Development of a Four-Enzyme Magnetic Nanobiocatalyst for Multi-Step Cascade Reactions
Abstract
1. Introduction
2. Results and Discussion
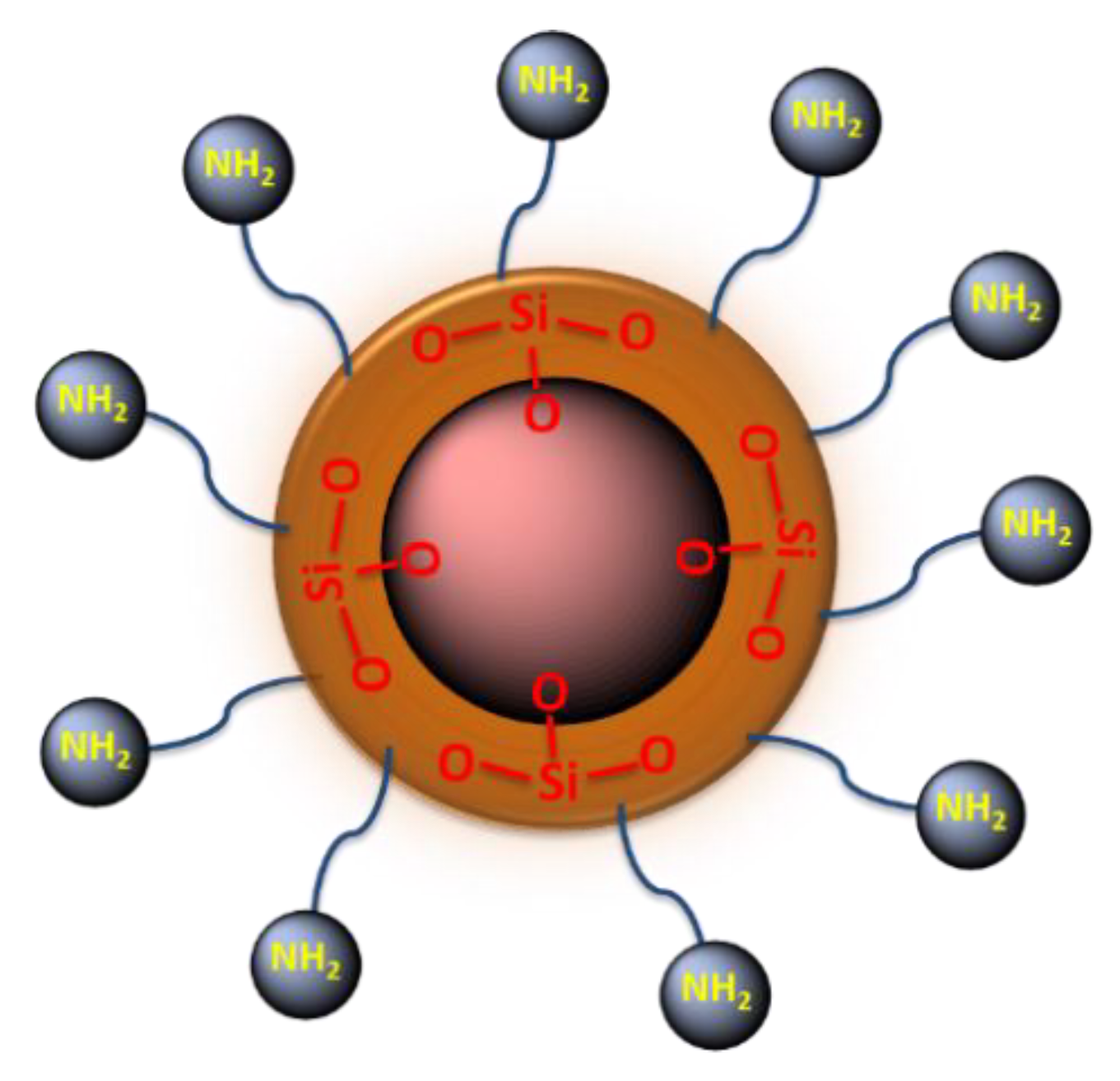
2.1. Synthesis and Characterization of the Magnetic Nanoparticles
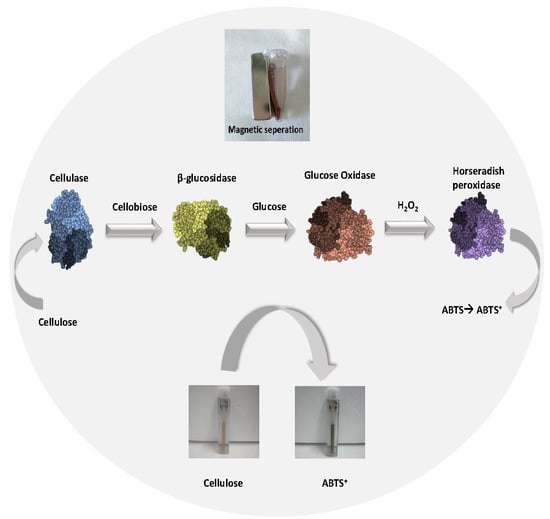
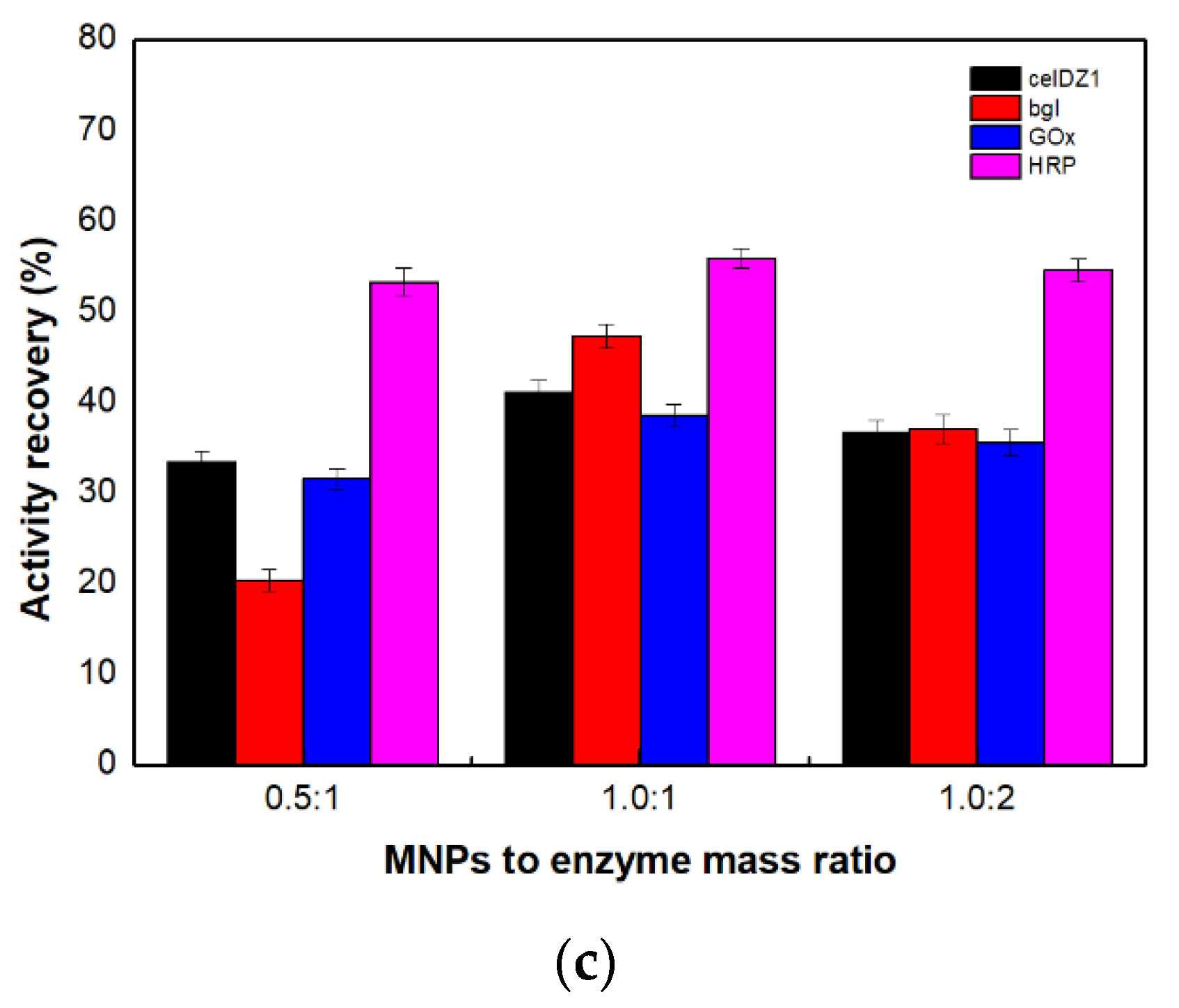
2.2. Preparation of the Magnetic Four-Enzyme Nanobiocatalyst
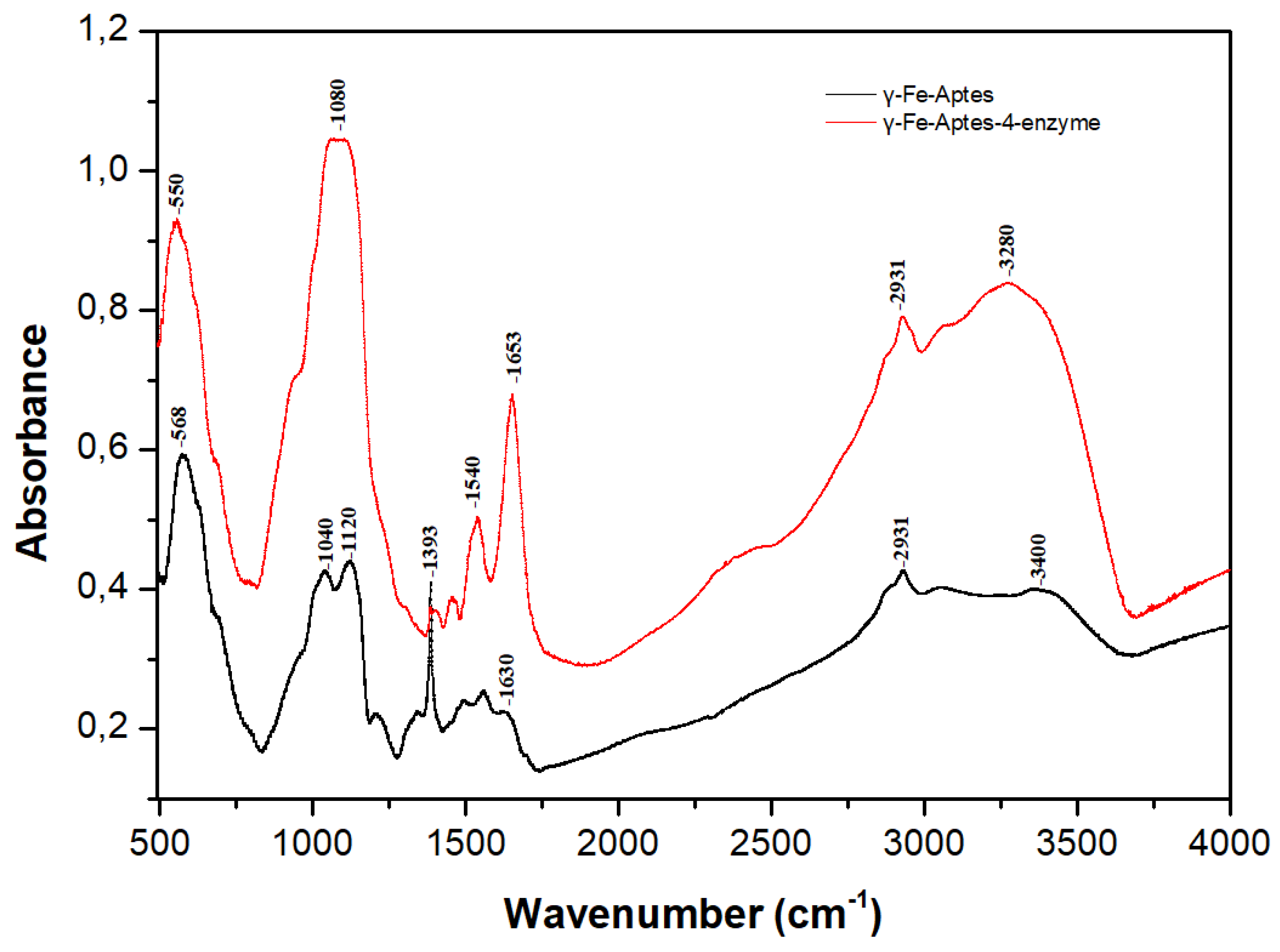
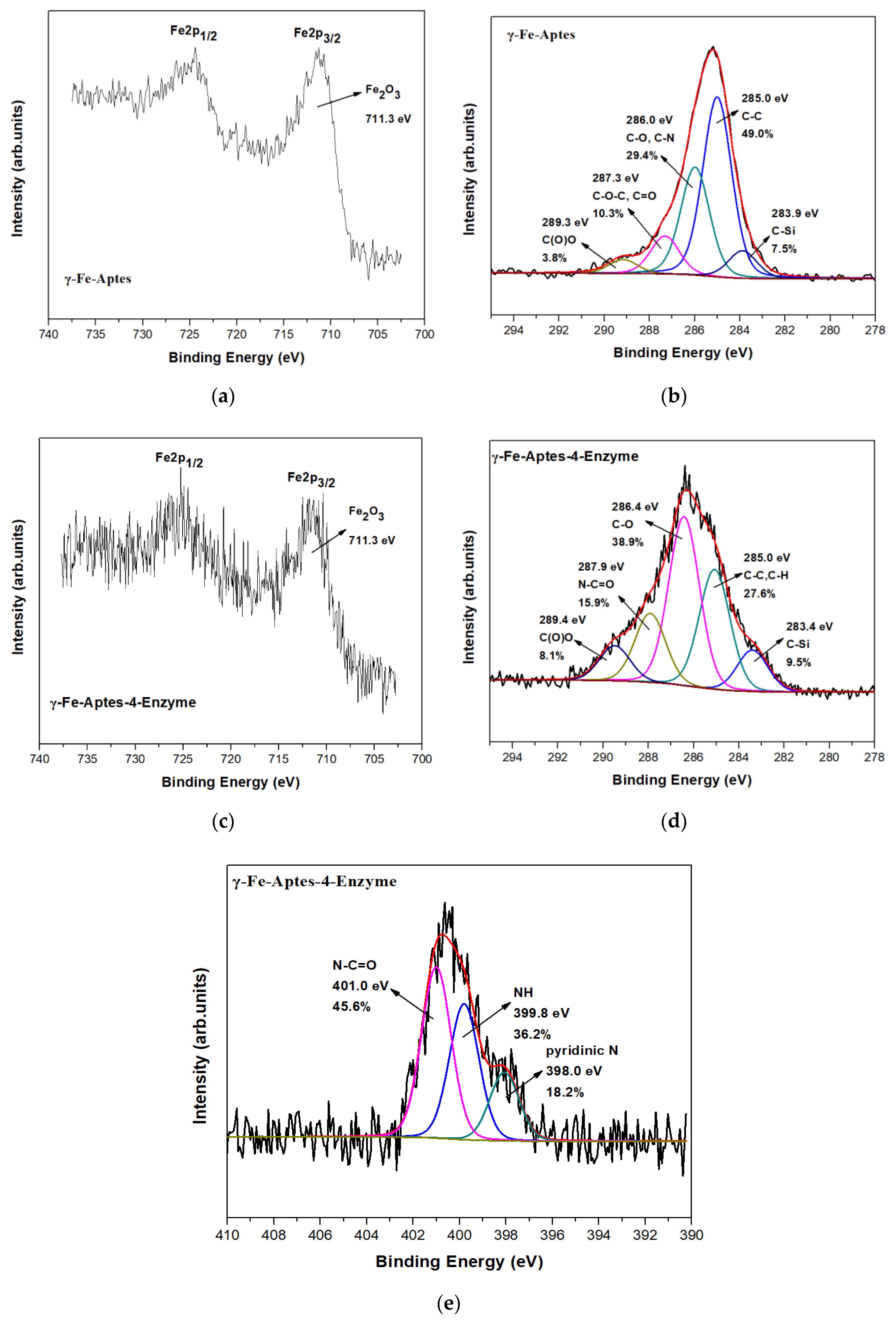
2.3. Characterization of the Four-Enzyme Magnetic Nanobiocatalyst
2.3.1. Fourier Transform Infrared Spectroscopy (FTIR)
2.3.2. X-ray Photoelectron Spectroscopy (XPS)
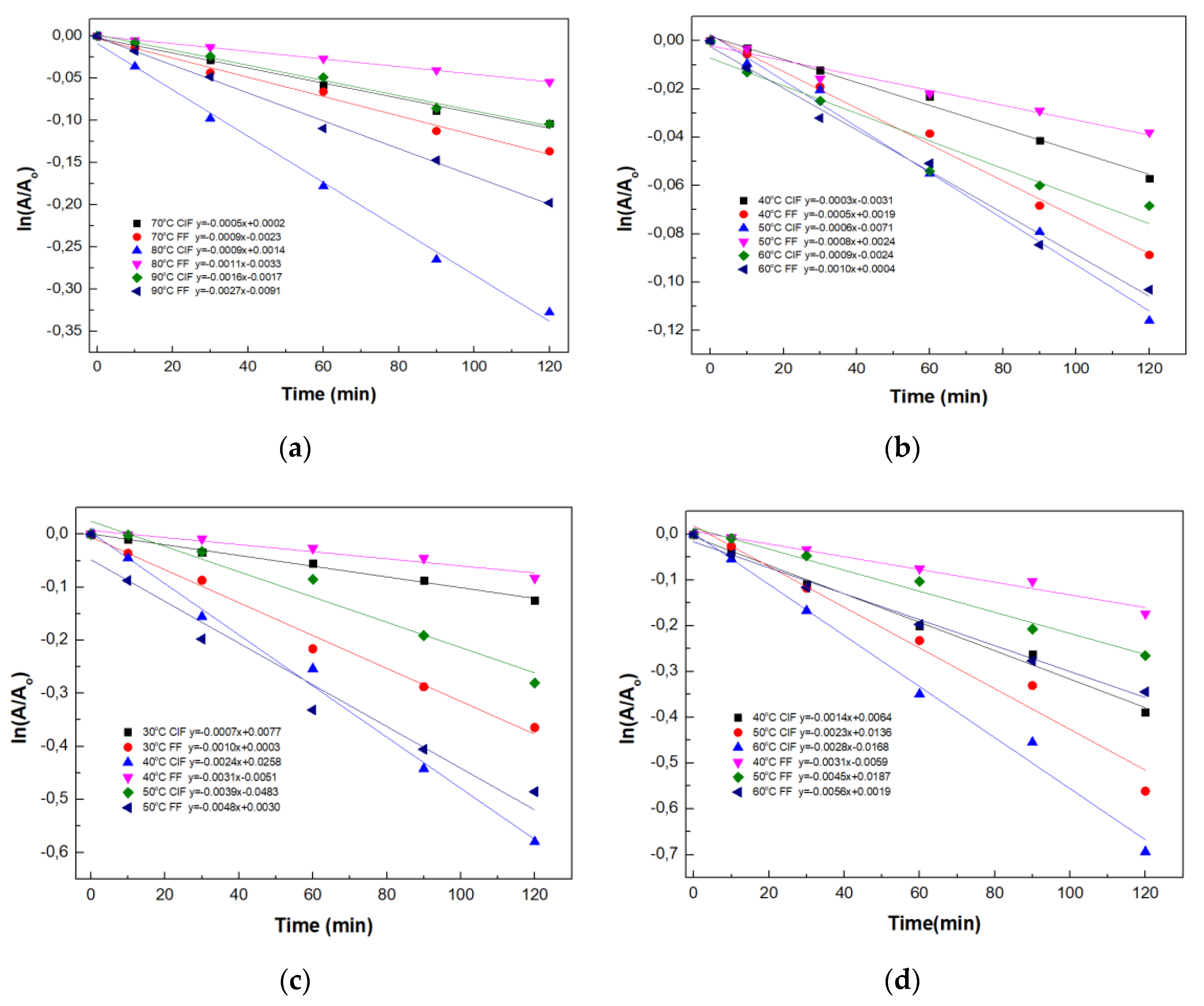
2.4. Thermal Stability Studies of Magnetic Four-Enzyme Nanobiocatalyst
2.5. Determination of Thermodynamic Parameters
2.6. Kinetic Parameters
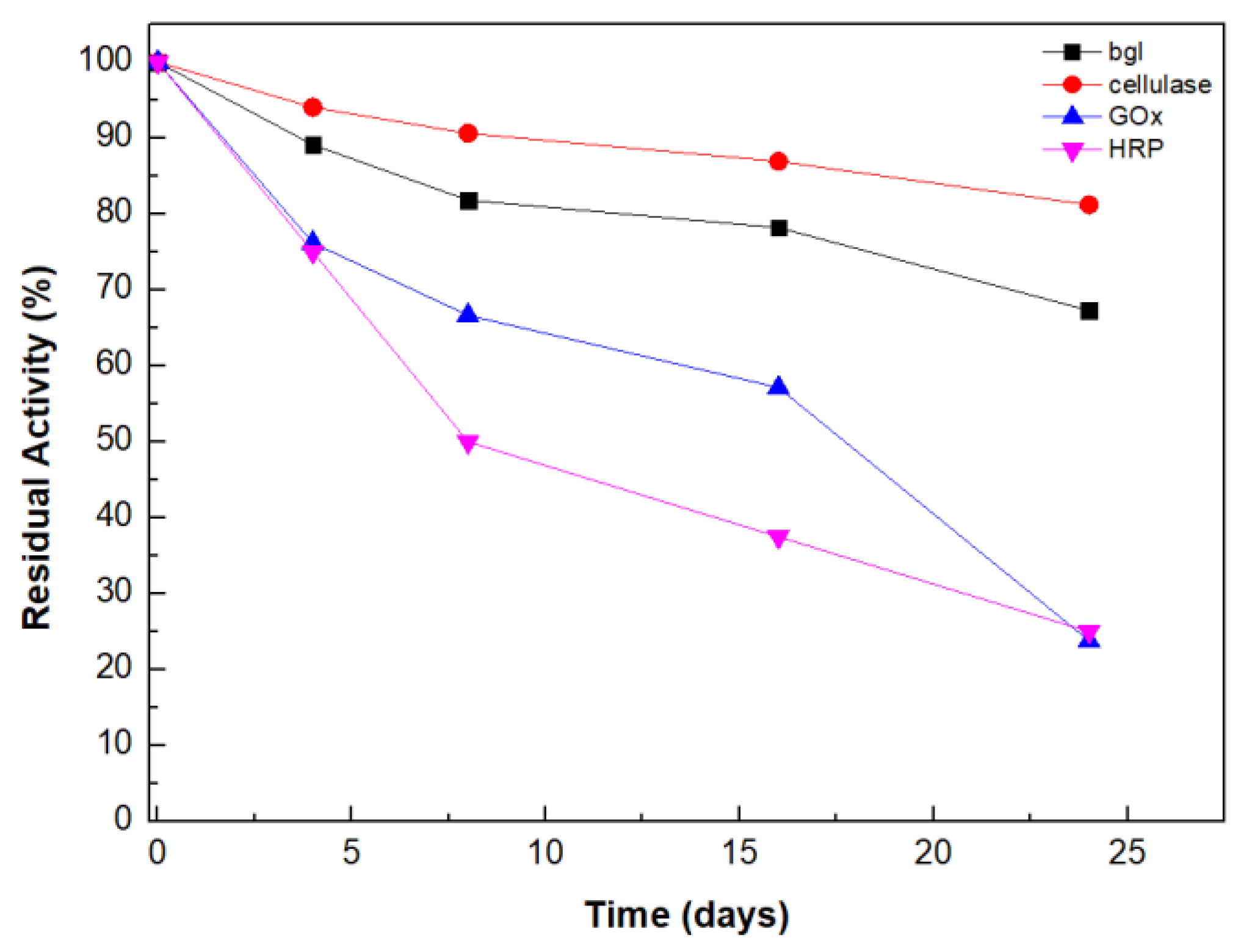
2.7. Storage Stability Studies
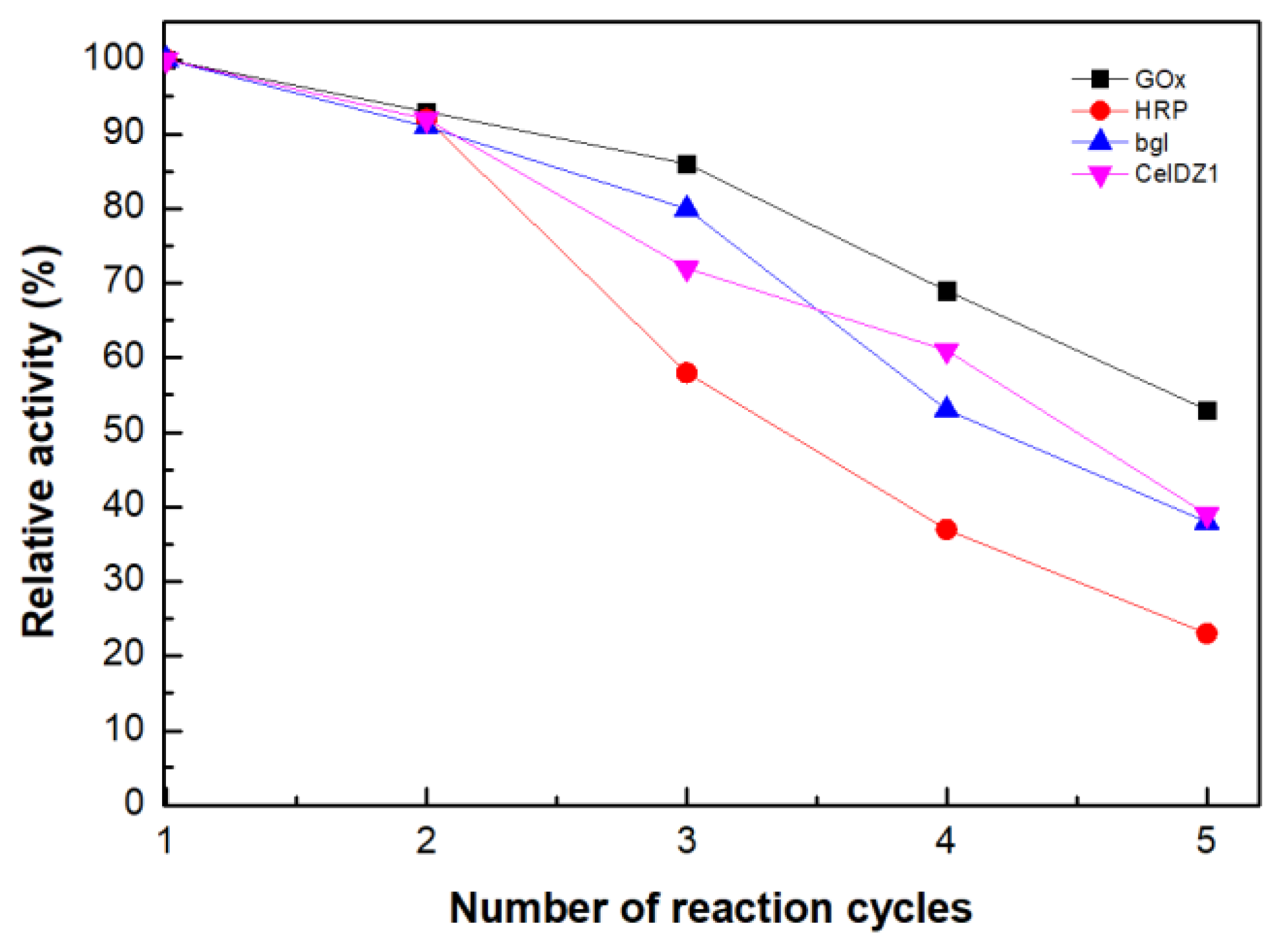
2.8. Reusability Studies
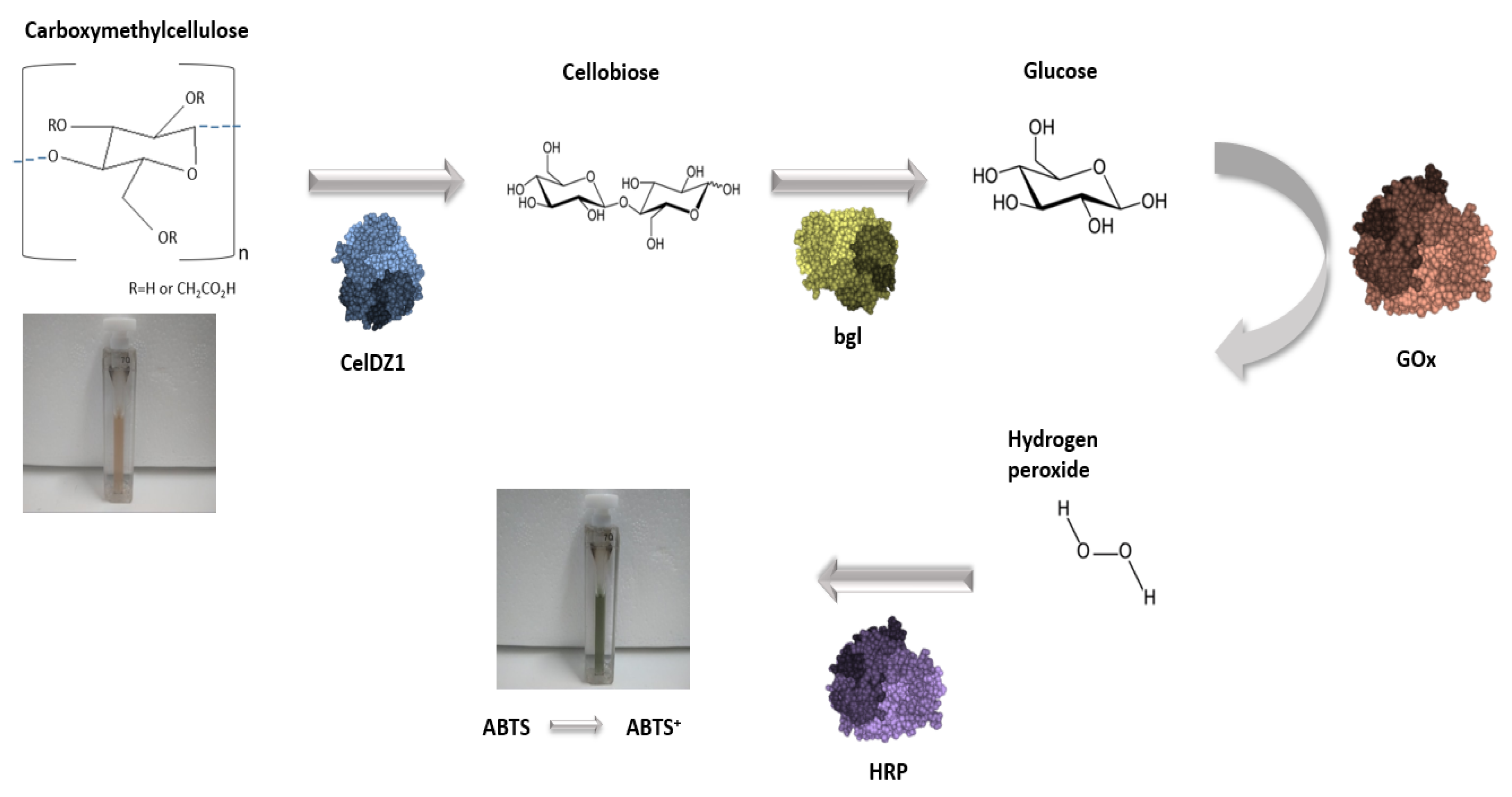
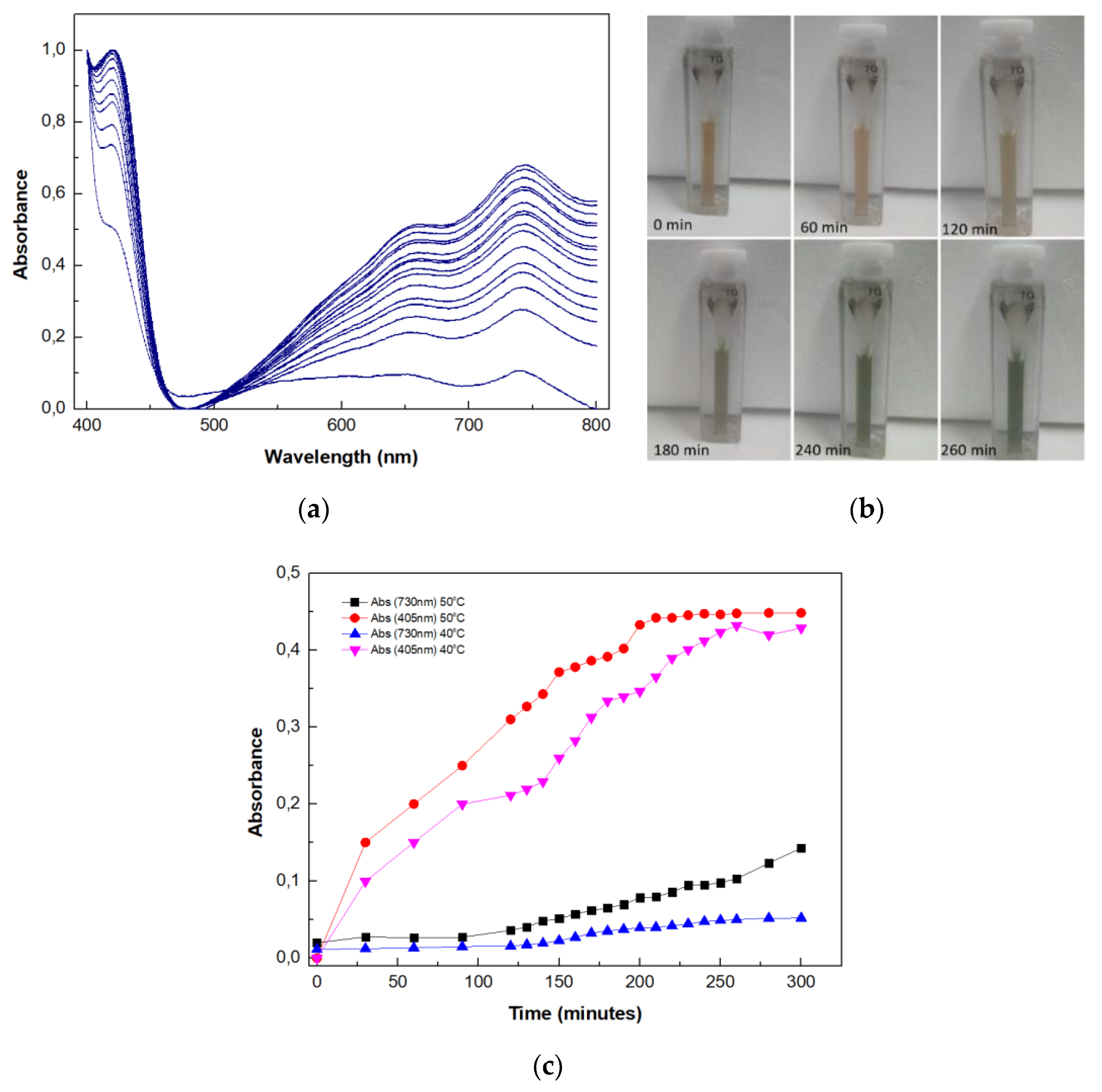
2.9. Use of the Multi-Enzyme Magnetic Nanobiocatalyst in a Four-Step Cascade Reaction
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Cellulase Isolation, Purification and Lyophilization
3.2.2. Synthesis of Magnetic Nanoparticles
3.2.3. Synthesis of Amino Functionalized Magnetic Nanoparticles
3.2.4. Characterization of the Amino-Functionalized Magnetic Nanoparticles
X-ray Diffraction (XRD)
Atomic Force Microscopy (AFM)
3.2.5. Preparation of Four-Enzyme Magnetic Nanobiocatalyst
3.2.6. Characterization of the Four-Enzyme Magnetic Nanobiocatalyst
Fourier-Transform Infrared Spectroscopy (FTIR)
X-ray Photoelectron Spectroscopy (XPS)
3.2.7. Enzyme Assays
3.2.8. Thermal Inactivation Kinetics Studies
3.2.9. Determination of Thermodynamic Parameters
3.2.10. Determination of Michaelis-Menten Kinetic Parameters
3.2.11. Reusability Studies
3.2.12. Storage Stability Studies
3.2.13. Application of the Magnetic Nanobiocatalyst for Cellulose Hydrolysis in a Four-Step Cascade Reaction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Min, K.; Yoo, Y.J. Recent progress in nanobiocatalysis for enzyme immobilization and its application. Biotechnol. Bioprocess Eng. 2014, 19, 553–567. [Google Scholar] [CrossRef]
- Choi, J.; Han, S.; Kim, H. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Narasimhan, B.; Mallapragada, S. Materials-Based Strategies for Multi-Enzyme Immobilization and Co-Localization: A Review. Biotechnol. Bioeng. 2014, 111, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Filice, M.; Palomo, J.M. Cascade reactions catalyzed by bionanostructures. ACS Catal. 2014, 4, 1588–1598. [Google Scholar] [CrossRef]
- Sperl, J.M.; Sieber, V. Multienzyme Cascade Reactions—Status and Recent Advances. ACS Catal. 2018, 8, 2385–2396. [Google Scholar] [CrossRef]
- Velasco-Lozano, S.; López-Gallego, F. Wiring step-wise reactions with immobilized multi-enzyme systems. Biocatal. Biotransform. 2018, 36, 184–194. [Google Scholar] [CrossRef]
- Giannakopoulou, A.; Gkantzou, E.; Polydera, Α.; Stamatis, H. Multienzymatic Nanoassemblies: Recent Progress and Applications. Trends Biotechnol. 2019. [Google Scholar] [CrossRef]
- Ji, Q.; Wang, B.; Tan, J.; Zhu, L.; Li, L. Immobilized multienzymatic systems for catalysis of cascade reactions. Process Biochem. 2016, 51, 1193–1203. [Google Scholar] [CrossRef]
- Hwang, E.T.; Lee, S. Multienzymatic Cascade Reactions via Enzyme Complex by Immobilization. ACS Catal. 2019, 9, 4402–4425. [Google Scholar] [CrossRef]
- Meryam Sardar, R.A. Enzyme Immobilization: An Overview on Nanoparticles as Immobilization Matrix. Biochem. Anal. Biochem. 2015, 4, 178. [Google Scholar] [CrossRef]
- Gupta, M.N.; Kaloti, M.; Kapoor, M.; Solanki, K. Nanomaterials as matrices for enzyme immobilization. Artif. Cells Blood Substit. Biotechnol. 2011, 39, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Patila, M.; Kouloumpis, A.; Gournis, D.; Rudolf, P.; Stamatis, H. Laccase-functionalized graphene oxide assemblies as efficient nanobiocatalysts for oxidation reactions. Sensors (Switz.) 2016, 16, 287. [Google Scholar] [CrossRef] [PubMed]
- Chatzikonstantinou, A.V.; Gkantzou, E.; Gournis, D.; Patila, M.; Stamatis, H. Stabilization of Laccase through Immobilization on Functionalized GO-Derivatives. Enzyme Nanoarchitectures: Enzymes Armored with Graphene, 1st ed.; Kumar, C.V., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 609, pp. 47–81. [Google Scholar]
- Chen, M.; Zeng, G.; Xu, P.; Lai, C.; Tang, L. How Do Enzymes ‘Meet’ Nanoparticles and Nanomaterials? Trends Biochem. Sci. 2017, 42, 914–930. [Google Scholar] [CrossRef] [PubMed]
- Pavlidis, I.V.; Patila, M.; Bornscheuer, U.T.; Gournis, D.; Stamatis, H. Graphene-based nanobiocatalytic systems: Recent advances and future prospects. Trends Biotechnol. 2014, 32, 312–320. [Google Scholar] [CrossRef]
- Bilal, M.; Zhao, Y.; Rasheed, T.; Iqbal, H.M.N. Magnetic nanoparticles as versatile carriers for enzymes immobilization: A review. Int. J. Biol. Macromol. 2018, 120, 2530–2544. [Google Scholar] [CrossRef]
- Sojitra, U.V.; Nadar, S.S.; Rathod, V.K. A magnetic tri-enzyme nanobiocatalyst for fruit juice clarification. Food Chem. 2016, 213, 296–305. [Google Scholar] [CrossRef]
- Muley, A.B.; Thorat, A.S.; Singhal, R.S.; Harinath Babu, K. A tri-enzyme co-immobilized magnetic complex: Process details, kinetics, thermodynamics and applications. Int. J. Biol. Macromol. 2018, 118, 1781–1795. [Google Scholar] [CrossRef]
- Orfanakis, G.; Patila, M.; Catzikonstantinou, A.V.; Lyra, K.-M.; Kouloumpis, A.; Spyrou, K.; Katapodis, P.; Paipetis, A.; Rudolf, P.; Gournis, D.; et al. Hybrid Nanomaterials of Magnetic Iron Nanoparticles and Graphene Oxide as Matrices for the Immobilization of β-Glucosidase: Synthesis, Characterization, and Biocatalytic Properties. Front. Mater. 2018, 5, 11. [Google Scholar] [CrossRef]
- Jia, F.; Mallapragada, S.K.; Narasimhan, B. Multienzyme Immobilization and Colocalization on Nanoparticles Enabled by DNA Hybridization. Ind. Eng. Chem. Res. 2015, 54, 10212–10220. [Google Scholar] [CrossRef]
- Wang, S.Z.; Zhang, Y.H.; Ren, H.; Wang, Y.L.; Jiang, W.; Fang, B.S. Strategies and perspectives of assembling multi-enzyme systems. Crit. Rev. Biotechnol. 2017, 37, 1024–1037. [Google Scholar] [CrossRef]
- Wheeldon, I.; Minteer, S.D.; Banta, S.; Barton, S.C.; Atanassov, P.; Sigman, M. Substrate channelling as an approach to cascade reactions. Nat. Chem. 2016, 8, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.P. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol. Adv. 2011, 29, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Poshyvailo, L.; Von Lieres, E.; Kondrat, S. Does metabolite channeling accelerate enzyme-catalyzed cascade reactions? PLoS ONE 2017, 12, e0172673. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.J.; Jung, S.; Kim, H.J.; Lee, Y.G.; Nam, K.C.; Lee, H.J.; Bae, H.J. Co-immobilization of three cellulases on Au-doped magnetic silica nanoparticles for the degradation of cellulose. Chem. Commun. 2012, 48, 886–888. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, D.; Wu, C.; Yao, K.; Li, Z.; Shi, N.; Wen, F.; Gates, I.D. Co-immobilization of cellulase and lysozyme on amino-functionalized magnetic nanoparticles: An activity-tunable biocatalyst for extraction of lipids from microalgae. Bioresour. Technol. 2018, 263, 317–324. [Google Scholar] [CrossRef]
- Zarafeta, D.; Kissas, D.; Sayer, C.; Gudbergsdottir, S.R. Discovery and Characterization of a Thermostable and Highly Halotolerant GH5 Cellulase from an Icelandic Hot Spring Isolate. PLoS ONE 2016, 11, e0146454. [Google Scholar] [CrossRef]
- Chen, S.; Wen, L.; Svec, F.; Lv, Y. Magnetic metal-organic frameworks as scaffolds for spatial co-location and positional assembly of multi-enzyme systems enabling enhanced cascade biocatalysis. RSC Adv. 2017, 7, 21205–21213. [Google Scholar] [CrossRef]
- Pitzalis, F.; Monduzzi, M.; Salis, A. A bienzymatic biocatalyst constituted by glucose oxidase and Horseradish peroxidase immobilized on ordered mesoporous silica. Microporous Mesoporous Mater. 2017, 241, 145–154. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, Y.; Ma, W.; Wang, S. A Fluorescent Sensor for Zinc Detection and Removal Based on Core-Shell Functionalized Fe3O4 @SiO2 Nanoparticles. J. Nanomater. 2013, 2013, 178138. [Google Scholar] [CrossRef]
- Bayat, A.; Shakourian-fard, M.; Ehyaei, N. A magnetic supported iron complex for selective oxidation of sulfides to sulfoxides using 30% hydrogen peroxide at room temperature. RSC Adv. 2014, 4, 44274–44281. [Google Scholar] [CrossRef]
- Majoul, N.; Aouida, S.; Bessaïs, B. Progress of porous silicon APTES-functionalization by FTIR investigations. Appl. Surf. Sci. 2015, 331, 388–391. [Google Scholar] [CrossRef]
- Tawil, N.; Sacher, E.; Boulais, E.; Mandeville, R.; Meunier, M. X-ray Photoelectron Spectroscopic and Transmission Electron Microscopic Characterizations of Bacteriophage–Nanoparticle Complexes for Pathogen Detection. J. Phys. Chem. C 2013, 117, 20656–20665. [Google Scholar] [CrossRef]
- Gupta, P.; Verma, S.; Vakhlu, J. Comparative Analysis of Β -Glucosidases Thermostability: Differences in Amino Acids Composition and Distribution among Mesostable and Thermostable Β -Glucosidases. J. Adv. Bioinform. Appl. Res. 2014, 5, 215–227. [Google Scholar]
- Salgaonkar, M.; Nadar, S.S.; Rathod, V.K. Combi-metal organic framework (Combi-MOF) of α-amylase and glucoamylase for one pot starch hydrolysis. Int. J. Biol. Macromol. 2018, 113, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.O.; Bork, J.A.; Fernandez-Lorente, G.; Guisan, J.M.; Furigo, A.; de Oliveira, D.; Pessela, B.C. Co-immobilization of lipases and Β-D-galactosidase onto magnetic nanoparticle supports: Biochemical characterization. Mol. Catal. 2018, 453, 12–21. [Google Scholar] [CrossRef]
- Eze, S.O.O.; Chilaka, F.C.; Nwanguma, B.C. Studies on Thermodynamics and Kinetics of Thermo-Inactivation of Some Quality-Related Enzymes in White Yam (Dioscorea rotundata). J. Thermodyn. Catal. 2010. [Google Scholar] [CrossRef]
- Monteiro, P.; Aliakbarian, B.; Ximenes, E.; Filho, F.; Oliveira, P.; Pessoa, A.; Converti, A.; Perego, P. Kinetic and thermodynamic studies of a novel acid protease from Aspergillus foetidus. Int. J. Biol. Macromol. 2015, 81, 17–21. [Google Scholar] [CrossRef]
- De Oliveira, R.L.; da Silva, O.S.; Converti, A.; Porto, T.S. Thermodynamic and kinetic studies on pectinase extracted from Aspergillus aculeatus: Free and immobilized enzyme entrapped in alginate beads. J. Biological. Macromol. 2018, 115, 1088–1093. [Google Scholar] [CrossRef]
- Farrugia, T.; Perriman, A.W.; Sharma, K.P.; Mann, S. Multi-enzyme cascade reactions using protein -polymer surfactant self-standing films. Chem. Commun. 2017, 53, 2094–2097. [Google Scholar] [CrossRef]
- Rezaei, S.; Landarani-Isfahani, A.; Moghadam, M.; Tangestaninejad, S.; Mirkhani, V.; Mohammadpoor-Baltork, I. Development of a novel bi-enzymatic silver dendritic hierarchical nanostructure cascade catalytic system for efficient conversion of starch into gluconic acid. Chem. Eng. J. 2019, 356, 423–435. [Google Scholar] [CrossRef]
- Kouassi, G.K.; Irudayaraj, J.; Mccarty, G. Activity of glucose oxidase functionalized onto magnetic nanoparticles. Biomagn. Res. Technol. 2005, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.E.; Verma, M.L.; Barrow, C.J.; Puri, M. Suitability of magnetic nanoparticle immobilised cellulases in enhancing enzymatic saccharification of pretreated hemp biomass. Biotechnol. Biofuels 2014, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wu, R.; Fu, Q.; Wang, H.; Zhang, Z.; Haji, Z.; Li, X.; Lian, X.; An, Y. Immobilization of β-Glucosidase BglC on Decanedioic Acid-Modified Magnetic Nanoparticles. J. Chem. Eng. Technol. 2018, 41, 1949–1955. [Google Scholar] [CrossRef]
- Chouyyok, W.; Panpranot, J.; Thanachayanant, C.; Prichanont, S. Effects of pH and pore characters of mesoporous silicas on horseradish peroxidase immobilization. J. Mol. Catal. B Enzym. 2009, 56, 246–252. [Google Scholar] [CrossRef]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial Applications of Enzymes: Recent Advances, Techniques, and Outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Hsin Chi, H.; Radu, D.R.; Dezayas, G.; Penney, M.; Yu Lai, C. Stellate MSN-based Dual-enzyme Nano-Biocatalyst for the Cascade Conversion of Non-Food Feedstocks to Food Products. J. Thermodyn. Catal. 2017, 8, 2. [Google Scholar] [CrossRef]
- Altun, S.; Çakıroğlu, B.; Özacar, M. A facile and effective immobilization of glucose oxidase on tannic acid modified CoFe2O4 magnetic nanoparticles. Colloids Surf. B Biointerfaces 2015, 136, 963–970. [Google Scholar] [CrossRef]
- Chang, Q.; Tang, H. Immobilization of horseradish peroxidase on NH2-modified magnetic Fe3O4/SiO2 particles and its application in removal of 2,4-dichlorophenol. Molecules 2014, 19, 15768–15782. [Google Scholar] [CrossRef]
- López-Gallego, F.; Guisán, J.M.; Betancor, L. Glutaraldehyde-mediated protein immobilization. Methods Mol. Biol. 2008, 1051, 33–41. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miller, G.L. Use of DinitrosaIicyIic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bowers, G.N.; Mccomb, R.B.; Christensen, R.C.; Schaffer, R. High-purity 4-nitrophenol: Purification, characterization, and specifications for use as a spectrophotometric reference material. Clin. Chem. 1980, 26, 724–729. [Google Scholar] [PubMed]
- Scott, S.L.; Chen, W.; Bakac, A.; Espenson, J.H. Spectroscopic parameters, electrode potentials, acid ionization constants, and electron exchange rates of the 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonate)radicals and ions. J. Phys. Chem. 1993, 97, 6710–6714. [Google Scholar] [CrossRef]

| Enzyme | M.M. (kDa) | pI |
|---|---|---|
| Cellulase, CelDZ1, from a Thermoanaerobacterium hot spring isolate [27] | 42 | 5.7 |
| β-Glucosidase from Thermotoga maritima | 53.7 | 6.0 |
| Glucose oxidase from Aspergillus niger | 160 | 4.2 |
| Horseradish peroxidase | 40 | 7.2 |
| Enzyme | t1/2 (h) | Ed (kJ/mol) |
|---|---|---|
| Free form of CelDZ1 | 2.5 | 25.7 |
| Co-immobilized form of CelDZ1 | 5.0 | 30.1 |
| Free form of bgl | 12.8 | 57.5 |
| Co-immobilized form of bgl | 23.1 | 60.2 |
| Free form of GOx | 14.4 | 30.1 |
| Co-immobilized form of GOx | 19.2 | 47.7 |
| Free form of HRP | 2.4 | 64.1 |
| Co-immobilized form of HRP | 2.9 | 70.1 |
| Enzyme | ΔΗ° (kJ/mol) | ΔG° (kJ/mol) | ΔS° (J/mol/K) |
|---|---|---|---|
| Free form of CelDZ1 | 23.00 | 92.45 | −215.01 |
| Co-immobilized form of CelDZ1 | 27.47 | 94.25 | −208.13 |
| Free form of bgl | 54.65 | 102.91 | −140.69 |
| Co-immobilized form of bgl | 57.35 | 104.60 | −137.75 |
| Free form of GOx | 27.42 | 97.07 | −215.63 |
| Co-immobilized form of GOx | 45.02 | 97.85 | −163.56 |
| Free form of HRP | 61.42 | 92.27 | −95.51 |
| Co-immobilized form of HRP | 67.42 | 92.83 | −78.66 |
| Forms | Km | Vmax (μmol/min) |
|---|---|---|
| Free CelDZ1 | 6.09 ± 0.78 | 1.41 ± 0.16 |
| CelDZ1 in co-immobilized form | 6.78 ± 0.21 | 0.40 ± 0.02 |
| CelDZ1 in individually immobilized form | 8.15 ± 2.47 | 1.22 ± 0.28 |
| Free bgl | 0.25 ± 0.01 | 37.54 ± 0.56 |
| bgl in co-immobilized form | 0.36 ± 0.05 | 9.46 ± 0.26 |
| bgl in individually immobilized form | 0.46 ± 0.04 | 2.36 ± 0.06 |
| Free GOx | 1.72 ± 0.11 | 12.07 ± 1.38 |
| GOx in co-immobilized form | 2.76 ± 0.98 | 5.30 ± 0.28 |
| GOx in individually immobilized form | 11.93 ± 1.77 | 3.70 ± 0.17 |
| Free HRP | 0.014 ± 0.001 | 20.74 ± 1.05 |
| HRP in co-immobilized form | 0.017 ± 0.013 | 5.45 ± 0.57 |
| HRP in individually immobilized form | 0.044 ± 0.010 | 3.66 ± 0.21 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannakopoulou, A.; Patila, M.; Spyrou, K.; Chalmpes, N.; Zarafeta, D.; Skretas, G.; Gournis, D.; Stamatis, H. Development of a Four-Enzyme Magnetic Nanobiocatalyst for Multi-Step Cascade Reactions. Catalysts 2019, 9, 995. https://doi.org/10.3390/catal9120995
Giannakopoulou A, Patila M, Spyrou K, Chalmpes N, Zarafeta D, Skretas G, Gournis D, Stamatis H. Development of a Four-Enzyme Magnetic Nanobiocatalyst for Multi-Step Cascade Reactions. Catalysts. 2019; 9(12):995. https://doi.org/10.3390/catal9120995
Chicago/Turabian StyleGiannakopoulou, Archontoula, Michaela Patila, Konstantinos Spyrou, Nikolaos Chalmpes, Dimitra Zarafeta, Georgios Skretas, Dimitrios Gournis, and Haralambos Stamatis. 2019. "Development of a Four-Enzyme Magnetic Nanobiocatalyst for Multi-Step Cascade Reactions" Catalysts 9, no. 12: 995. https://doi.org/10.3390/catal9120995
APA StyleGiannakopoulou, A., Patila, M., Spyrou, K., Chalmpes, N., Zarafeta, D., Skretas, G., Gournis, D., & Stamatis, H. (2019). Development of a Four-Enzyme Magnetic Nanobiocatalyst for Multi-Step Cascade Reactions. Catalysts, 9(12), 995. https://doi.org/10.3390/catal9120995






_Stamatis.png)