Detail Analysis for Energy Transfer and Pigment Assembling in C−Phycocyanin through Time−Resolved Spectroscopy and AlphaFold2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth of Red Algae and Isolation of PBSs
2.2. Determination and Analysis of Time-Resolved Spectra
2.3. Protein Structure Prediction of C−PC Complex Based on AlphaFold2
3. Results
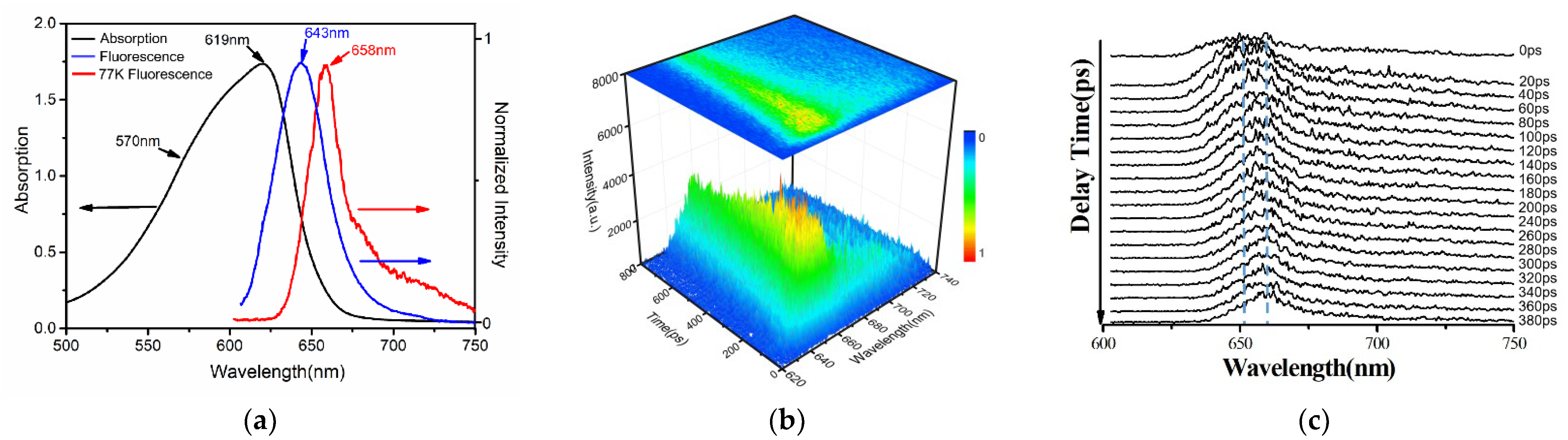
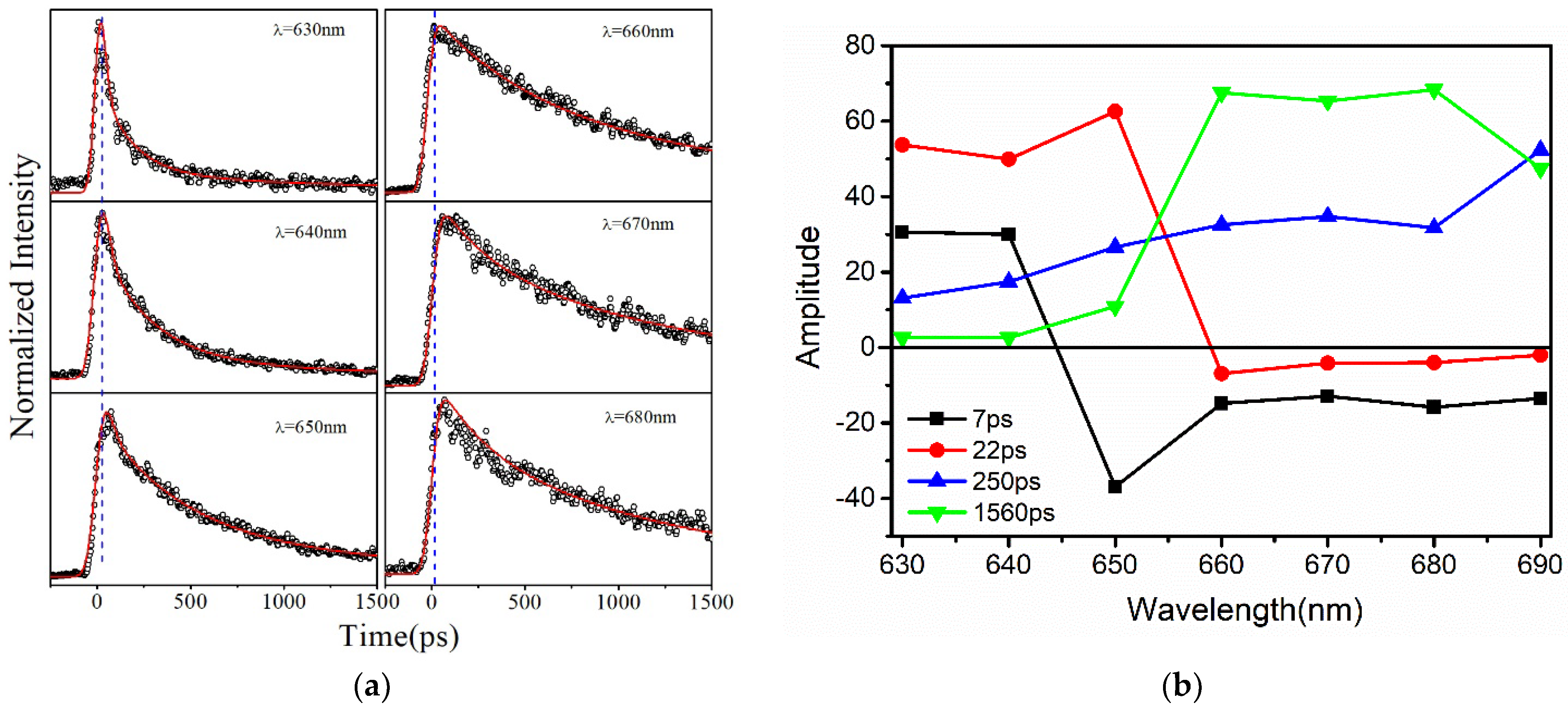
3.1. Steady State and Time−Resolved Spectroscopy Results
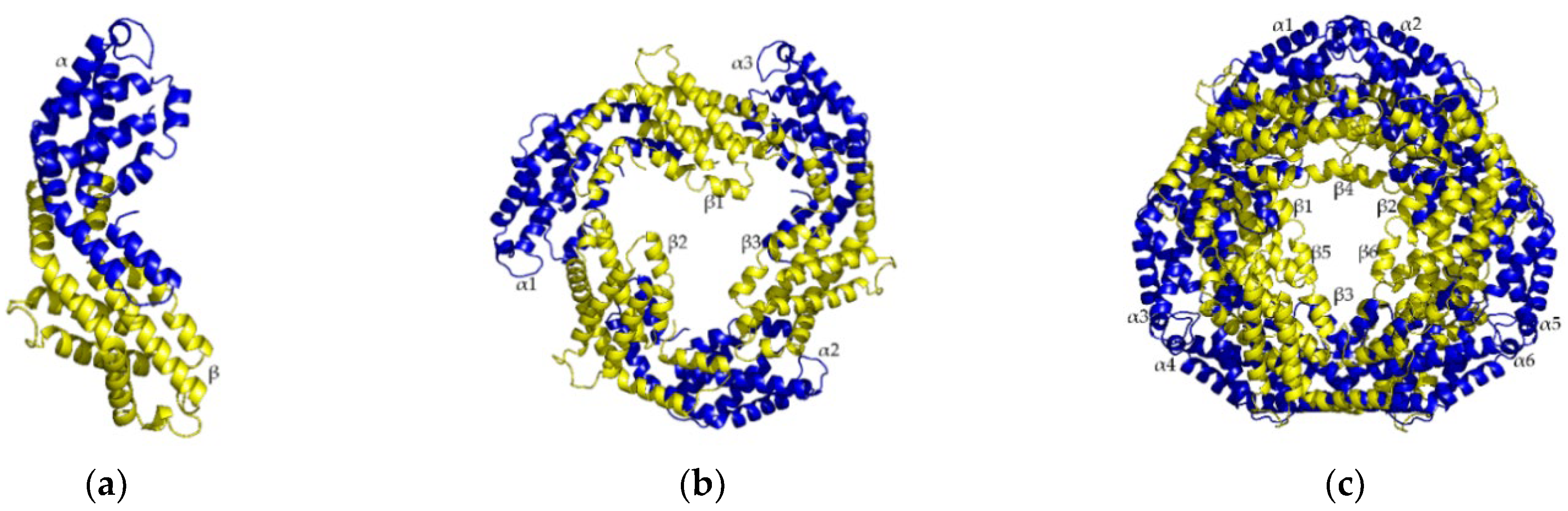
3.2. Protein Structure Prediction Results
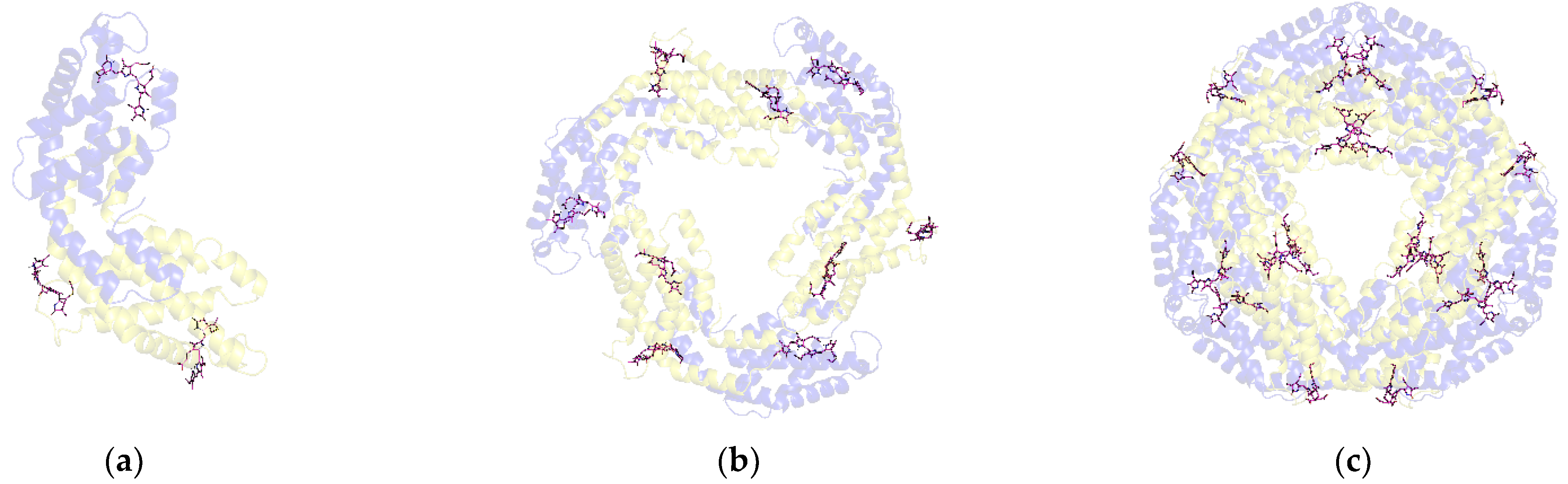
3.3. Chromophore Arrangement Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blankenship, R.E. Molecular Mechanisms of Photosynthesis, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2014. [Google Scholar]
- Wiltbank, L.B.; Kehoe, D.M. Diverse light response of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol. 2019, 17, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Scholes, G.D.; Fleming, G.R.; Olaya-Castro, A.; van Grondelle, R. Lessons from nature about solar light harvesting. Nat. Chem. 2011, 3, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.A. The Photosynthetic Apparatus: Molecular Biology and Operation. In Cell Culture and Somatic Cell Genetics of Plants, 1st ed.; Bogorad, L., Vasil, I.K., Eds.; Academic Press: New York, NY, USA, 1991; pp. 255–298. [Google Scholar]
- Glazer, A.N. Light harvesting by phycobilisomes. Annu. Rev. Biophys. Biophys. Chem. 1985, 14, 47–77. [Google Scholar] [PubMed]
- van Grondelle, R. Excitation energy transfer, trapping and annihilation in photosynthetic systems. Biochim. Et Biophys-Ica Acta (BBA)—Rev. Bioenerg. 1985, 811, 147–195. [Google Scholar] [CrossRef]
- Gao, X.; Wei, T.-D.; Zhang, N.; Xie, B.-B.; Su, H.-N.; Zhang, X.-Y.; Chen, X.-L.; Zhou, B.-C.; Wang, Z.-X.; Wu, J.-W.; et al. Molecular insights into the terminal energy acceptor in cyanobacterial phycobilisome. Mol. Microbiol. 2012, 85, 907–915. [Google Scholar] [CrossRef]
- Watanabe, M.; Ikeuchi, M. Phycobilisome: Architecture of a light-harvesting supercomplex. Photosynth. Res. 2013, 116, 265–276. [Google Scholar] [CrossRef]
- Peng, P.-P.; Dong, L.-L.; Sun, Y.-F.; Zeng, X.-L.; Ding, W.-L.; Scheer, H.; Yang, X.; Zhao, K.-H. The structure of allophycocyanin B fromSynechocystisPCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 2558–2569. [Google Scholar] [CrossRef]
- Kuzminov, F.; Bolychevtseva, Y.; Elanskaya, I.; Karapetyan, N. Effect of APCD and APCF subunits depletion on phycobilisome fluorescence of the cyanobacterium Synechocystis PCC 6803. J. Photochem. Photobiol. B Biol. 2014, 133, 153–160. [Google Scholar] [CrossRef]
- Chang, L.; Liu, X.; Li, Y.; Liu, C.-C.; Yang, F.; Zhao, J.; Sui, S.-F. Structural organization of an intact phycobilisome and its association with photosystem II. Cell Res. 2015, 25, 726–737. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, L.; Chang, W.; Zhang, J.; Gui, L.; Guo, B.; Liang, D. Structure of C-phycocyanin from Spirulina platensis at 2.2 A resolution: A novel monoclinic crystal form for phycobiliproteins in phycobilisomes. Acta Cryst. 2001, D57, 784–792. [Google Scholar]
- Padyana, A.; Bhat, V.; Madyastha, K.; Rajashankar, K.; Ramakumar, S. Crystal Structure of a Light-Harvesting Protein C-Phycocyanin from Spirulina platensis. Biochem. Biophys. Res. Commun. 2001, 282, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Debreczeny, M.P.; Sauer, K. Comparison of Calculated and Experimentally Resolved Rate Constants for Excitation Energy Transfer in C-Phycocyanin. 1. Monomers. J. Phys. Chem. 1995, 99, 8412–8419. [Google Scholar] [CrossRef]
- Debreczeny, M.P.; Sauer, K.; Zhou, J.; Bryant, D.A. Comparison of Calculated and Experimentally Resolved Rate Constants for Excitation Energy Transfer in C-Phycocyanin. 2. Trimers. J. Phys. Chem. 1994, 99, 8420–8431. [Google Scholar] [CrossRef]
- Zhao, J.; Zhu, J.; Jiang, L. Computer simulation on kinetics of primary process in photosynthesis of algae (V)—Excitation energy transfer in phycoerythrocyanins. China Ser. B-Chem. 1996, 5, 73–80. [Google Scholar]
- Zhao, J.; Zhu, J.; Jiang, L. Computer simulation on kinetics of primary process in photosynthesis (III). Energy transfer in the trimers and hexamers of C-Phycocyanins. China Ser. B-Chem. 1994, 7, 35–42. [Google Scholar]
- Zhao, J.; Zhu, J.; Jiang, L. Study on the energy transfer processes in phycobilisomes from blue-green algae by the use of stochastic simulation approach. BBA—Bioenerg. 1995, 1229, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhu, J.; Jiang, L. Computer Simulation on Kinetics of Primary Processes in Photosynthesis I-Mechanism of Exciton Annihilation of Excited State Decay in Phycobiliproteins. S China Ser. B-Chem. 1993, 36, 1186–1197. [Google Scholar]
- Matamala, A.R.; Almonacid, D.E.; Figueroa, M.F.; Martínez-Oyanedel, J.; Bunster, M.C. A semiempirical approach to the intraphycocyanin and inter-phycocyanin fluorescence resonance energy-transfer. J. Comput Chem. 2007, 28, 1200–1207. [Google Scholar] [CrossRef] [Green Version]
- Schneider, S.; Geiselhart, P.; Siebzehnrübl, S. Energy transfer within trimers of Mastigocladus laminosus studied by picosecond time-resolved transient absorption spectroscopy. Z. Nat. C 2014, 43, 55–62. [Google Scholar] [CrossRef]
- Xie, M.; Li, W.; Lin, H.; Wang, X.; Dong, J.; Qin, S.; Zhao, F. Difference in light use strategy in red alga between Griffithsia pacifica and Porphyridium purpureum. Sci. Rep. 2021, 11, 14367. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Li, W.; Xiao, C.; Zhen, Z.; Ma, J.; Lin, H.; Qin, S.; Zhao, F. Time-Resolved Fluorescence Spectroscopy Study of Energy Transfer Dynamics in Phycobilisomes from Cyanobacteria Thermosynechococcus vulcanus NIES 2134 and Synechocystis sp. PCC 6803. Crystals 2021, 11, 1233. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, J.; Jiang, L. Energy transfer processes in phycobilisome model complex at 77 K. China Ser. B-Chem. 2000, 43, 233–239. [Google Scholar] [CrossRef]
- Zhao, F.; Zheng, X.; Zhang, J.; Wang, H.; Yu, Z.; Zhao, J.; Jiang, L. Energy transfer in allophycocyanin hexamer from Anabaena variabilis by time-resolved spectroscopy. J. Photochem. Photobiol. B 1998, 45, 144–149. [Google Scholar] [CrossRef]
- Bianco, G.; Forli, S.; Goodsell, D.S.; Olson, A.J. Covalent docking using autodock: Two-point attractor and flexible side chain methods. Protein Sci. 2015, 25, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Forli, S.; Huey, R.; Pique, M.; Sanner, M.; Goodsell, D.; Olson, A. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [Green Version]
- Seeliger, D.; de-Groot, B. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided. Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Xia, A.D.; Zhang, X.Y.; Hong, Q.A.; Meng, J.; Hou, S.J.; Sudha, M.; Maruthi Sai, P.S.; Jha, I.B. Spectroscopic Characteristics and Energy-Transfer Processes in C-phycocyanin from Cyanobacterium Westiellopsis-Prolifica. Sci. China-Chem. 1992, 35, 811–821. [Google Scholar]
- Ren, Y.; Chi, B.; Melhem, O.; Wei, K.; Feng, L.; Li, Y.; Han, X.; Li, D.; Zhang, Y.; Wan, J.; et al. Understanding the electronic energy transfer pathways in the trimeric and hexameric aggregation state of cyanobacteria phycocyanin within the framework of Förster theory. J. Comput. Chem. 2013, 34, 1005–1012. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, F.; Zheng, X.; Wang, H.Z.; Yang, T.-S.; Hayashi, M.; Lin, S. Excitation energy-transfer processes between two trimers in C-phycocyanin hexamer from the cyanobacterium Anabaena variabilis. Investigation by group theory and time-resolved fluorescence spectroscopy. J. Photochem. Photobiol. B-Biol. 1999, 53, 128–135. [Google Scholar] [CrossRef]
- Rajesh, S.; Overduin, M.; Bonev, B.B. NMR of membrane proteins: Beyond crystals. Adv. Exp. Med. Biol. 2016, 922, 29–42. [Google Scholar] [PubMed]
- Ma, J.; You, X.; Sun, S.; Wang, X.; Qin, S.; Sui, S. Structural basis of energy transfer in Porphyridium purpureum phycobili-some. Nature 2020, 579, 146–151. [Google Scholar] [CrossRef] [PubMed]
| C-PC Complex | Energy Transfer Pairs | Distance (Å) |
|---|---|---|
| αβ monomer | β1–84 to β1–155 | 35 |
| β2–84 to β2–155 | ||
| β3–84 to β3–155 | ||
| β4–84 to β4–155 | ||
| β5–84 to β5–155 | ||
| β6–84 to β6–155 | ||
| (αβ)3 trimer | α1–84 to β2–84 | 20 |
| α2–84 to β3–84 | ||
| α3–84 to β1–84 | ||
| α4–84 to β5–84 | ||
| α5–84 to β6–84 | ||
| α6–84 to β4–84 | ||
| β1–84 to β2–84 | 36 | |
| β2–84 to β3–84 | ||
| β3–84 to β1–84 | ||
| β4–84 to β5–84 | ||
| β5–84 to β6–84 | ||
| β6–84 to β4–84 | ||
| α1–84 to β3–155 | 39 | |
| α2–84 to β1–155 | ||
| α3–84 to β2–155 | ||
| α4–84 to β5–155 | ||
| α5–84 to β6–155 | ||
| α6–84 to β4–155 | ||
| (αβ)6 hexamer | α1–84 to α4–84 | 26 |
| α2–84 to α5–84 | ||
| α3–84 to α6–84 | ||
| β1–155 to β5–155 | 28 | |
| β2–155 to β6–155 | ||
| β3–155 to β4–155 | ||
| β1–84 to β6–84 | 35 | |
| β2–84 to β4–84 | ||
| β3–84 to β5–84 | ||
| α1–84 to β5–84 | 35 | |
| α2–84 to β6–84 | ||
| α3–84 to β4–84 | ||
| α4–84 to β3–84 | ||
| α5–84 to β1–84 | ||
| α6–84 to β2–84 | ||
| α1–84 to β5–155 | 37 | |
| α2–84 to β6–155 | ||
| α3–84 to β4–155 | ||
| α4–84 to β3–155 | ||
| α5–84 to β1–155 | ||
| α6–84 to β2–155 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, C.; Guo, N.; Liang, Z.; Zhao, F.; Xie, M. Detail Analysis for Energy Transfer and Pigment Assembling in C−Phycocyanin through Time−Resolved Spectroscopy and AlphaFold2. Crystals 2022, 12, 1595. https://doi.org/10.3390/cryst12111595
Xiao C, Guo N, Liang Z, Zhao F, Xie M. Detail Analysis for Energy Transfer and Pigment Assembling in C−Phycocyanin through Time−Resolved Spectroscopy and AlphaFold2. Crystals. 2022; 12(11):1595. https://doi.org/10.3390/cryst12111595
Chicago/Turabian StyleXiao, Chao, Na Guo, Zidong Liang, Fuli Zhao, and Mingyuan Xie. 2022. "Detail Analysis for Energy Transfer and Pigment Assembling in C−Phycocyanin through Time−Resolved Spectroscopy and AlphaFold2" Crystals 12, no. 11: 1595. https://doi.org/10.3390/cryst12111595






