Depicting the DNA Binding and Cytotoxicity Studies against Human Colorectal Cancer of Aquabis (1-Formyl-2-Naphtholato-k2O,O′) Copper(II): A Biophysical and Molecular Docking Perspective
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods and Instrumentation
2.3. Synthesis of Aquabis (1-Formyl-2-Naphtholato-k2O,O′) Copper(II) (1)
2.4. Crystallography
2.5. DNA Binding Studies
2.6. Cytotoxicity Studies
2.7. Molecular Docking
3. Results and Discussion
3.1. Synthesis and Characterization
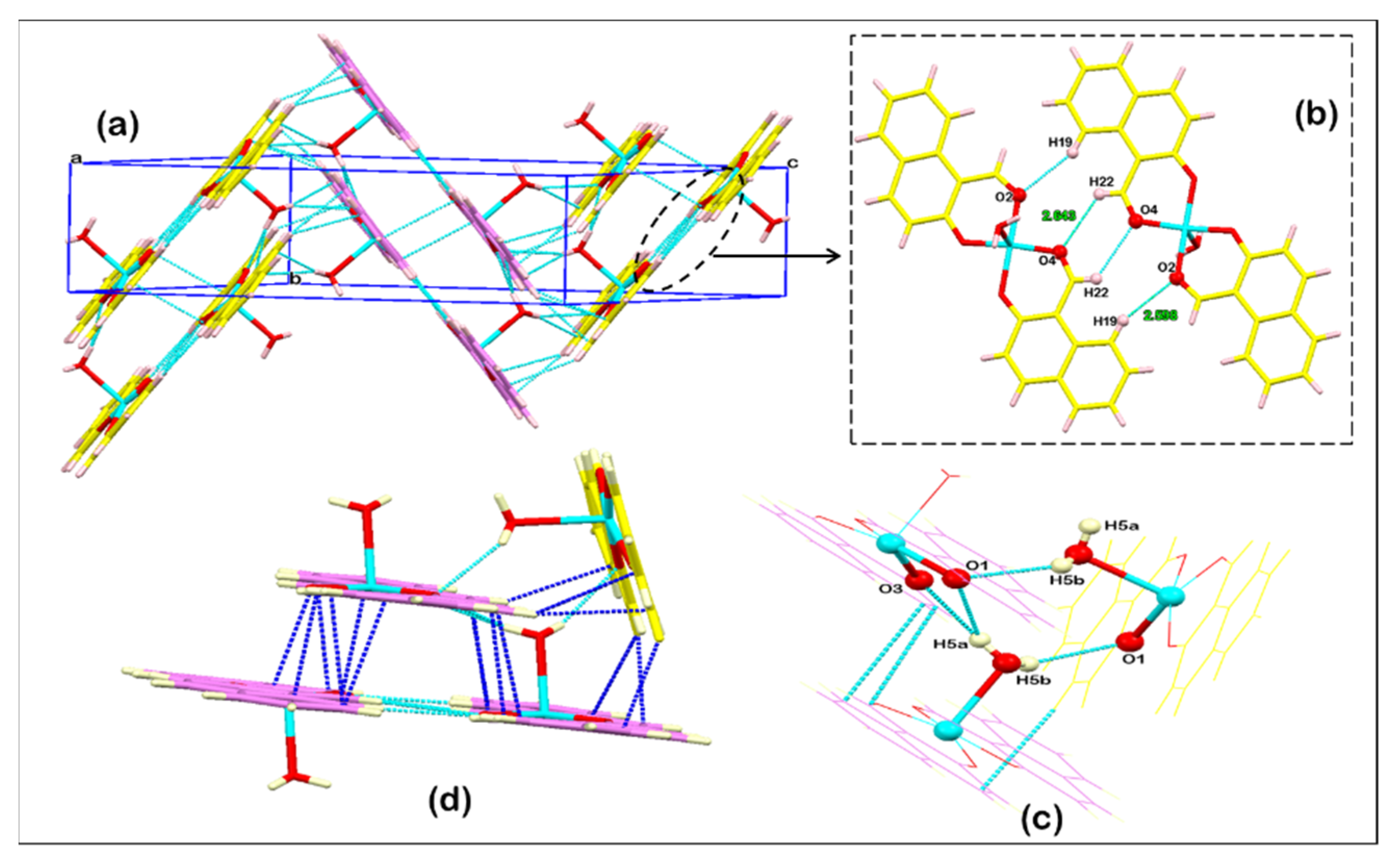
3.2. Structure Description
3.3. Hirshfeld Surface Analysis
3.4. DNA Binding
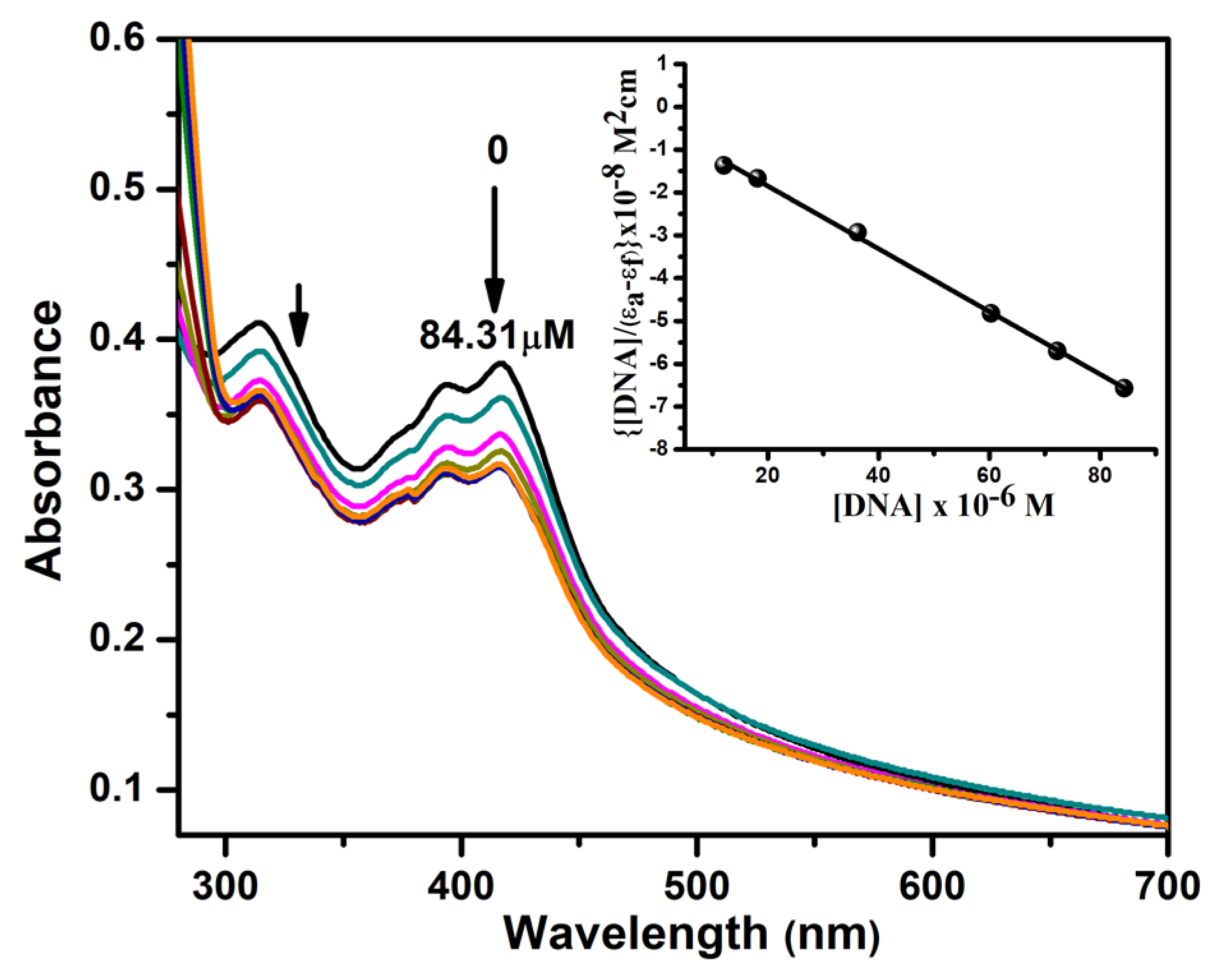
3.4.1. Absorption Titrations
3.4.2. Ethidium Bromide Displacement
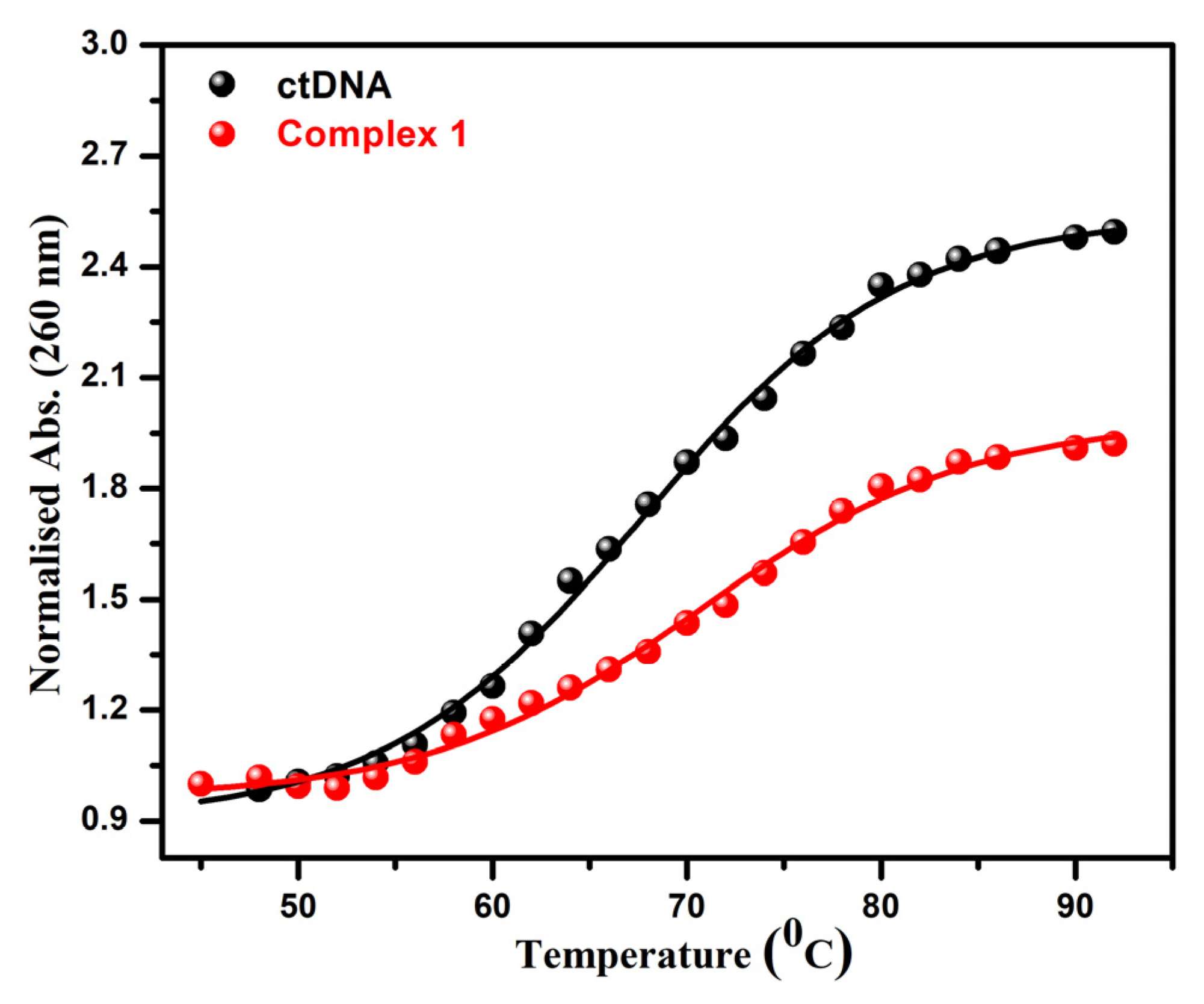
3.4.3. UV Optical Melting
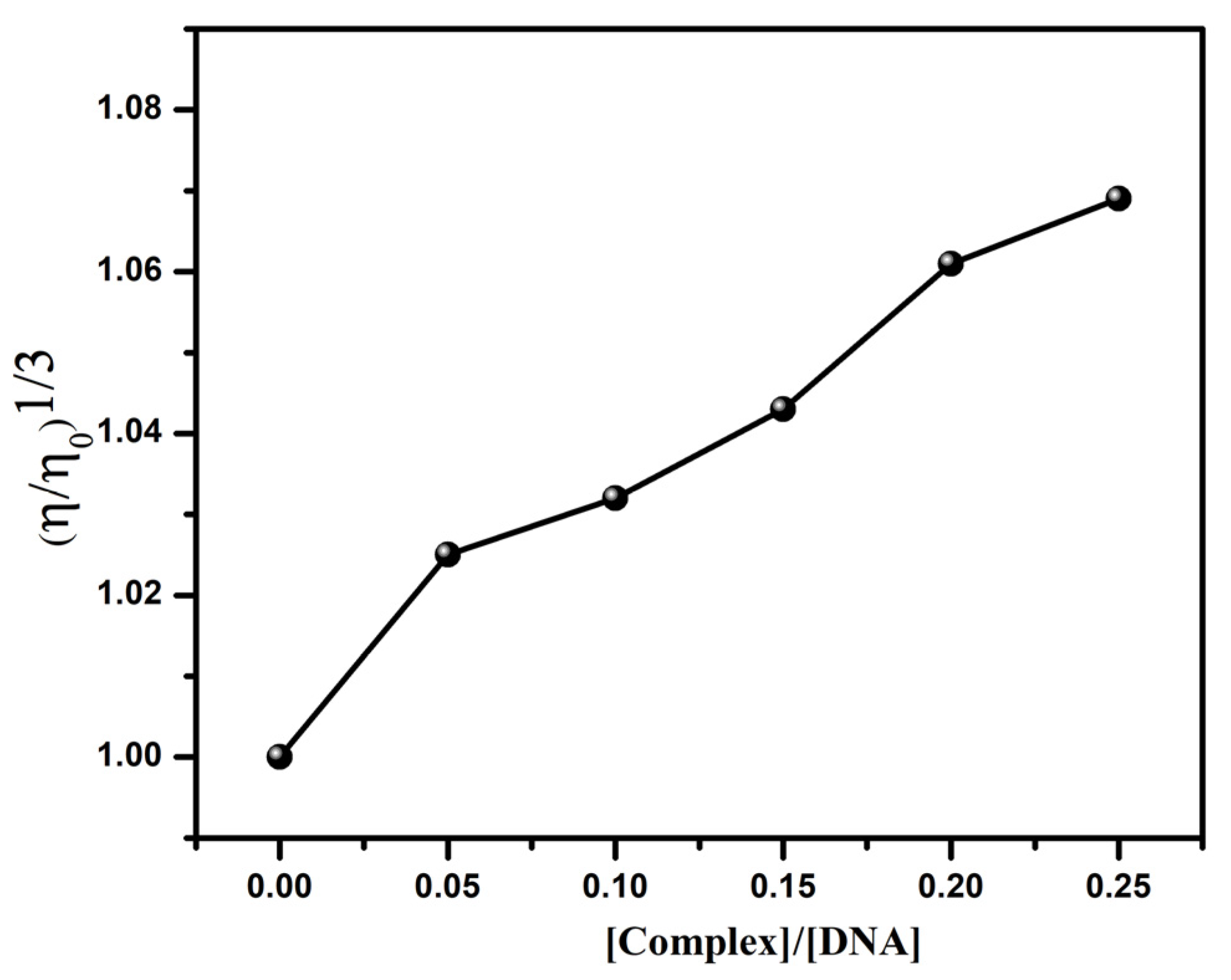
3.4.4. Viscosity Measurements
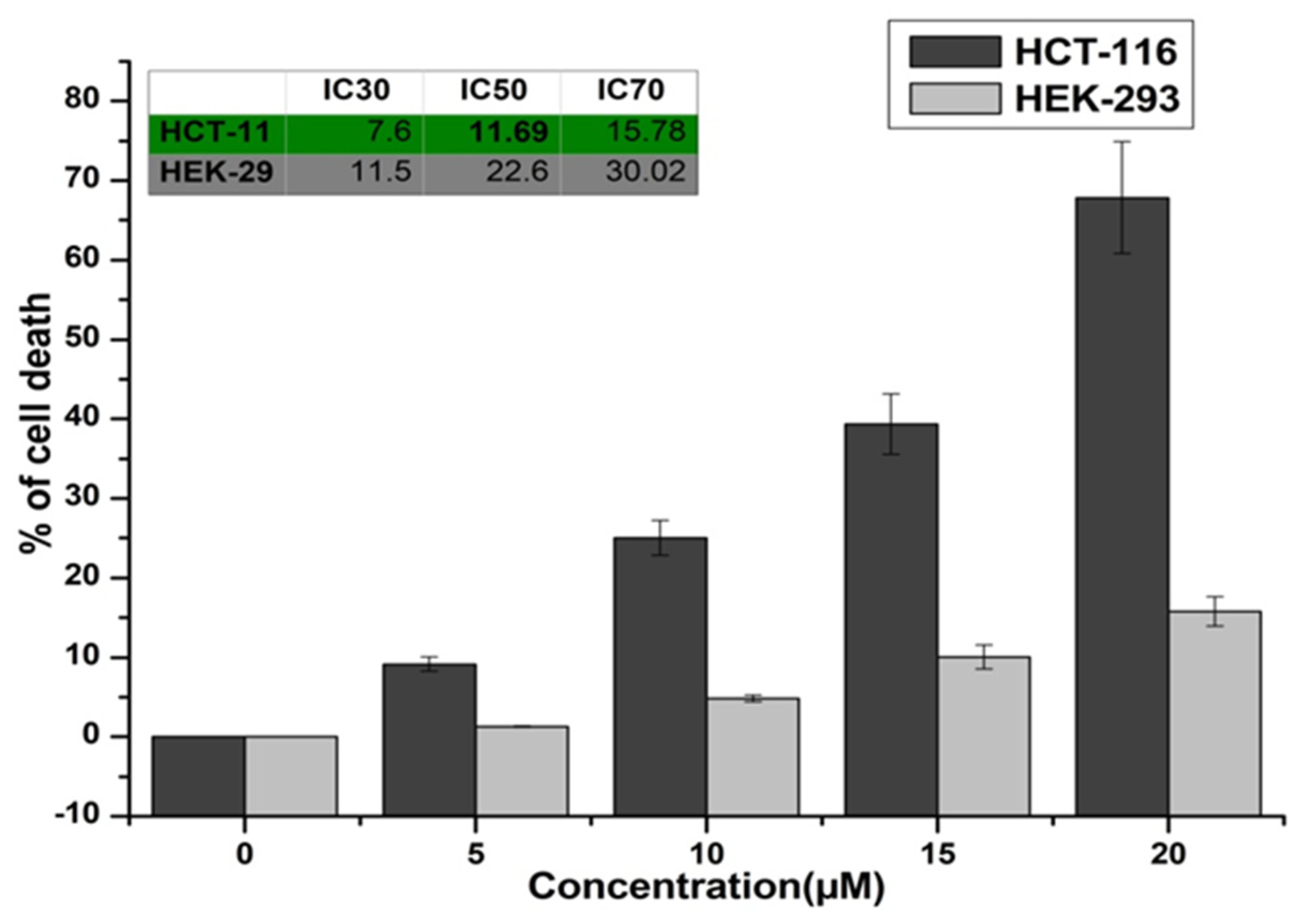
3.5. In Vitro Cytotoxicity
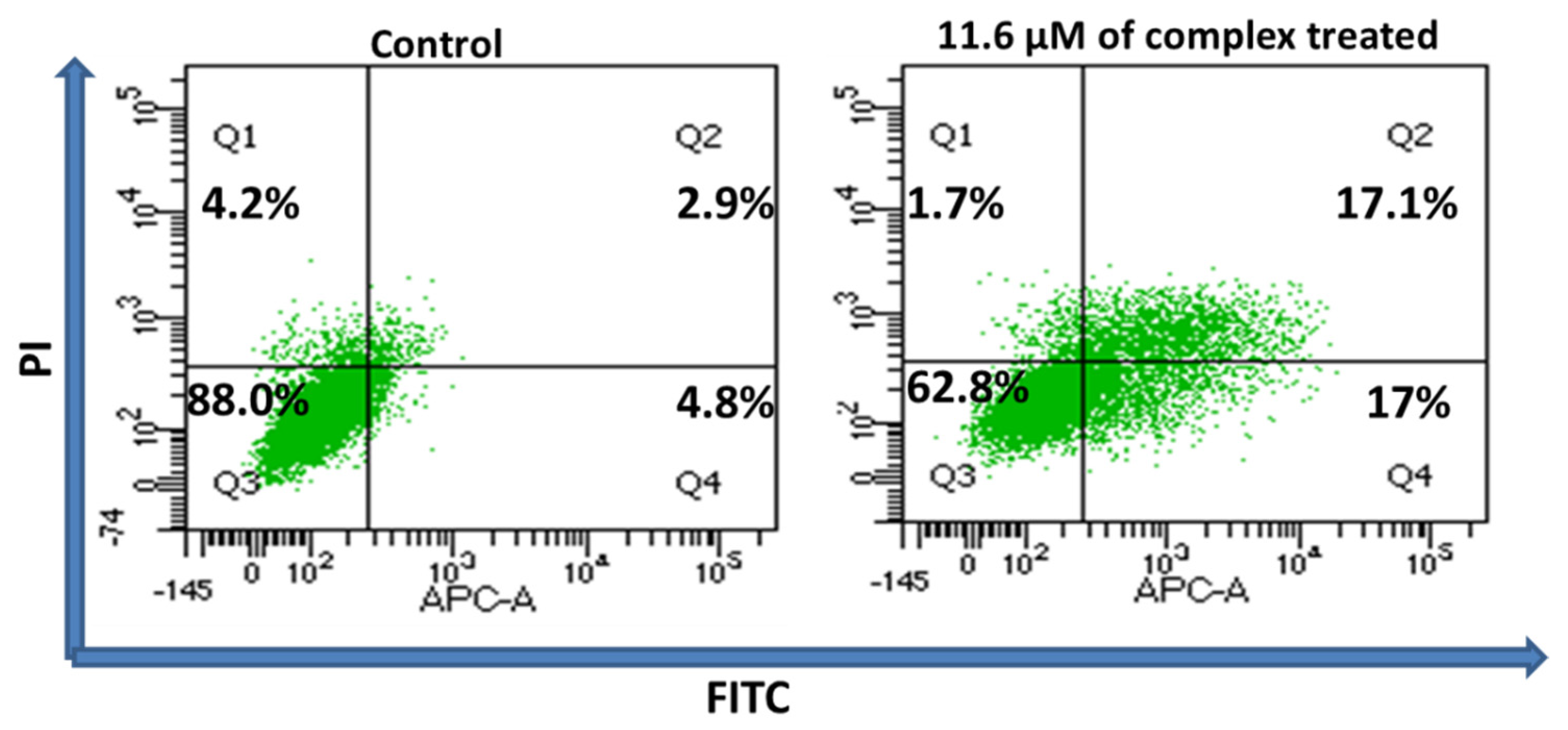
3.6. Quantification of Apoptosis and Necrosis Using Flow Cytometry
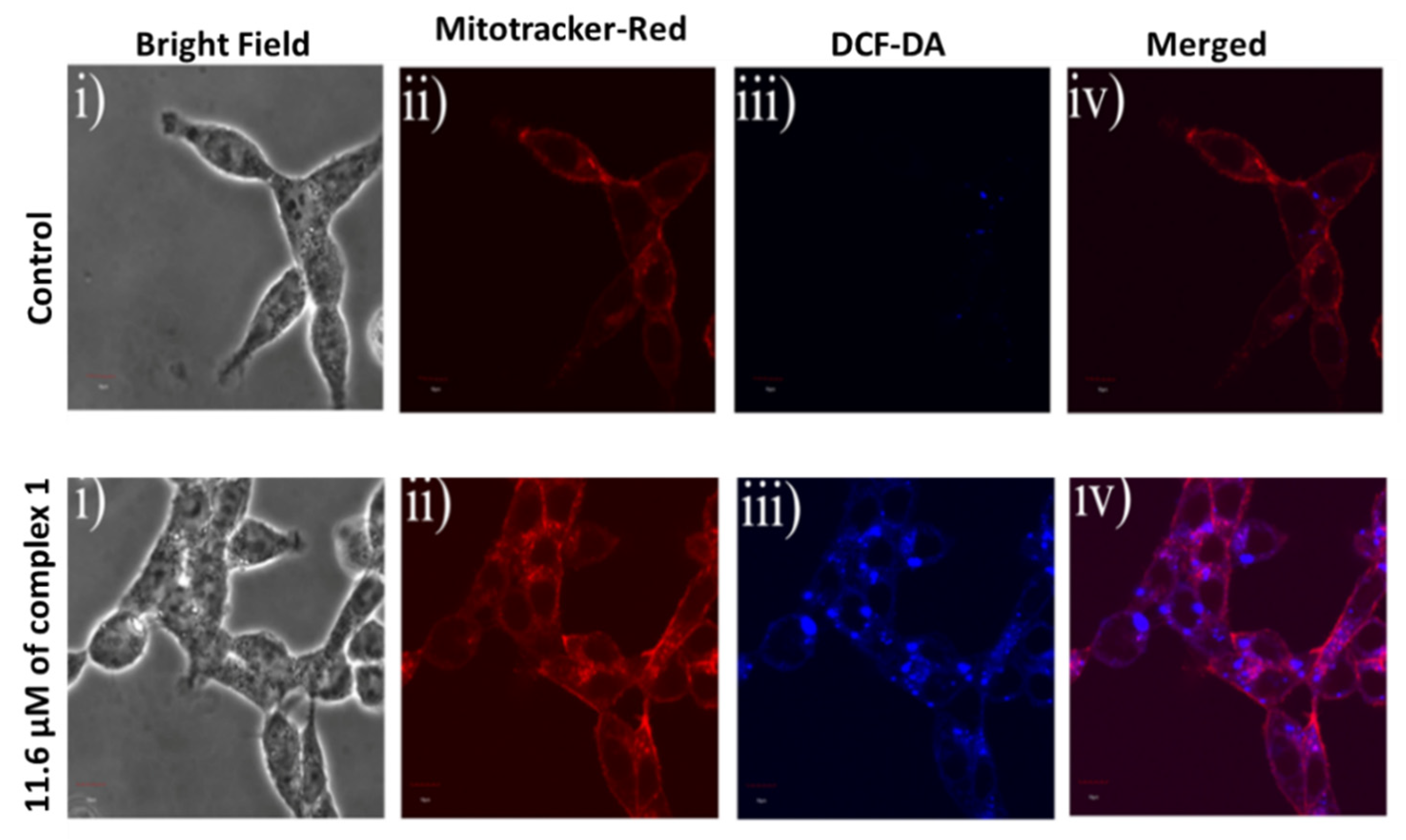
3.7. Quantification of Mitochondrial Reactive Oxygen Species (ROS)
3.8. DNA Fragmentation Assay
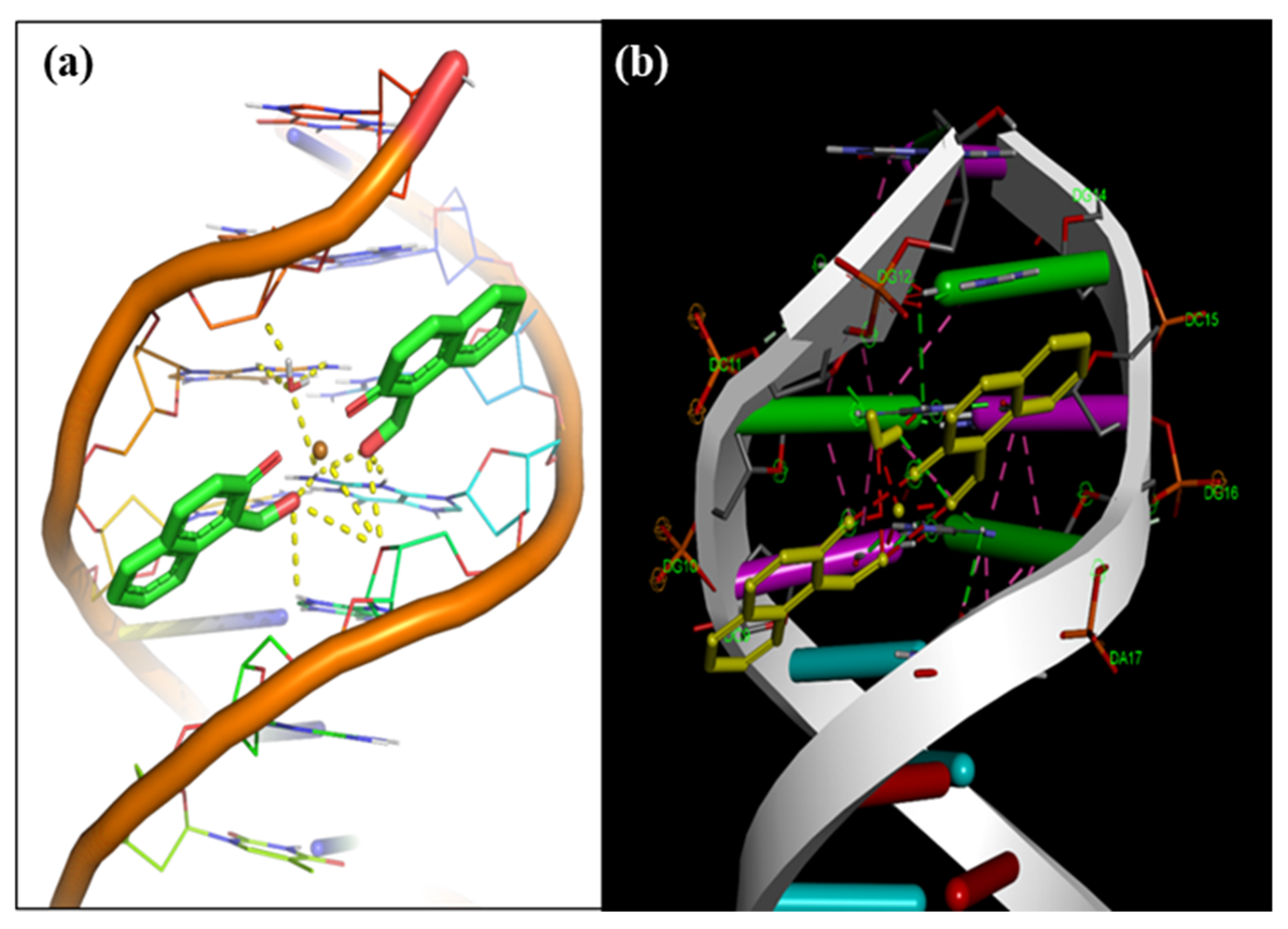
3.9. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Araghi, M.; Soerjomataram, I.; Jenkins, M.; Brierley, J.; Morris, E.; Bray, F.; Arnold, M. Global trends in colorectal cancer mortality: Projections to the year 2035. Int. J. Cancer 2019, 144, 2992–3000. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- González-Ballesteros, N.; Pérez-Álvarez, D.; Rodríguez-Argüelles, M.C.; Henriques, M.S.C.; Paixão, J.A.; Prado-López, S. Synthesis, spectral characterization and X-ray crystallographic study of new copper(I) complexes. Antitumor activity in colon cancer. Polyhedron 2016, 119, 112–119. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Sun, Z.; Cui, Y.; Zhang, H.; Zhang, S.; Wang, X.; Liu, S.; Gao, Q. Oxaliplatin derived monofunctional triazole-containing platinum(II) complex counteracts oxaliplatin-induced drug resistance in colorectal cancer. Bioorg. Chem. 2021, 107, 104636. [Google Scholar] [CrossRef] [PubMed]
- Golshani, G.; Zhang, Y. Advances in immunotherapy for colorectal cancer: A review. Therap. Adv. Gastroenterol. 2020, 13, 175628482091752. [Google Scholar] [CrossRef]
- Wahab, S.; Alshahrani, M.Y.; Ahmad, M.F.; Abbas, H. Current trends and future perspectives of nanomedicine for the management of colon cancer. Eur. J. Pharmacol. 2021, 910, 174464. [Google Scholar] [CrossRef]
- Park, J.; Baik, H.; Kang, S.H.; Seo, S.H.; Kim, K.H.; Oh, M.K.; Lee, H.S.; Lee, S.H.; Kim, K.H.; An, M.S. Comparison between oxaliplatin therapy and capecitabine monotherapy for high-risk stage II–III elderly colon cancer patients. Asian J. Surg. 2021. [Google Scholar] [CrossRef] [PubMed]
- Rottenberg, S.; Disler, C.; Perego, P. The rediscovery of platinum-based cancer therapy. Nat. Rev. Cancer 2021, 21, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.; Das, S.; Das, B.; Roy, P. Intercalative DNA binding, protein binding, antibacterial activities and cytotoxicity studies of a mononuclear copper(II) complex. Inorganica Chim. Acta 2021, 514, 119961. [Google Scholar] [CrossRef]
- Bhattacharjee, M.; Boruah, S.R.; Dutta Purkayastha, R.N.; Ganguly, R.; Maiti, D.; Franconetti, A.; Frontera, A.; Kirillov, A.M.; Chowdhury, S.; Roy, S.; et al. Synthesis, characterization, DNA binding ability, in vitro cytotoxicity, electrochemical properties and theoretical studies of copper(II) carboxylate complexes. Inorganica Chim. Acta 2021, 518, 120235. [Google Scholar] [CrossRef]
- Liccardi, G.; Hartley, J.A.; Hochhauser, D. EGFR Nuclear Translocation Modulates DNA Repair following Cisplatin and Ionizing Radiation Treatment. Cancer Res. 2011, 71, 1103–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharathi, S.; Mahendiran, D.; Kumar, R.S.; Kim, Y.G.; Gajendiran, M.; Kim, K.; Rahiman, A.K. Biocompatibility, in Vitro Antiproliferative, and in Silico EGFR/VEGFR2 Studies of Heteroleptic Metal(II) Complexes of Thiosemicarbazones and Naproxen. Chem. Res. Toxicol. 2019, 32, 1554–1571. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Tatsuishi, H.; Banno, T.; Fujie, T.; Yamamoto, C.; Naka, H.; Kaji, T. Copper(II) Bis(diethyldithiocarbamate) Induces the Expression of Syndecan-4, a Transmembrane Heparan Sulfate Proteoglycan, via p38 MAPK Activation in Vascular Endothelial Cells. Int. J. Mol. Sci. 2018, 19, 3302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, C.H.; Kong, S.M.; Tiong, Y.L.; Maah, M.J.; Sukram, N.; Ahmad, M.; Khoo, A.S.B. Selective anticancer copper(II)-mixed ligand complexes: Targeting of ROS and proteasomes. Metallomics 2014, 6, 892–906. [Google Scholar] [CrossRef]
- Annaraj, B.; Balakrishnan, C.; Neelakantan, M.A. Synthesis, structure information, DNA/BSA binding affinity and in vitro cytotoxic studies of mixed ligand copper(II) complexes containing a phenylalanine derivative and diimine co-ligands. J. Photochem. Photobiol. B Biol. 2016, 160, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Baskaran, S.; Krishnan, M.M.; Arumugham, M.N.; Kumar, R. Synthesis, crystal structure, DNA interaction, DFT analysis and molecular docking studies of copper(II) complexes with 1-methyl-l-tryptophan and phenanthroline units. J. Mol. Struct. 2021, 1224, 129236. [Google Scholar] [CrossRef]
- Shanbhag, V.C.; Gudekar, N.; Jasmer, K.; Papageorgiou, C.; Singh, K.; Petris, M.J. Copper metabolism as a unique vulnerability in cancer. Biochim. Biophys. Acta-Mol. Cell Res. 2021, 1868, 118893. [Google Scholar] [CrossRef]
- Denoyer, D.; Masaldan, S.; La Fontaine, S.; Cater, M.A. Targeting copper in cancer therapy: ‘Copper That Cancer. ’ Metallomics 2015, 7, 1459–1476. [Google Scholar] [CrossRef]
- Zarei, L.; Asadi, Z.; Dusek, M.; Eigner, V. Homodinuclear Ni(II) and Cu(II) Schiff base complexes derived from O-vanillin with a pyrazole bridge: Preparation, crystal structures, DNA and protein (BSA) binding, DNA cleavage, molecular docking and cytotoxicity study. J. Photochem. Photobiol. A Chem. 2019, 374, 145–160. [Google Scholar] [CrossRef]
- Paul, A.; Mistri, S.; Bertolasi, V.; Manna, S.C. DNA/protein binding and molecular docking studies of two tetranuclear Cu(II) complexes with double-open-cubane core like structure. Inorganica Chim. Acta 2019, 495, 119005. [Google Scholar] [CrossRef]
- Liu, H.-K.; Sadler, P.J. Metal Complexes as DNA Intercalators. Acc. Chem. Res. 2011, 44, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Mohideen, M.; Kamaruzaman, N.A.; Hamali, M.A.; Mordi, M.N.; Mansor, S.M.; Rahman, A.F.M.M. N,N-Bis(Substituted benzyl)-β-Carbolineum Bromides as Potential Anticancer Therapeutics: Design, Synthesis, Cytotoxicity, Drug-DNA Intercalation and In-Silico Binding Properties. J. Mol. Struct. 2021, 1243, 130771. [Google Scholar] [CrossRef]
- Brissos, R.F.; Torrents, E.; Mariana dos Santos Mello, F.; Carvalho Pires, W.; de Paula Silveira-Lacerda, E.; Caballero, A.B.; Caubet, A.; Massera, C.; Roubeau, O.; Teat, S.J.; et al. Highly cytotoxic DNA-interacting copper(II) coordination compounds. Metallomics 2014, 6, 1853–1868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigamonti, L.; Reginato, F.; Ferrari, E.; Pigani, L.; Gigli, L.; Demitri, N.; Kopel, P.; Tesarova, B.; Heger, Z. From solid state to in vitro anticancer activity of copper(II) compounds with electronically-modulated NNO Schiff base ligands. Dalt. Trans. 2020, 49, 14626–14639. [Google Scholar] [CrossRef]
- Mondal, S.S.; Jaiswal, N.; Bera, P.S.; Tiwari, R.K.; Behera, J.N.; Chanda, N.; Ghosal, S.; Saha, T.K. Cu(II) and Co(II/III) complexes of N,O-chelated Schiff base ligands: DNA interaction, protein binding, cytotoxicity, cell death mechanism and reactive oxygen species generation studies. Appl. Organomet. Chem. 2021, 35. [Google Scholar] [CrossRef]
- Zianna, A.; Psomas, G.; Hatzidimitriou, A.; Lalia-Kantouri, M. Copper(II) complexes of salicylaldehydes and 2-hydroxyphenones: Synthesis, structure, thermal decomposition study and interaction with calf-thymus DNA and albumins. RSC Adv. 2015, 5, 37495–37511. [Google Scholar] [CrossRef]
- Tabassum, S.; Afzal, M.; Arjmand, F. New heterobimetallic complex as potential topoisomerase I inhibitor: In vitro DNA binding, cleavage and cytotoxicity against human cancer cell lines. J. Photochem. Photobiol. B Biol. 2012, 115, 63–72. [Google Scholar] [CrossRef]
- Festa, R.A.; Thiele, D.J. Copper: An essential metal in biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Li, H.; Wang, X.; Liu, Q.; Ni, J.; Sun, H. Kinetics and thermodynamics of metal binding to the N-terminus of a human copper transporter, hCTR1. Chem. Commun. 2013, 49, 9134. [Google Scholar] [CrossRef]
- Ivy, K.D.; Kaplan, J.H. A Re-Evaluation of the Role of hCTR1, the Human High-Affinity Copper Transporter, in Platinum-Drug Entry into Human Cells. Mol. Pharmacol. 2013, 83, 1237–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Lutsenko, S. Human copper transporters: Mechanism, role in human diseases and therapeutic potential. Future Med. Chem. 2009, 1, 1125–1142. [Google Scholar] [CrossRef] [Green Version]
- Holden, L.; Burke, C.S.; Cullinane, D.; Keyes, T.E. Strategies to promote permeation and vectorization, and reduce cytotoxicity of metal complex luminophores for bioimaging and intracellular sensing. RSC Chem. Biol. 2021, 2, 1021–1049. [Google Scholar] [CrossRef]
- Zhang, Z.; Buffle, J.; van Leeuwen, H.P.; Wojciechowski, K. Roles of Metal Ion Complexation and Membrane Permeability in the Metal Flux through Lipophilic Membranes. Labile Complexes at Permeation Liquid Membranes. Anal. Chem. 2006, 78, 5693–5703. [Google Scholar] [CrossRef]
- Liu, C. DNA damage by copper(II) complexes: Coordination-structural dependence of reactivities. J. Inorg. Biochem. 1999, 75, 233–240. [Google Scholar] [CrossRef]
- De Medeiros, W.M.T.Q.; de Medeiros, M.J.C.; Carvalho, E.M.; de Lima, J.A.; da S. Oliveira, V.; de B. Pontes, A.C.F.; da Silva, F.O.N.; Ellena, J.A.; de O. Rocha, H.A.; de Sousa, E.H.S.; et al. A vanillin-based copper(II) metal complex with a DNA-mediated apoptotic activity. RSC Adv. 2018, 8, 16873–16886. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Chowdhury, S.; Ghorai, A.; Ghosh, U.; Stoeckli-Evans, H. Synthesis, structure, interaction with DNA and cytotoxicity of a luminescent copper(II) complex with a hydrazone ligand. Polyhedron 2013, 51, 228–234. [Google Scholar] [CrossRef]
- Khan, M.H.; Cai, M.; Deng, J.; Yu, P.; Liang, H.; Yang, F. Anticancer Function and ROS-Mediated Multi-Targeting Anticancer Mechanisms of Copper(II) 2-hydroxy-1-naphthaldehyde Complexes. Molecules 2019, 24, 2544. [Google Scholar] [CrossRef] [Green Version]
- Abbasi, Z.; Salehi, M.; Khaleghian, A.; Kubicki, M. In vitro cytotoxic activity of a novel Schiff base ligand derived from 2-hydroxy-1-naphthaldehyde and its mononuclear metal complexes. J. Mol. Struct. 2018, 1173, 213–220. [Google Scholar] [CrossRef]
- Li, J.; Dong, J.; Cui, H.; Xu, T.; Li, L. A copper(II) complex of the Schiff base from l-valine and 2-hydroxy-1-naphthalidene plus 1,10-phenanthroline: Synthesis, crystal structure, and DNA interaction. Transit. Met. Chem. 2012, 37, 175–182. [Google Scholar] [CrossRef]
- Nagesh, G.Y.; Mruthyunjayaswamy, B.H.M. Synthesis, characterization and biological relevance of some metal(II) complexes with oxygen, nitrogen and oxygen (ONO) donor Schiff base ligand derived from thiazole and 2-hydroxy-1-naphthaldehyde. J. Mol. Struct. 2015, 1085, 198–206. [Google Scholar] [CrossRef]
- Hajrezaie, M.; Paydar, M.; Zorofchian Moghadamtousi, S.; Hassandarvish, P.; Gwaram, N.S.; Zahedifard, M.; Rouhollahi, E.; Karimian, H.; Looi, C.Y.; Ali, H.M.; et al. A Schiff Base-Derived Copper(II) Complex Is a Potent Inducer of Apoptosis in Colon Cancer Cells by Activating the Intrinsic Pathway. Sci. World J. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Yu, Q.; Li, C.-Y.; Yang, X.-E.; He-Dong, B.; Liang, H. Aquabis(1-formyl-2-naphtholato-κ2 O,O′)copper(II). Acta Crystallogr. Sect. E Struct. Reports Online 2006, 62, m391–m393. [Google Scholar] [CrossRef]
- Afzal, M.; Alarifi, A.; Hasnain, M.S.; Muddassir, M. Elucidation of DNA binding interaction of new Cu(II)/Zn(II) complexes derived from Schiff base and L-tryptophan amino acid: A multispectroscopic and molecular docking approach. Environ. Sci. Pollut. Res. 2021, 28, 44039–44050. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.; Shimer, G.H.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Qais, F.A.; Alam, M.M.; Naseem, I.; Ahmad, I. Understanding the mechanism of non-enzymatic glycation inhibition by cinnamic acid: An in vitro interaction and molecular modelling study. RSC Adv. 2016, 6, 65322–65337. [Google Scholar] [CrossRef]
- Pandya, P.; Agarwal, L.K.; Gupta, N.; Pal, S. Molecular recognition pattern of cytotoxic alkaloid vinblastine with multiple targets. J. Mol. Graph. Model. 2014, 54, 1–9. [Google Scholar] [CrossRef]
- Ebead, Y.H.; Salman, H.M.A.; Shebany, Y.M.; Abdellah, M.A. Synthesis, Spectroscopic, and Biological Investigations of the Ternary Metal Complexes of Schiff Bases Derived From 2-hydroxy-1-naphthaldehyde and Some Amino Acids. Synth. React. Inorganic, Met. Nano-Metal Chem. 2016, 46, 1819–1828. [Google Scholar] [CrossRef]
- Murukan, B.; Mohanan, K. Synthesis, Characterization, Electrochemical Properties and Antibacterial Activity of Some Transiton Metal Complexes with [(2-hydroxy-1-naphthaldehyde)-3-isatin]-bishydrazone. Transit. Met. Chem. 2006, 31, 441–446. [Google Scholar] [CrossRef]
- Shi, S.; Yu, S.; Quan, L.; Mansoor, M.; Chen, Z.; Hu, H.; Liu, D.; Liang, Y.; Liang, F. Synthesis and antitumor activities of transition metal complexes of a bis-Schiff base of 2-hydroxy-1-naphthalenecarboxaldehyde. J. Inorg. Biochem. 2020, 210, 111173. [Google Scholar] [CrossRef]
- Niu, M.; Cao, Z.; Xue, R.; Wang, S.; Dou, J.; Wang, D. Structural diversity of Cu(II) compounds of Schiff bases derived from 2-hydroxy-1-naphthaldehyde and a series of aminobenzoic acid. J. Mol. Struct. 2011, 996, 101–109. [Google Scholar] [CrossRef]
- Efthimiadou, E.K.; Thomadaki, H.; Sanakis, Y.; Raptopoulou, C.P.; Katsaros, N.; Scorilas, A.; Karaliota, A.; Psomas, G. Structure and biological properties of the copper(II) complex with the quinolone antibacterial drug N-propyl-norfloxacin and 2,2′-bipyridine. J. Inorg. Biochem. 2007, 101, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Serezhkin, V.N.; Vologzhanina, A.V.; Serezhkina, L.B.; Smirnova, E.S.; Grachova, E.V.; Ostrova, P.V.; Antipin, M.Y. Crystallochemical formula as a tool for describing metal–ligand complexes – a pyridine-2,6-dicarboxylate example. Acta Crystallogr. Sect. B Struct. Sci. 2009, 65, 45–53. [Google Scholar] [CrossRef]
- Alexandrov, E.V.; Shevchenko, A.P.; Asiri, A.A.; Blatov, V.A. New knowledge and tools for crystal design: Local coordination versus overall network topology and much more. CrystEngComm 2015, 17, 2913–2924. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Spackman, M.A.; Mitchell, A.S. Novel tools for visualizing and exploring intermolecular interactions in molecular crystals. Acta Crystallogr. Sect. B Struct. Sci. 2004, 60, 627–668. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 3814–3816. [Google Scholar] [CrossRef]
- Patel, M.N.; Parmar, P.A.; Gandhi, D.S. Square pyramidal copper(II) complexes with forth generation fluoroquinolone and neutral bidentate ligand: Structure, antibacterial, SOD mimic and DNA-interaction studies. Bioorg. Med. Chem. 2010, 18, 1227–1235. [Google Scholar] [CrossRef]
- Wu, J.-Z.; Yuan, L. Synthesis and DNA interaction studies of a binuclear ruthenium(II) complex with 2,9-bis(2-imidazo[4,5- f ][1,10]phenanthroline)-1,10-phenanthroline as bridging and intercalating ligand. J. Inorg. Biochem. 2004, 98, 41–45. [Google Scholar] [CrossRef]
- Darabi, F.; Hadadzadeh, H.; Ebrahimi, M.; Khayamian, T.; Rudbari, H.A. The piroxicam complex of cobalt(II): Synthesis in two different ionic liquids, structure, DNA- and BSA interaction and molecular modeling. Inorganica Chim. Acta 2014, 409, 379–389. [Google Scholar] [CrossRef]
- Jyothi, P.; Koppu, S.; Sumalatha, V.; Ushaiah, B.; Gyana Kumari, C. Synthesis, characterisation, cytotoxicity, DNA binding and antimicrobial studies of binary and ternary metal complexes of Co(II). Inorg. Chem. Commun. 2019, 110, 107590. [Google Scholar] [CrossRef]
- Lincoln, P.; Broo, A.; Nordén, B. Diastereomeric DNA-Binding Geometries of Intercalated Ruthenium(II) Trischelates Probed by Linear Dichroism: [Ru(phen)2DPPZ]2+ and [Ru(phen)2BDPPZ]2+. J. Am. Chem. Soc. 1996, 118, 2644–2653. [Google Scholar] [CrossRef]
- Hirohama, T.; Kuranuki, Y.; Ebina, E.; Sugizaki, T.; Arii, H.; Chikira, M.; Tamil Selvi, P.; Palaniandavar, M. Copper(II) complexes of 1,10-phenanthroline-derived ligands: Studies on DNA binding properties and nuclease activity. J. Inorg. Biochem. 2005, 99, 1205–1219. [Google Scholar] [CrossRef]
- Qiu, L.; Lv, G.; Guo, L.; Chen, L.; Luo, S.; Zou, M.; Lin, J. Synthesis, crystal structure and antitumor effect of a novel copper(II) complex bearing zoledronic acid derivative. Eur. J. Med. Chem. 2015, 89, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Almes, F.J.; Porschke, D. Mechanism of intercalation into the DNA double helix by ethidium. Biochemistry 1993, 32, 4246–4253. [Google Scholar] [CrossRef]
- Chakraborty, A.; Panda, A.K.; Ghosh, R.; Biswas, A. DNA minor groove binding of a well known anti-mycobacterial drug dapsone: A spectroscopic, viscometric and molecular docking study. Arch. Biochem. Biophys. 2019, 665, 107–113. [Google Scholar] [CrossRef]
- Yılmaz, Z.T.; Odabaşoğlu, H.Y.; Şenel, P.; Adımcılar, V.; Erdoğan, T.; Özdemir, A.D.; Gölcü, A.; Odabaşoğlu, M. A novel 3-((5-methylpyridin-2-yl)amino)isobenzofuran-1(3H)-one: Molecular structure describe, X-ray diffractions and DFT calculations, antioxidant activity, DNA binding and molecular docking studies. J. Mol. Struct. 2020, 1205, 127585. [Google Scholar] [CrossRef]
- Mirzaei-Kalar, Z.; Yavari, A.; Jouyban, A. Increasing DNA binding affinity of doxorubicin by loading on Fe3O4 nanoparticles: A multi-spectroscopic study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 229, 117985. [Google Scholar] [CrossRef]
- Joksimović, N.; Petronijević, J.; Janković, N.; Baskić, D.; Popović, S.; Todorović, D.; Matić, S.; Bogdanović, G.A.; Vraneš, M.; Tot, A.; et al. Synthesis, characterization, anticancer evaluation and mechanisms of cytotoxic activity of novel 3-hydroxy-3-pyrrolin-2-ones bearing thenoyl fragment: DNA, BSA interactions and molecular docking study. Bioorg. Chem. 2019, 88, 102954. [Google Scholar] [CrossRef] [PubMed]
- Bhanja, P.; Mishra, S.; Manna, K.; Mallick, A.; Das Saha, K.; Bhaumik, A. Covalent Organic Framework Material Bearing Phloroglucinol Building Units as a Potent Anticancer Agent. ACS Appl. Mater. Interfaces 2017, 9, 31411–31423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandi, R.; Mishra, S.; Maji, T.K.; Manna, K.; Kar, P.; Banerjee, S.; Dutta, S.; Sharma, S.K.; Lemmens, P.; Das Saha, K.; et al. A novel nanohybrid for cancer theranostics: Folate sensitized Fe2O3 nanoparticles for colorectal cancer diagnosis and photodynamic therapy. J. Mater. Chem. B 2017, 5, 3927–3939. [Google Scholar] [CrossRef] [PubMed]




| CCDC Number | 1986064 |
|---|---|
| Empirical formula | C22H16CuO5 |
| Formula weight | 423.91 |
| Temperature/K | 100 (2) |
| Crystal system | monoclinic |
| Space group | P21/c |
| a/Å | 16.1069 (14) |
| b/Å | 5.4478 (5) |
| c/Å | 19.531 (2) |
| α/° | 90 |
| β/° | 95.936 (3) |
| γ/° | 90 |
| Volume/Å3 | 1704.6 (3) |
| Z | 4 |
| ρcalcg/cm3 | 1.6517 |
| μ/mm−1 | 1.315 |
| F (000) | 869.8 |
| Crystal size/mm3 | 0.4 × 0.31 × 0.18 |
| Radiation | Mo Kα (λ = 0.71073) |
| 2Θ range for data collection/° | 5.08 to 50.1 |
| Index ranges | −21 ≤ h ≤ 21, −7 ≤ k ≤ 7, −26 ≤ l ≤ 26 |
| Reflections collected | 17315 |
| Independent reflections | 3017 [Rint = 0.0820, Rsigma = 0.0744] |
| Data/restraints/parameters | 3017/0/255 |
| Goodness of fit on F2 | 1.106 |
| Final R indexes [I > = 2σ (I)] | R1 = 0.0731, wR2 = 0.2061 |
| Final R indexes [all data] | R1 = 0.0896, wR2 = 0.2262 |
| Largest diff. peak/hole / e Å−3 | 2.46/−0.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alolayqi, E.; Afzal, M.; Alarifi, A.; Beagan, A.; Muddassir, M. Depicting the DNA Binding and Cytotoxicity Studies against Human Colorectal Cancer of Aquabis (1-Formyl-2-Naphtholato-k2O,O′) Copper(II): A Biophysical and Molecular Docking Perspective. Crystals 2022, 12, 15. https://doi.org/10.3390/cryst12010015
Alolayqi E, Afzal M, Alarifi A, Beagan A, Muddassir M. Depicting the DNA Binding and Cytotoxicity Studies against Human Colorectal Cancer of Aquabis (1-Formyl-2-Naphtholato-k2O,O′) Copper(II): A Biophysical and Molecular Docking Perspective. Crystals. 2022; 12(1):15. https://doi.org/10.3390/cryst12010015
Chicago/Turabian StyleAlolayqi, Ebtisam, Mohd Afzal, Abdullah Alarifi, Abeer Beagan, and Mohd Muddassir. 2022. "Depicting the DNA Binding and Cytotoxicity Studies against Human Colorectal Cancer of Aquabis (1-Formyl-2-Naphtholato-k2O,O′) Copper(II): A Biophysical and Molecular Docking Perspective" Crystals 12, no. 1: 15. https://doi.org/10.3390/cryst12010015
APA StyleAlolayqi, E., Afzal, M., Alarifi, A., Beagan, A., & Muddassir, M. (2022). Depicting the DNA Binding and Cytotoxicity Studies against Human Colorectal Cancer of Aquabis (1-Formyl-2-Naphtholato-k2O,O′) Copper(II): A Biophysical and Molecular Docking Perspective. Crystals, 12(1), 15. https://doi.org/10.3390/cryst12010015







