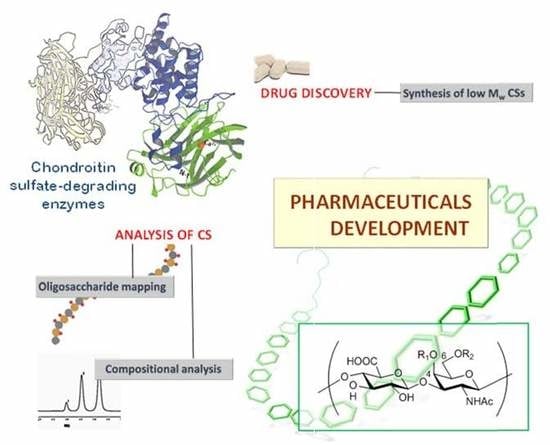
Chondroitin Sulfate-Degrading Enzymes as Tools for the Development of New Pharmaceuticals
Abstract
:1. Introduction
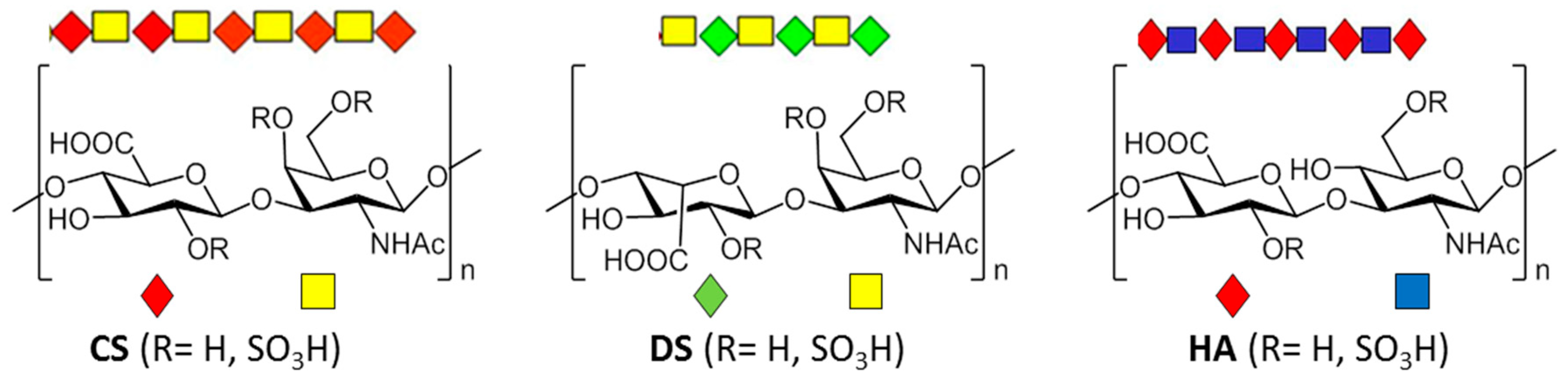
2. Types, Mechanism and Structure of CS Lyases
2.1. Chondriotinases ABC
2.1.1. CSase ABC I Endolyase
2.1.2. CSase ABC II Exolyase from Bacteroides thetaiotaomicron
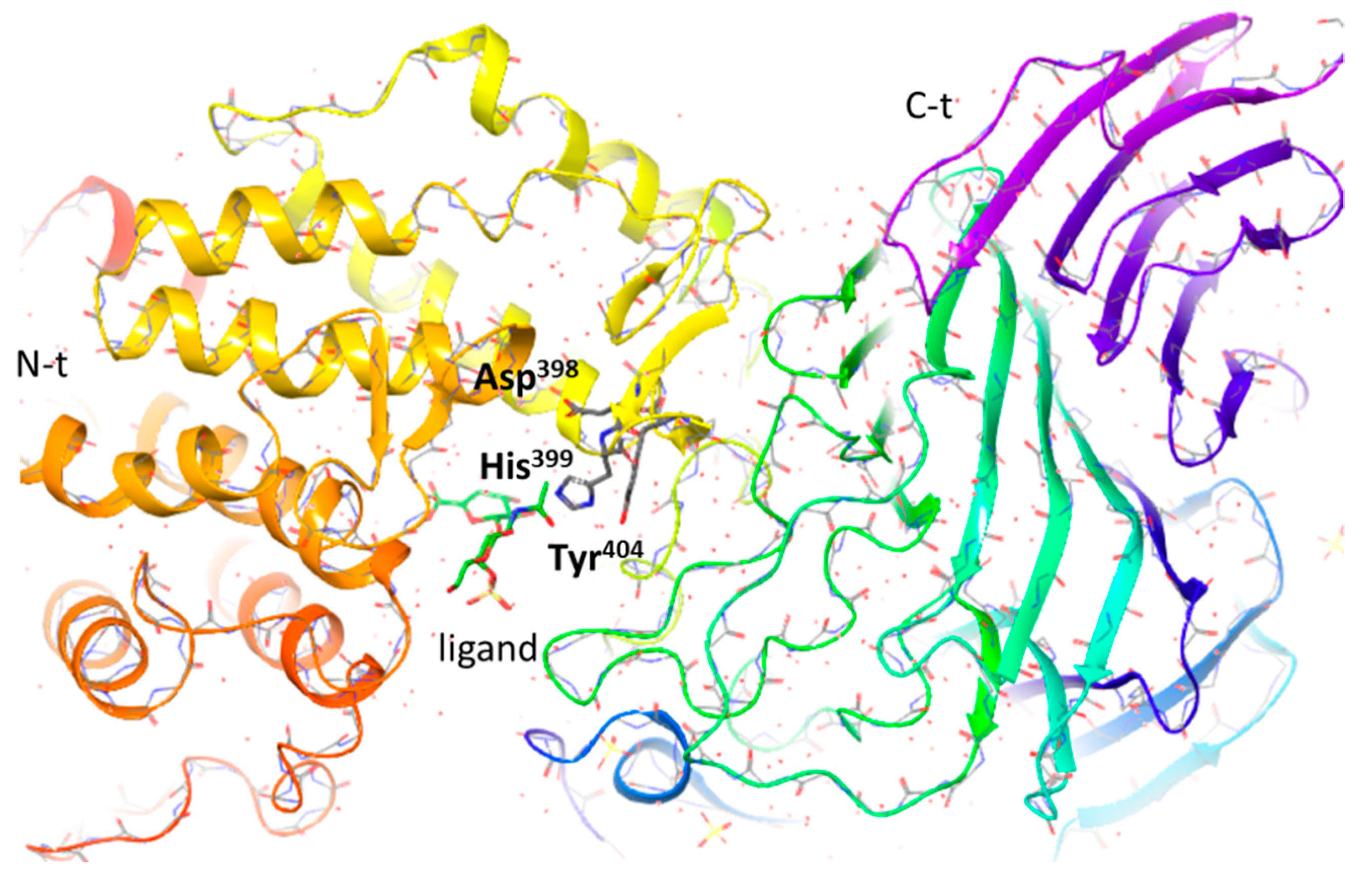
2.1.3. CSase ABC II Exolyase from Proteus vulgaris
2.1.4. Other CSases
2.2. CSases AC
2.3. CSase B
2.4. HAases
3. Applications of CSases
3.1. Synthetic Applications: Preparation of Low Molecular Weight Chondroitin Sulphate (LMWCS) as Therapeutic Agents
3.2. Analytical Applications
3.2.1. Oligosaccharide Mapping
3.2.2. Compositional Analysis of GAGs
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Volpi, N. Therapeutic applications of glycosaminoglycans. Curr. Med. Chem. 2006, 13, 1799–1810. [Google Scholar] [CrossRef] [PubMed]
- Stick, R.V.; Williams, S. Glycoproteins and proteoglycans. In Carbohydrates: The Essential Molecules of Life; Stick, R.V., Williams, S., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2008; pp. 369–412. [Google Scholar]
- Hermann, W.; Lambova, S.; Muller-Ladner, U. Current treatment options for osteoarthritis. Curr. Rheumatol. Rev. 2018, 14, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Mourão, A.P. Perspective on the use of sulfated polysaccharides from marine organisms as a source of new antithrombotic drugs. Mar. Drugs 2015, 13, 2770–2784. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Sugahara, K. Potential therapeutic application of chondroitin sulfate/dermatan sulfate. Curr. Drug Discov. Tech. 2008, 5, 289–301. [Google Scholar] [CrossRef]
- Milstone, L.M.; Houghmonroe, L.; Kugelman, L.C.; Bender, J.R.; Haggerty, J.G. Epican, a heparan/chondroitin sulfate proteoglycan form of CD44, mediates cell-cell adhesion. J. Cell Sci. 1994, 107, 3183–3190. [Google Scholar] [PubMed]
- Stabler, T.V.; Huang, Z.; Montell, E.; Verges, J.; Kraus, V.B. Chondroitin sulphate inhibits NF-κB activity induced by interaction of pathogenic and damage associated molecules. Osteoarthritis Cartilage 2017, 25, 166–174. [Google Scholar] [CrossRef]
- Wu, F.F.; Zhou, C.H.; Zhou, D.D.; Ou, S.Y.; Liu, Z.J.; Huang, H.H. Immune-enhancing activities of chondroitin sulfate in murine macrophage RAW 264.7 cells. Carbohydr. Polym. 2018, 198, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Rogerson, S.J.; Chaiyaroj, S.C.; Ng, K.; Reeder, J.C.; Brown, G.V. Chondroitin sulfate A is a cell-surface receptor for Plamodium-falciparum infected erythrocytes. J. Exp. Med. 1995, 182, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.A.; Clausen, T.M.; Pehrson, C.; Mao, Y.; Resende, M.; Daugaard, M.; Kristensen, A.R.; Spliid, C.; Mathiesen, L.; Knudsen, L.E.; et al. Placental Sequestration of Plasmodium falciparum malaria parasites is mediated by the interaction between VAR2CSA and Chondroitin sulfate A on Syndecan-1. PloS Pathog. 2016, 12, e1005831. [Google Scholar] [CrossRef]
- Mourao, P.A.S.; Pereira, M.S.; Pavao, M.S.G.; Mulloy, B.; Tollefsen, D.M.; Mowinckel, M.C.; Abildgaard, U. Structure and anticoagulant activity of a fucosylated chondroitin sulfate from echinoderm.Sulfated fucose branches on the polysaccharide account for its high anticoagulant action. J. Biol. Chem. 1996, 271, 23973–23984. [Google Scholar] [CrossRef]
- Glauser, B.F.; Pereira, M.S.; Monteiro, R.Q.; Mourao, P.A.S. Serpin-independent anticoagulant activity of a fucosylated chondroitin sulfate. Thromb. Haemos. 2008, 100, 420–428. [Google Scholar] [CrossRef]
- Ustyuzhanina, N.E.; Bilan, M.I.; Panina, E.G.; Sanamyan, N.P.; Dmitrenok, A.S.; Tsvetkova, E.A.; Ushakova, N.A.; Shashkov, A.S.; Nifantiev, N.E.; Usov, A.I. Structure and anti-inflammatory activity of a new unusual fucosylated chondroitin sulfate from Cucumaria djakonovi. Mar. Drugs 2018, 16, 389. [Google Scholar] [CrossRef] [PubMed]
- Lovu, M.; Dumais, G.; du Souich, P. Anti-inflammatory activity of chondroitin sulfate. Osteoarthritis Cartilage 2008, 16, S14–S18. [Google Scholar]
- Bergefall, K.; Trybala, E.; Johansson, M.; Uyama, T.; Naito, S.; Yamada, S.; Kitagawa, H.; Sugahara, K.; Bergström, T. Chondroitin sulfate characterized by the E-disaccharide unit is a potent inhibitor of herpes simplex virus infectivity and provides the virus binding sites on gro2C cells. J. Biol. Chem. 2005, 280, 32193–32199. [Google Scholar] [CrossRef] [PubMed]
- Mycroft-West, C.J.; Yates, E.A.; Skidmore, M.A. Marine glycosaminoglycan-like carbohydrates as potential drug candidates for infectious disease. Biochem. Soc. Trans. 2018, 46, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Goel, S.; Muthusamy, A.; Miao, J.; Cui, L.W.; Salanti, A.; Winzeler, E.A.; Gowda, D.C. Targeted disruption of a ring-infected erythrocyte surface antigen (RESA)-like export protein gene in Plasmodium falciparum confers stable chondroitin 4-sulfate cytoadherence Capacity. J. Biol.l Chem. 2014, 289, 34408–34421. [Google Scholar] [CrossRef] [PubMed]
- Alkhalil, A.; Achur, R.N.; Valiyaveettil, M.; Ockenhouse, C.F.; Gowda, D.C. Structural requirements for the adherence of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate proteoglycans of human placenta. J. Biol. Chem. 2000, 275, 40357–40364. [Google Scholar] [CrossRef] [PubMed]
- Asimakopoulou, A.P.; Theocharis, A.D.; Tzanakakis, G.N.; Karamanos, N.K. The biological role of chondroitin sulfate in cancer and chondroitin-based anticancer agents. In Vivo 2008, 22, 385–389. [Google Scholar] [PubMed]
- Borsig, L.; Wang, L.; Cavalcante, M.C.M.; Cardilo-Reis, L.; Ferreira, P.L.; Mourão, P.A.S.; Esko, J.D.; Pavão, M.S.G. Selectin blocking activity of a fucosylated chondroitin sulfate glycosaminoglycan from sea cucumber: Effect on tumor metastasis and neutrophil recruitment. J. Biol. Chem. 2007, 282, 14984–14991. [Google Scholar] [CrossRef]
- Merida-de-Barros, D.A.; Chaves, S.P.; Belmiro, C.L.R.; Wanderley, J.L.M. Leishmaniasis and glycosaminoglycans: a future therapeutic strategy? Parasit. Vectors 2018, 11, 536. [Google Scholar] [CrossRef] [PubMed]
- Vallen, M.J.E.; van Tilborg, A.A.G.; Tesselaar, M.H.; ten Dam, G.B.; Bulten, J.; van Kuppevelt, T.H.; Massuger, L. Novel single-chain antibody GD3A10 defines a chondroitin sulfate biomarker for ovarian cancer. Biomark. Med. 2014, 8, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Tamaki, H.; Fukui, S. Detection of oligosaccharide ligands for Hepatocyte growth factor/Scatter factor (HGF/SF), Keratinocyte growth factor (KGF/FGF-7), RANTES and Heparin cofactor II by neoglycolipid microarrays of glycosaminoglycan-derived oligosaccharide fragments. Glycoconjugate J. 2006, 23, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Djerbal, L.; Lortat-Jacob, H.; Kwok, J. Chondroitin sulfates and their binding molecules in the central nervous system. Glycoconjugate J. 2017, 34, 363–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G.M.; Hsieh-Wilson, L.C. Sugar-dependent modulation of neuronal development, regeneration, and plasticity by chondroitin sulfate proteoglycans. Exp. Neurol. 2015, 274, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Gao, J.; Hou, L.; Wang, L.; Zhang, F.; Sun, F.; Zhang, T.; Xu, P.; Shi, Z.; Hu, F.; et al. Neuroprotective effect of chondroitin sulfate on SH-SY5Y cells overexpressing wild-type or A53T mutant α-synuclein. Mol. Med. Rep. 2017, 16, 8721–8728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, X.H.; Jiang, Y.Z.; Zhang, G.R.; Jin, H.M.; Hieu, N.T.M.; Ouyang, H.W. Specific interactions between human fibroblasts and particular chondroitin sulfate molecules for wound healing. Acta Biomater. 2009, 5, 1588–1595. [Google Scholar] [CrossRef]
- Kowitsch, A.; Zhou, G.Y.; Groth, T. Medical application of glycosaminoglycans: A review. J. Tissue Eng. Regen. Med. 2018, 12, E23–E41. [Google Scholar] [CrossRef] [PubMed]
- Altgarde, N.; Nileback, E.; de Battice, L.; Pashkuleva, I.; Reis, R.L.; Becher, J.; Moller, S.; Schnabelrauch, M.; Svedhem, S. Probing the biofunctionality of biotinylated hyaluronan and chondroitin sulfate by hyaluronidase degradation and aggrecan interaction. Acta Biomater 2013, 9, 8158–8166. [Google Scholar] [CrossRef] [PubMed]
- Kliemt, S.; Lange, C.; Otto, W.; Hintze, V.; Möller, S.; von Bergen, M.; Hempel, U.; Kalkhof, S. Sulfated Hyaluronan Containing Collagen Matrices Enhance Cell-Matrix-Interaction, Endocytosis, and Osteogenic Differentiation of Human Mesenchymal Stromal Cells. J. Proteome Res. 2013, 12, 378–389. [Google Scholar] [CrossRef]
- Korn, P.; Schulz, M.C.; Hintze, V.; Range, U.; Mai, R.; Eckelt, U.; Schnabelrauch, M.; Moller, S.; Becher, J.; Scharnweber, D.; Stadlinger, B. Chondroitin sulfate and sulfated hyaluronan-containing collagen coatings of titanium implants influence peri-implant bone formation in a minipig model. J. Biomed. Mater. Res. Part A 2014, 102, 2334–2344. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.A.; Melchels, F.P.W.; Schrobback, K.; Hutmacher, D.W.; Malda, J.; Klein, T.J. A biomimetic extracellular matrix for cartilage tissue engineering centered on photocurable gelatin, hyaluronic acid and chondroitin sulfate. Acta Biomater. 2014, 10, 214–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, S.S.; Liu, Y.C.; Shu, X.Z.; Prestwich, G.D. Injectable glycosaminoglycan hydrogels for controlled release of human basic fibroblast growth factor. Biomaterials 2005, 26, 6054–6067. [Google Scholar] [CrossRef] [PubMed]
- Elia, R.; Newhide, D.R.; Pedevillano, P.D.; Reiss, G.R.; Firpo, M.A.; Hsu, E.W.; Kaplan, D.L.; Prestwich, G.D.; Peattie, R.A. Silk-hyaluronan-based composite hydrogels: A novel, securable vehicle for drug delivery. J. Biomater. Appl. 2013, 27, 749–762. [Google Scholar] [CrossRef]
- Shu, X.Z.; Ahmad, S.; Liu, Y.C.; Prestwich, G.D. Synthesis and evaluation of injectable, in situ crosslinkable synthetic extracellular matrices for tissue engineering. J. Biomed. Mater. Res. Part A 2006, 79A, 902–912. [Google Scholar] [CrossRef]
- Jin, R.; Lou, B.; Lin, C. Tyrosinase-mediated in situ forming hydrogels from biodegradable chondroitin sulfate-tyramine conjugates. Polym. Int. 2013, 62, 353–361. [Google Scholar] [CrossRef]
- Ni, Y.L.; Tang, Z.R.; Cao, W.X.; Lin, H.; Fan, Y.J.; Guo, L.K.; Zhang, X.D. Tough and elastic hydrogel of hyaluronic acid and chondroitin sulfate as potential cell scaffold materials. Int. J. Biol. Macromol. 2015, 74, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.Q.; Yeo, Y.; Clifton, R.J.; Jiao, T.; Kohane, D.S.; Kobler, J.B.; Zeitels, S.M.; Langer, R. Hyaluronic acid-based microgels and microgel networks for vocal fold regeneration. Biomacromolecules 2006, 7, 3336–3344. [Google Scholar] [CrossRef]
- Kirker, K.R.; Luo, Y.; Nielson, J.H.; Shelby, J.; Prestwich, G.D. Glycosaminoglycan hydrogel films as bio-interactive dressings for wound healing. Biomaterials 2002, 23, 3661–3671. [Google Scholar] [CrossRef]
- Philandrianos, C.; Andrac-Meyer, L.; Mordon, S.; Feuerstein, J.-M.; Sabatier, F.; Veran, J.; Magalon, G.; Casanova, D. Comparison of five dermal substitutes in full-thickness skin wound healing in a porcine model. Burns 2012, 38, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zhang, Q.; Wang, J.; Liu, Y.; Lu, S.; Li, M.; Kaplan, D.L. Silk fibroin/chondroitin sulfate/hyaluronic acid ternary scaffolds for dermal tissue reconstruction. Acta Biomater. 2013, 9, 6771–6782. [Google Scholar] [CrossRef] [PubMed]
- Kesti, M.; Mueller, M.; Becher, J.; Schnabelrauch, M.; D’Este, M.; Eglin, D.; Zenobi-Wong, M. A versatile bioink for three-dimensional printing of cellular scaffolds based on thermally and photo-triggered tandem gelation. Acta Biomater. 2015, 11, 162–172. [Google Scholar] [CrossRef]
- Daamen, W.F.; van Moerkerk, H.T.B.; Hafmans, T.; Buttafoco, L.; Poot, A.A.; Veerkamp, J.H.; van Kuppevelt, T.H. Preparation and evaluation of molecularly-defined collagen-elastin-glycosaminoglycan scaffolds for tissue engineering. Biomaterials 2003, 24, 4001–4009. [Google Scholar] [CrossRef]
- McFadden, T.M.; Duffy, G.P.; Allen, A.B.; Stevens, H.Y.; Schwarzmaier, S.M.; Plesnila, N.; Murphy, J.M.; Barry, F.P.; Guldberg, R.E.; O’Brien, F.J. The delayed addition of human mesenchymal stem cells to pre-formed endothelial cell networks results in functional vascularization of a collagen-glycosaminoglycan scaffold in vivo. Acta Biomater. 2013, 9, 9303–9316. [Google Scholar] [CrossRef]
- Ko, C.-S.; Huang, J.-P.; Huang, C.-W.; Chu, I.M. Type II collagen-chondroitin sulfate-hyaluronan scaffold cross-linked by genipin for cartilage tissue engineering. J. Biosci. Bioeng. 2009, 107, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-L.; Lee, H.-P.; Chan, H.-Y.; Sung, L.-Y.; Chen, H.-C.; Hu, Y.-C. Composite chondroitin-6-sulfate/dermatan sulfate/chitosan scaffolds for cartilage tissue engineering. Biomaterials 2007, 28, 2294–2305. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-Y.; Chen, C.-H.; Hsiao, C.-Y.; Chen, J.-P. Incorporation of chitosan in biomimetic gelatin/chondroitin-6-sulfate/hyaluronan cryogel for cartilage tissue engineering. Carbohyd. Polym. 2015, 117, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.L.; McCoy, M.G.; Grant, S.A. Electrospinning collagen and hyaluronic acid nanofiber meshes. J. Mater. Sci. Mater. Med. 2012, 23, 1645–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Y.; Ghosh, K.; Li, B.; Sokolov, J.C.; Clark, R.A.F.; Rafailovich, M.H. Dual-syringe reactive electrospinning of cross-linked hyaluronic acid hydrogel nanofibers for tissue engineering applications. Macromol. Biosci. 2006, 6, 811–817. [Google Scholar] [CrossRef]
- Zhong, S.P.; Teo, W.E.; Zhu, X.; Beuertnan, R.; Ramakrishna, S.; Yung, L.Y.L. Development of a novel collagen-GAG nanofibrous scaffold via electrospinning. Mater. Sci. Eng. C Biomim. Supramol. Syst. 2007, 27, 262–266. [Google Scholar] [CrossRef]
- Santo, V.E.; Gomes, M.E.; Mano, J.F.; Reis, R.L. Chitosan-chondroitin sulphate nanoparticles for controlled delivery of platelet lysates in bone regenerative medicine. J. Tissue Eng. Regen. Med. 2012, 6, s47–s59. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H.; Wang, X. Synthetic oligosaccharide libraries and microarray technology: A powerful combination for the success of current glycosaminoglycan interactomics. ChemMedChem 2018, 13, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Avci, F.Y.; DeAngelis, P.L.; Liu, J.; Linhardt, R.J. Enzymatic synthesis of glycosaminoglycans: improving on nature. In Frontiers in Modern Carbohydrate Chemistry; Demchenko, A.V., Ed.; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2007; Volume 960, pp. 253–284. [Google Scholar]
- Linhardt, R.J.; Avci, F.Y.; Toida, T.; Kim, Y.S.; Cygler, M. CS lyases: Structure, activity, and applications in analysis and the treatment of diseases. Adv. Pharmacol. 2006, 53, 187–215. [Google Scholar] [PubMed]
- Kasinathan, N.; Volety, S.M.; Josyula, V.R. Chondroitinase: A promising therapeutic enzyme. Crit. Rev. Microbiol. 2016, 42, 474–484. [Google Scholar] [CrossRef]
- Orr, M.B.; Gensel, J.C. Spinal cord injury scarring and inflammation: Therapies targeting glial and inflammatory responses. Neurotherapeutics 2018, 15, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Denholm, E.M.; Lin, Y.Q.; Silver, P.J. Anti-tumor activities of chondroitinase AC and chondroitinase B: inhibition of angiogenesis, proliferation and invasion. Eur. J. Pharmacol. 2001, 416, 213–221. [Google Scholar] [CrossRef]
- Yao, X.Y.; Hageman, G.S.; Marmor, M.F. Recovery of retinal adhesion after enzymatic perturbation of the interphotoreceptor matrix. Invest. Ophthalmol. Vis. Sci. 1992, 33, 498–503. [Google Scholar]
- Kato, F.; Iwata, H.; Mimatsu, K.; Miura, T. Experimental chemonucleolysis with chondroitinase ABC. Clin. Orthop. Relat. Res. 1990, 253, 301–308. [Google Scholar] [CrossRef]
- Buhren, B.A.; Schrumpf, H.; Hoff, N.P.; Bolke, E.; Hilton, S.; Gerber, P.A. Hyaluronidase: From clinical applications to molecular and cellular mechanisms. Eur. J. Med. Res. 2016, 21, 5. [Google Scholar] [CrossRef] [PubMed]
- Rzany, B.; Becker-Wegerich, P.; Bachmann, F.; Erdmann, R.; Wollina, U. Hyaluronidase in the correction of hyaluronic acid-based fillers: A review and a recommendation for use. J. Cosmet. Dermatol. 2009, 8, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Niazi, Z.R.; Rehman, F.U.; Akhtar, A.; Khan, M.M.; Khan, S.; Baloch, N.; Khan, S. Hyaluronidases: A Therapeutic Enzyme. Protein Pept. Lett. 2018, 25, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Zechel, D.L.; Withers, S.G. Glycosidase mechanisms: Anatomy of a finely tuned catalyst. Acc. Chem. Res. 2000, 33, 11–18. [Google Scholar] [PubMed]
- Linhardt, R.J.; Galliher, P.M.; Cooney, C.L. Polysaccharide lyases. Appl. Biochem. Biotechnol. 1986, 12, 135–176. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Bernard, T.; Rancurel, C.; Brumer, H.; Coutinho, P.M.; Henrissat, B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 2010, 432, 437–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garron, M.L.; Cygler, M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 2010, 20, 1547–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Wang, J.; Li, F. Hyaluronidase and chondroitinase. Adv. Exp. Med. Biol. 2017, 925, 75–87. [Google Scholar] [PubMed]
- Stern, R.; Jedrzejas, M.J. Hyaluronidases: Their genomics, structures, and mechanisms of action. Chem. Rev. 2006, 106, 818–839. [Google Scholar] [CrossRef]
- Hovingh, P.; Linker, A. Hyaluronidase activity in leeches (Hirudinea). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1999, 124, 319–326. [Google Scholar] [CrossRef]
- Shaya, D.; Hahn, B.S.; Bjerkan, T.M.; Kim, W.S.; Park, N.Y.; Sim, J.S.; Kim, Y.S.; Cygler, M. Composite active site of chondroitin lyase ABC accepting both epimers of uronic acid. Glycobiology 2008, 18, 270–277. [Google Scholar] [CrossRef] [Green Version]
- Linn, S.; Chan, T.; Lipeski, L.; Salyers, A.A. Isolation and characterization of two chondroitin lyases from Bacteroides thetaiotaomicron. J. Bacteriol. 1983, 156, 859–866. [Google Scholar]
- Yamagata, T.; Saito, H.; Habuchi, O.; Suzuki, S. Purification and properties of bacterial chondroitinases and chondrosulfatases. J. Biol. Chem. 1968, 243, 1523–1535. [Google Scholar]
- Huang, W.; Lunin, V.V.; Li, Y.; Suzuki, S.; Sugiura, N.; Miyazono, H.; Cygler, M. Crystal structure of Proteus vulgaris chondroitin sulfate ABC lyase I at 1.9A resolution. J. Mol. Biol. 2003, 328, 623–634. [Google Scholar] [CrossRef]
- Prabhakar, V.; Raman, R.; Capila, I.; Bosques, C.J.; Pojasek, K.; Sasisekharan, R. Biochemical characterization of the chondroitinase ABC I active site. Biochem. J. 2005, 390, 395–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunin, V.V.; Li, Y.; Linhardt, R.J.; Miyazono, H.; Kyogashima, M.; Kaneko, T.; Bell, A.W.; Cygler, M. High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism. J. Mol. Biol. 2004, 337, 367–386. [Google Scholar] [CrossRef] [PubMed]
- Elmabrouk, Z.H.; Vincent, F.; Zhang, M.; Smith, N.L.; Turkenburg, J.P.; Charnock, S.J.; Black, G.W.; Taylor, E.J. Crystal structures of a family 8 polysaccharide lyase reveal open and highly occluded substrate-binding cleft conformations. Proteins 2011, 79, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Fethiere, J.; Eggimann, B.; Cygler, M. Crystal structure of chondroitin AC lyase, a representative of a family of glycosaminoglycan degrading enzymes. J. Mol. Biol. 1999, 288, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Boju, L.; Tkalec, L.; Su, H.; Yang, H.O.; Gunay, N.S.; Linhardt, R.J.; Kim, Y.S.; Matte, A.; Cygler, M. Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis. Biochemistry 2001, 40, 2359–2372. [Google Scholar] [CrossRef] [PubMed]
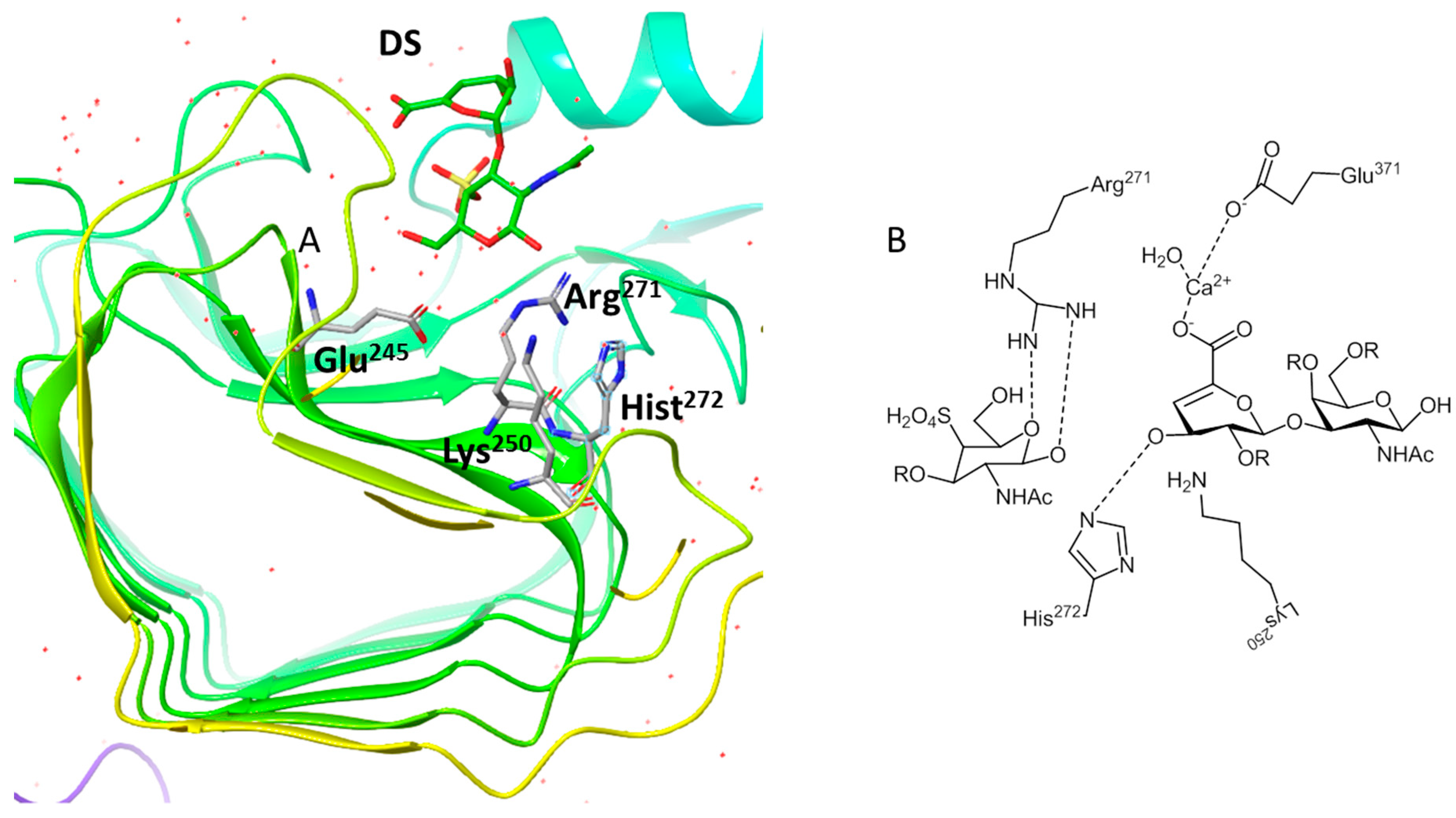
- Pojasek, K.; Raman, R.; Kiley, P.; Venkataraman, G.; Sasisekharan, R. Biochemical characterization of the chondroitinase B active site. J. Biol. Chem. 2002, 277, 31179–31186. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Matte, A.; Li, Y.; Kim, Y.S.; Linhardt, R.J.; Su, H.; Cygler, M. Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution. J. Mol. Biol. 1999, 294, 1257–1269. [Google Scholar] [CrossRef]
- Rigden, D.J.; Jedrzejas, M.J. Structures of Streptococcus pneumoniae hyaluronate lyase in complex with chondroitin and chondroitin sulfate disaccharides. Insights into specificity and mechanism of action. J. Biol. Chem. 2003, 278, 50596–50606. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, J.; Zhang, J.; Jiang, Y.; Shen, Z.; Guan, H.; Jiang, X. Purification and characterization of chondroitinase ABC from Acinetobacter sp. C26. Int. J. Biol. Macromol. 2017, 95, 80–86. [Google Scholar] [CrossRef]
- Jandik, K.A.; Gu, K.; Linhardt, R.J. Action pattern of polysaccharide lyases on glycosaminoglycans. Glycobiology 1994, 4, 289–296. [Google Scholar] [CrossRef] [PubMed]
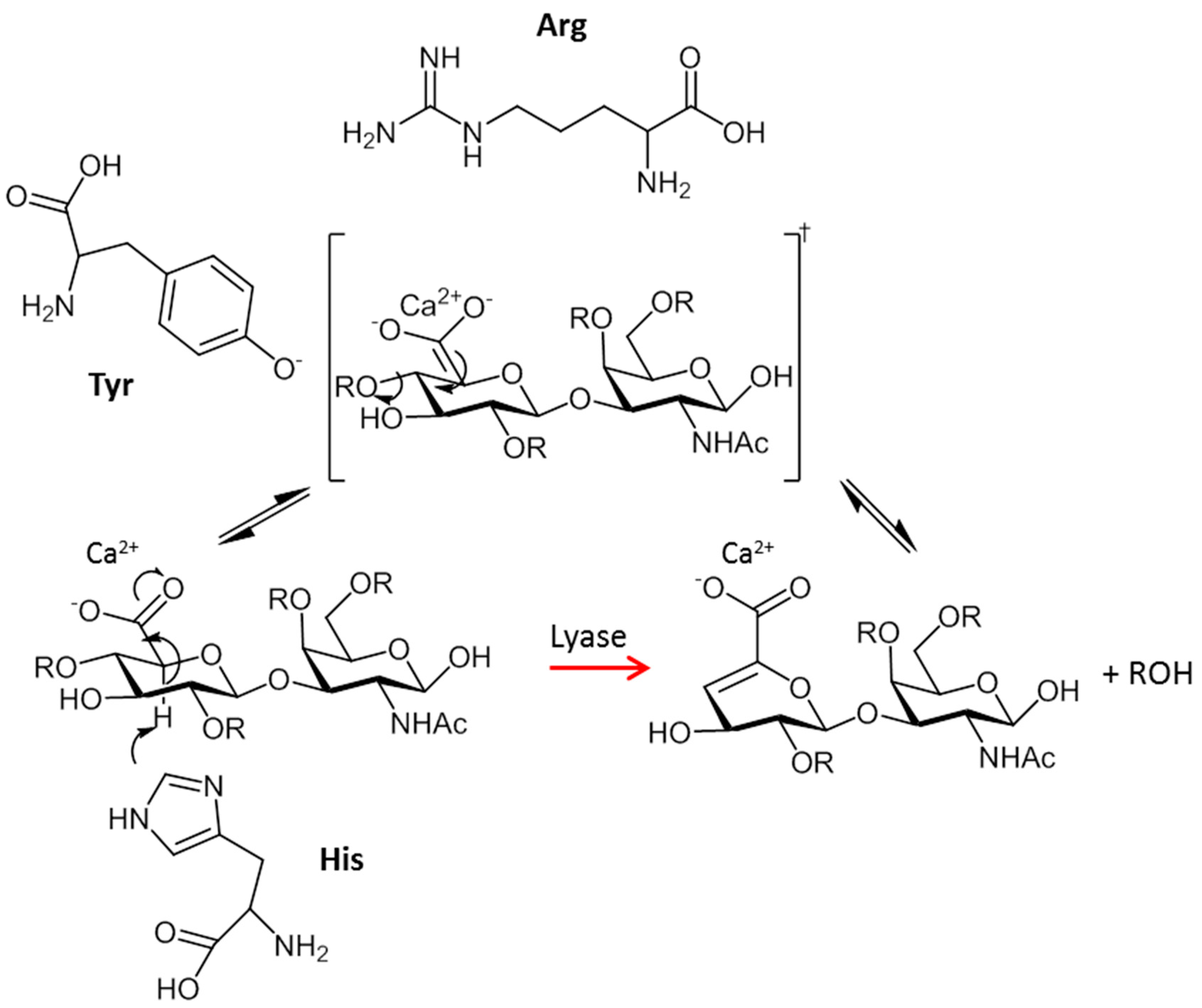
- Michel, G.; Pojasek, K.; Li, Y.; Sulea, T.; Linhardt, R.J.; Raman, R.; Prabhakar, V.; Sasisekharan, R.; Cygler, M. The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a calcium-dependent catalytic machinery. J. Biol. Chem. 2004, 279, 32882–32896. [Google Scholar] [CrossRef] [PubMed]
- Cordula, C.R.; Lima, M.A.; Shinjo, S.K.; Gesteira, T.F.; Pol-Fachin, L.; Coulson-Thomas, V.J.; Verli, H.; Yates, E.A.; Rudd, T.R.; Pinhal, M.A.; Toma, L.; Dietrich, C.P.; Nader, H.B.; Tersariol, I.L. On the catalytic mechanism of polysaccharide lyases: Evidence of His and Tyr involvement in heparin lysis by heparinase I and the role of Ca2+. Mol. BioSyst. 2014, 10, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.; Feng, D.; Xu, L.; Yin, F.; Zang, H.; Liu, C.; Wang, F. Preparation of low molecular weight chondroitin sulfates, screening of a high anti-complement capacity of low molecular weight chondroitin sulfate and its biological activity studies in attenuating osteoarthritis. Int. J. Mol. Sci. 2016, 17, 1685. [Google Scholar] [CrossRef] [PubMed]
- Buyue, Y.; Sheehan, J.P. Fucosylated chondroitin sulfate inhibits plasma thrombin generation via targeting of the factor IXa heparin-binding exosite. Blood 2009, 114, 3092–3100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tat, S.K.; Pelletier, J.P.; Mineau, F.; Duval, N.; Martel-Pelletier, J. Variable effects of 3 different chondroitin sulfate compounds on human osteoarthritic cartilage/chondrocytes: relevance of purity and Production Process. J. Rheumatol. 2010, 37, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.C. Isolation and purification of chondroitin sulfate. Adv. Pharmacol. 2006, 53, 21–31. [Google Scholar] [PubMed]
- Volpi, N. Analytical aspects of pharmaceutical grade chondroitin sulfates. J. Pharm. Sci. 2007, 96, 3168–3180. [Google Scholar] [CrossRef] [PubMed]
- Leeb, B.F.; Schweitzer, H.; Montag, K.; Smolen, J.S. A metaanalysis of chondroitin sulfate in the treatment of osteoarthritis. J. Rheumatol. 2000, 27, 205–211. [Google Scholar]
- Surapaneni, L.; Haley-Zitlin, V.; Bodine, A.; Jiang, X.P.; Brooks, J. Examination of chondroitin sulfate molecular weights on in vitro anti-inflammatory activity. FASEB J. 2013, 27, 1. [Google Scholar]
- Cho, S.Y.; Sim, J.S.; Jeong, C.S.; Chang, S.Y.; Choi, D.W.; Toida, T.; Kim, Y.S. Effects of low molecular weight chondroitin sulfate on type II collagen-induced arthritis in DBA/1J mice. Biol. Pharm. Bull. 2004, 27, 47–51. [Google Scholar] [CrossRef]
- Igarashi, N.; Takeguchi, A.; Sakai, S.; Akiyama, H.; Higashi, K.; Toida, T. Effect of molecular sizes of chondroitin sulfate on interaction with L-Selectin. Int. J. Carbohydr. Chem. 2013, 2013, 856142. [Google Scholar] [CrossRef]
- Renukuntla, J.; Vadlapudi, A.D.; Patel, A.; Boddu, S.H.S.; Mitra, A.K. Approaches for enhancing oral bioavailability of peptides and proteins. Int. J. Pharm. 2013, 447, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, P.; Cheng, Y.; Zhang, X.; Sheng, J.; Wang, D.; Li, J.; Zhang, Q.; Zhong, C.; Cao, R.; Wang, F. Enhancing the intestinal absorption of low molecular weight chondroitin sulfate by conjugation with alpha-linolenic acid and the transport mechanism of the conjugates. Int. J. Pharm. 2014, 465, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Martel-Pelletier, J.; Farran, A.; Montell, E.; Verges, J.; Pelletier, J.P. Discrepancies in composition and biological effects of different formulations of chondroitin sulfate. Molecules 2015, 20, 4277–4289. [Google Scholar] [CrossRef] [PubMed]
- Pecly, I.M.D.; Melo, N.M.; Mourao, P.A.S. Effects of molecular size and chemical structure on renal and hepatic removal of exogenously administered chondroitin sulfate in rats. Biochim. Biophys. Acta Gen. Subj. 2006, 1760, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Volpi, N. About oral absorption and human pharmacokinetics of chondroitin sulfate. Osteoarthritis Cartilage 2010, 18, 1104–1105. [Google Scholar] [CrossRef] [Green Version]
- Williams, A.; He, W.Q.; Cress, B.F.; Liu, X.Y.; Alexandria, J.; Yoshizawa, H.; Nishimura, K.; Toida, T.; Koffas, M.; Linhardt, R.J. Cloning and expression of recombinant chondroitinase acii and its comparison to the arthrobacter aurescens enzyme. Biotechnol. J. 2017, 12, 1700239. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, Y.; Yuan, Q. Expression, purification and thermostability of MBP-chondroitinase ABC I from Proteus vulgaris. Int. J. Biol. Macromol. 2015, 72, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Z.; Zhou, Z.; Yuan, Q. Expression, purification and characterization of GAPDH-ChSase ABC I from Proteus vulgaris in Escherichia coli. Protein Expr. Purif. 2016, 128, 36–41. [Google Scholar] [CrossRef]
- Wang, J.P.; Zhang, L.; Jin, Z.Y. Separation and purification of low-molecular-weight chondroitin sulfates and their anti-oxidant properties. Bangladesh J. Pharmacol. 2016, 11, S61–S67. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Jiang, Z.; Chang, J.; Han, B.; Liu, W.; Peng, Y. Purification, characterization of Chondroitinase ABC from Sphingomonas paucimobilis and in vitro cardiocytoprotection of the enzymatically degraded CS-A. Int. J. Biol. Macromol. 2018, 115, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, A.A.; Sucupira, I.D.; Guedes, A.L.; Queiroz, I.N.; Frattani, F.S.; Fonseca, R.J.; Pomin, V.H. Anticoagulant and antithrombotic properties of three structurally correlated sea urchin sulfated glycans and their low-molecular-weight derivatives. Mar. Drugs 2018, 16, 304. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wu, M.; Xiao, C.; Yang, L.; Zhou, L.; Gaoa, N.; Li, Z.; Chen, J.; Chen, J.; Liu, J.; et al. Discovery of an intrinsic tenase complex inhibitor: Pure nonasaccharide from fucosylated glycosaminoglycan. Proc. Natl. Acad. Sci. USA 2015, 112, 8284–8289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Wang, J.; Hu, S.; Wang, Y.; Xue, C.; Li, H. The effects of fucosylated chondroitin sulfate isolated from Isostichopus badionotus on antimetastatic activity via down-regulation of Hif-1α and Hpa. Food Technol. Biotechnol. 2014, 23, 1643–1651. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, D.; Wang, S.; Tao, L.; Wang, A.; Chen, W.; Zhu, Z.; Zheng, S.; Gao, X.; Lu, Y. Holothurian glycosaminoglycan inhibits metastasis and thrombosis via targeting of nuclear factor-κB/tissue factor/Factor Xa pathway in melanoma B16F10 cells. PLoS ONE 2013, 8, e56557. [Google Scholar] [CrossRef]
- Li, J.; Li, S.; Yan, L.; Ding, T.; Linhardt, R.J.; Yu, Y.; Liu, X.; Liu, D.; Ye, X.; Chen, S. Fucosylated chondroitin sulfate oligosaccharides exert anticoagulant activity by targeting at intrinsic tenase complex with low FXII activation: Importance of sulfation pattern and molecular size. Eur. J. Med. Chem. 2017, 139, 191–200. [Google Scholar] [CrossRef]
- Wu, N.; Zhang, Y.; Ye, X.; Hu, Y.; Ding, T.; Chen, S. Sulfation pattern of fucose branches affects the anti-hyperlipidemic activities of fucosylated chondroitin sulfate. Carbohyd. Polym. 2016, 147, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Li, G.; Wu, N.; Guo, X.; Liao, N.; Ye, X.; Liu, D.; Xue, C.; Chai, W. Sulfation pattern of the fucose branch is important for the anticoagulant and antithrombotic activities of fucosylated chondroitin sulfates. BBA Gen. Subjects 2013, 1830, 3054–3066. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, J.P.; Walke, E.N. Depolymerized holothurian glycosaminoglycan and heparin inhibit the intrinsic tenase complex by a common antithrombin-independent mechanism. Blood 2006, 107, 3876–3882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solakyildirim, K.; Zhang, Z.; Linhardt, R.J. Ultraperformance liquid chromatography with electrospray ionization ion trap mass spectrometry for chondroitin disaccharide analysis. Anal. Biochem. 2010, 397, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.N.; Wang, Q.B.; Wang, S.M.; Wang, W.S.; Jiao, R.M.; Han, W.J.; Li, F.C. A chondroitin sulfate and hyaluronic acid lyase with poor activity to glucuronyl 4,6-O-disulfated N-acetylgalactosamine (E-type)-containing structures. J. Biol. Chem. 2018, 293, 4230–4243. [Google Scholar] [CrossRef] [PubMed]
- Mizumoto, S.; Murakoshi, S.; Kalayanamitra, K.; Deepa, S.S.; Fukui, S.; Kongtawelert, P.; Yamada, S.; Sugahara, K. Highly sulfated hexasaccharide sequences isolated from chondroitin sulfate of shark fin cartilage: Insights into the sugar sequences with bioactivities. Glycobiology 2013, 23, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, Y.; Higashi, K.; Linhardt, R.J.; Toida, T. Comprehensive analysis of glycosaminoglycans from the edible shellfish. Carbohyd. Polym. 2018, 184, 269–276. [Google Scholar] [CrossRef]
- Benito-Arenas, R.; Doncel-Pérez, E.; Fernández-Gutiérrez, M.; Garrido, L.; García-Junceda, E.; Revuelta, J.; Bastida, A.; Fernández-Mayoralas, A. A holistic approach to unravelling chondroitin sulfation: Correlations between surface charge, structure and binding to growth factors. Carbohyd. Polym. 2018, 202, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Doncel-Pérez, E.; Aranaz, I.; Bastida, A.; Revuelta, J.; Camacho, C.; Acosta, N.; Garrido, L.; Civera, C.; García-Junceda, E.; Heras, A.; et al. Synthesis, physicochemical characterization and biological evaluation of chitosan sulfate as heparan sulfate mimics. Carbohyd. Polym. 2018, 191, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Pudełko, A.; Wisowski, G.; Olczyk, K.; Koźma, E.M. The dual role of the glycosaminoglycan chondroitin-6-sulfate in the development, progression and metastasis of cancer. FEBS J. 2019. [Google Scholar] [CrossRef]








| Physiological Function | Reference |
|---|---|
| Cell–cell/cell–matrix interactions | [6] |
| Immune modulation | [7,8] |
| Host–pathogen interactions | [9,10] |
| Anticoagulant activities | [11,12] |
| Potential Therapeutic Application | |
| Anti-inflammatory | [13,14] |
| Antiviral | [15,16] |
| Antimalarial vaccine | [17,18] |
| Anticancer | [19,20] |
| Antiparasitic | [21] |
| Biomarker | [22] |
| Liver regeneration | [23] |
| Repair of the central nervous system | [24,25] |
| Neuroprotective | [26] |
| Wound healing | [27] |
| Glycosaminoglycan Conjugate | Type of Application | Target Tissue/Application Method | Reference |
|---|---|---|---|
| Biotinylated Hyaluronic Acid(HA)CS | Material coating | In vitro biosensor | [29] |
| Collagen/CS | Implant coating | Osseointegration | [30,31] |
| Gelatin methaclylate/CS methacrylate | Hydrogel | Cartilage regeneration | [32] |
| Cross-linked thiolated HA, CS, Heparine, gelatin | Biodegradable hydrogel | Drug/Growth factor (GF) delivery, tissue regeneration | [33,34,35] |
| Cross-linked CS–tyramine | Hydrogel | Drug delivery | [36,37] |
| Chemically cross-linked HA/CS | Hydrogel matrix/particles | Wound dressing, skin regeneration | [38,39,40,41] |
| Photochemically cross-linked HA/CS | Hydrogel | Cell encapsulation, cartilage repair | [42] |
| Collagen-CS | Porous scaffold | Neovascularization, tissue (bone) regeneration | [43,44] |
| Genipin cross-linked HA, CS | Porous scaffold | Cartilage regeneration | [45] |
| Carbodiimide-cross-linked HA, CS, Dermatan sulfate (DS), Chitosan gelatin | Porous scaffold | Cartilage regeneration | [46,47] |
| Electrospun collagen–CS | Porous mesh | Artificial extracellular matrix(ECM), cartilage regeneration | [48,49,50] |
| Chitosan–CS | Nanoparticles | GF delivery, bone regeneration | [51] |
| Lyase | Substrate | PDB | Species | Catalytic tetrad | Ref. |
|---|---|---|---|---|---|
| CSase ABC II (exo) CSase ABC II (exo) CSase ABC I (endo) | CS, DS, HA Tetra-CS, Tetra-DS CS, DS, HA | 2Q1F 1HNO | Bacteroides thetaiotaomicron Proteus vulgaris Proteus vulgaris | Glu628–Tyr461–His454– Arg514 His453–Tyr 460 His501–Tyr508–Arg560– Glu653 | [70,71,72,73,74] |
| CSase AC II (Exo) CSase AC I (Endo) CSase ACY253A | CS, HA CS, HA | 1RWF;1RWG;1RWH;1RW9;1RWA;1RWC 2WA; 2XO3 1CB8 1HMW;1HM2;1HM3;1HMU | Arthrobacter aurescens Streptomyces coelicor A3 Flavobacterium heparinum Pedobacter heparinus | His225–Tyr234–Arg288–Glu371 | [75,76,77,78] |
| CSase B (Endo) | DS | 1DBO;1DBG;1OFL;1OFM | Flavobacterium heparinum | Lys250–Arg271–His272 | [79,80] |
| HAase | HA, CS | 1OJO;1OJM; 1OJN;1OJP | Streptococcus pneumoniae | [81] |
| SAMPLE a | MOLECULAR WEIGHT (Da) | ||||
|---|---|---|---|---|---|
| Natural | Depolymerized CS/Low Molecular Weight of CS | ||||
| HCl/H20/60 °C | NaOH/MAOS b/60 °C | Enzymatic | H2O2(30%) | ||
| CS-B | 18.150 | 1.826 | 8.085 | 2.583 | 1.561 |
| CS-S | 31.300 | 1.217 | 4.269 | 1.994 | 1.511 |
| CS-P | 10.070 | 2.690 | 3.663 | 2.920 | 2.191 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benito-Arenas, R.; Zárate, S.G.; Revuelta, J.; Bastida, A. Chondroitin Sulfate-Degrading Enzymes as Tools for the Development of New Pharmaceuticals. Catalysts 2019, 9, 322. https://doi.org/10.3390/catal9040322
Benito-Arenas R, Zárate SG, Revuelta J, Bastida A. Chondroitin Sulfate-Degrading Enzymes as Tools for the Development of New Pharmaceuticals. Catalysts. 2019; 9(4):322. https://doi.org/10.3390/catal9040322
Chicago/Turabian StyleBenito-Arenas, Raúl, Sandra G. Zárate, Julia Revuelta, and Agatha Bastida. 2019. "Chondroitin Sulfate-Degrading Enzymes as Tools for the Development of New Pharmaceuticals" Catalysts 9, no. 4: 322. https://doi.org/10.3390/catal9040322
APA StyleBenito-Arenas, R., Zárate, S. G., Revuelta, J., & Bastida, A. (2019). Chondroitin Sulfate-Degrading Enzymes as Tools for the Development of New Pharmaceuticals. Catalysts, 9(4), 322. https://doi.org/10.3390/catal9040322