.15 | 1 | 180.15 | 1.00 | 0.3461 |
| [IPTG], D | 1716.06 | 1 | 1716.06 | 9.55 | 0.0149 |
| AB | 1786.06 | 1 | 1786.60 | 9.94 | 0.0135 |
| AD | 478.86 | 1 | 478.86 | 2.66 | 0.1413 |
| BC | 693.44 | 1 | 693.44 | 3.86 | 0.0851 |
| BD | 666.31 | 1 | 666.31 | 3.71 | 0.0904 |
| CD | 3164.45 | 1 | 3164.45 | 17.60 | 0.0030 |
| B2 | 1759.00 | 1 | 1759.00 | 9.78 | 0.0141 |
| C2 | 5678.26 | 1 | 5678.26 | 31.58 | 0.0005 |
| D2 | 1914.23 | 1 | 1914.23 | 10.65 | 0.0115 |
| ABC | 1206.70 | 1 | 1206.70 | 6.71 | 0.0321 |
| Residual | 1438.26 | 8 | 179.78 | - | - |
| Lack-of-fit | 1397.71 | 6 | 232.95 | 11.49 | 0.0822 |
| Pure error | 40.56 | 2 | 20.28 | - | - |
| Corrected total | 71,172.56 | 23 | - | - | - |
| Run no. | Molasses, A (g/L) | Fish Waste, B (g/L) | NaCl, C (g/L) | IPTG Concentration, D (mM) | Lipase Activity (U/mL) | Difference | |
|---|---|---|---|---|---|---|---|
| Actual | Predicted | ||||||
| 1. | 1.00 | 2.29 | 3.46 | 0.03 | 164.37 | 172.89 | 8.52 |
| Variable | Levels | ||||
|---|---|---|---|---|---|
| –2 | –1 | 0 | +1 | +2 | |
| Molasses, A (g/L) | 0.4 | 1.0 | 2.50 | 4.0 | 4.6 |
| Fish waste, B (g/L) | 1.258 | 1.43 | 1.86 | 2.29 | 2.462 |
| NaCl, C (g/L) | 2.4 | 3.0 | 4.5 | 6.0 | 6.6 |
| [Inducer], D (mM) | 0.004 | 0.02 | 0.06 | 0.1 | 0.116 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nooh, H.M.; Masomian, M.; Salleh, A.B.; Mohamad, R.; Ali, M.S.M.; Rahman, R.N.Z.R.A. Production of Thermostable T1 Lipase Using Agroindustrial Waste Medium Formulation. Catalysts 2018, 8, 485. https://doi.org/10.3390/catal8110485
Nooh HM, Masomian M, Salleh AB, Mohamad R, Ali MSM, Rahman RNZRA. Production of Thermostable T1 Lipase Using Agroindustrial Waste Medium Formulation. Catalysts. 2018; 8(11):485. https://doi.org/10.3390/catal8110485
Chicago/Turabian StyleNooh, Hisham Mohd, Malihe Masomian, Abu Bakar Salleh, Rosfarizan Mohamad, Mohd Shukuri Mohamad Ali, and Raja Noor Zaliha Raja Abd Rahman. 2018. "Production of Thermostable T1 Lipase Using Agroindustrial Waste Medium Formulation" Catalysts 8, no. 11: 485. https://doi.org/10.3390/catal8110485





