Dominant Effects of Short-Chain Branching on the Initial Stage of Nucleation and Formation of Tie Chains for Bimodal Polyethylene as Revealed by Molecular Dynamics Simulation
Abstract
:1. Introduction
2. Models and Methods
3. Results and Discussion
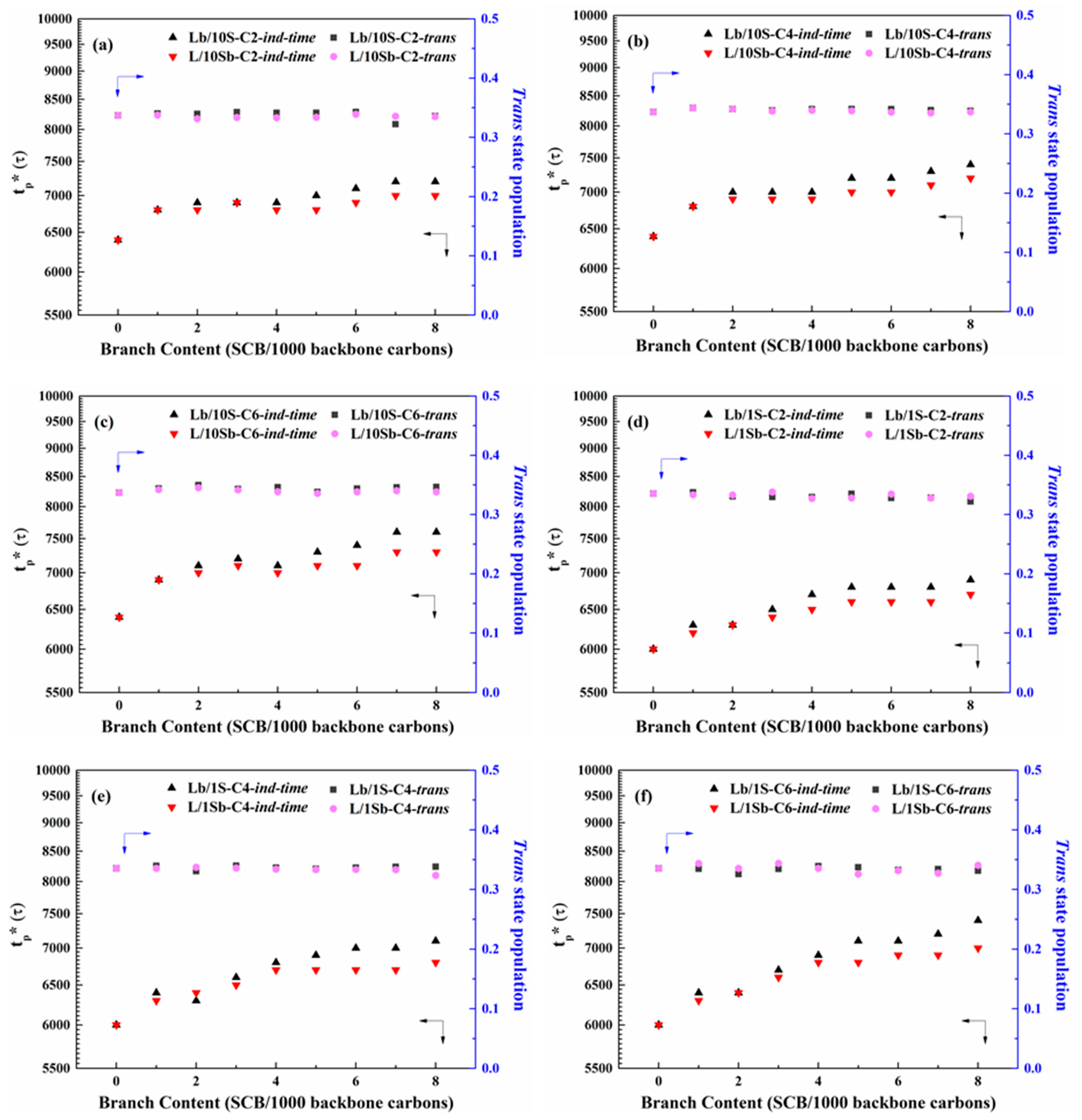
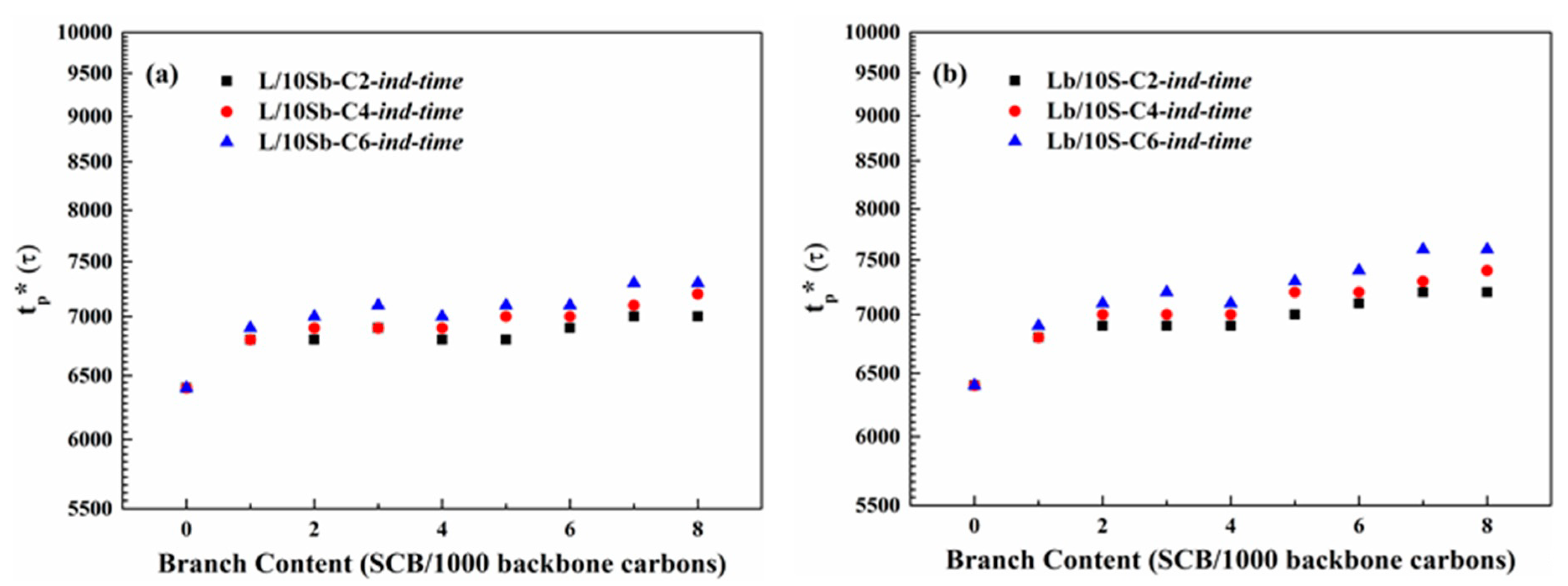
3.1. Induction Time of Precursor Formation
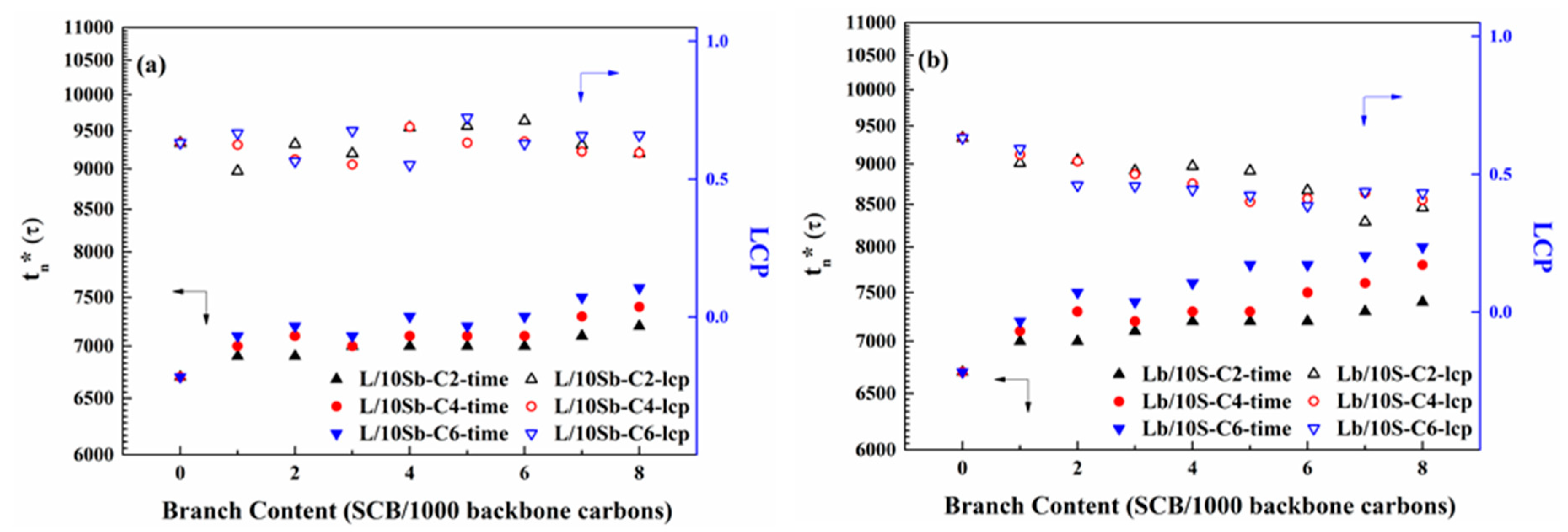
3.2. The Transformation of Precursors to Stable Nuclei
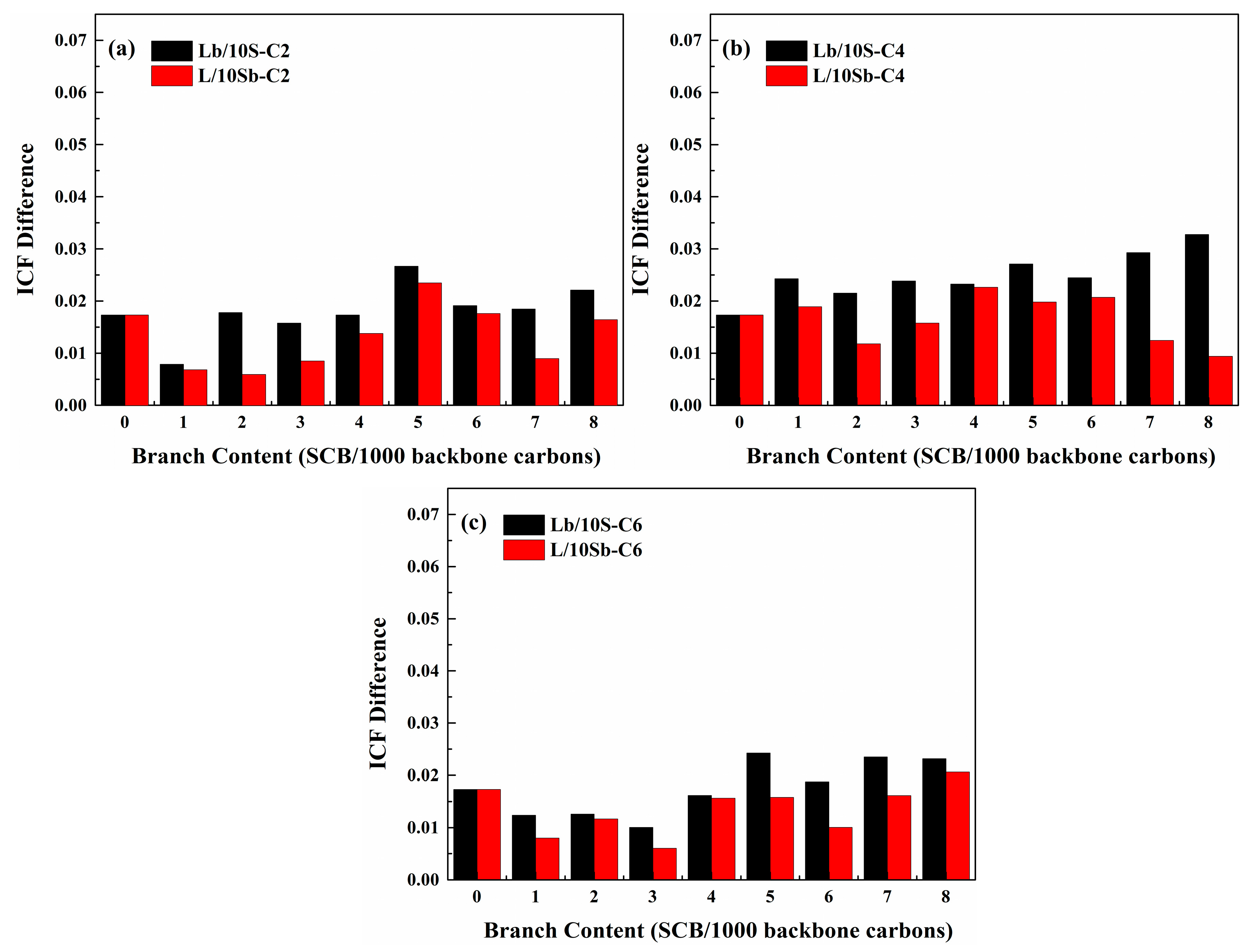
3.3. Crystal Structures
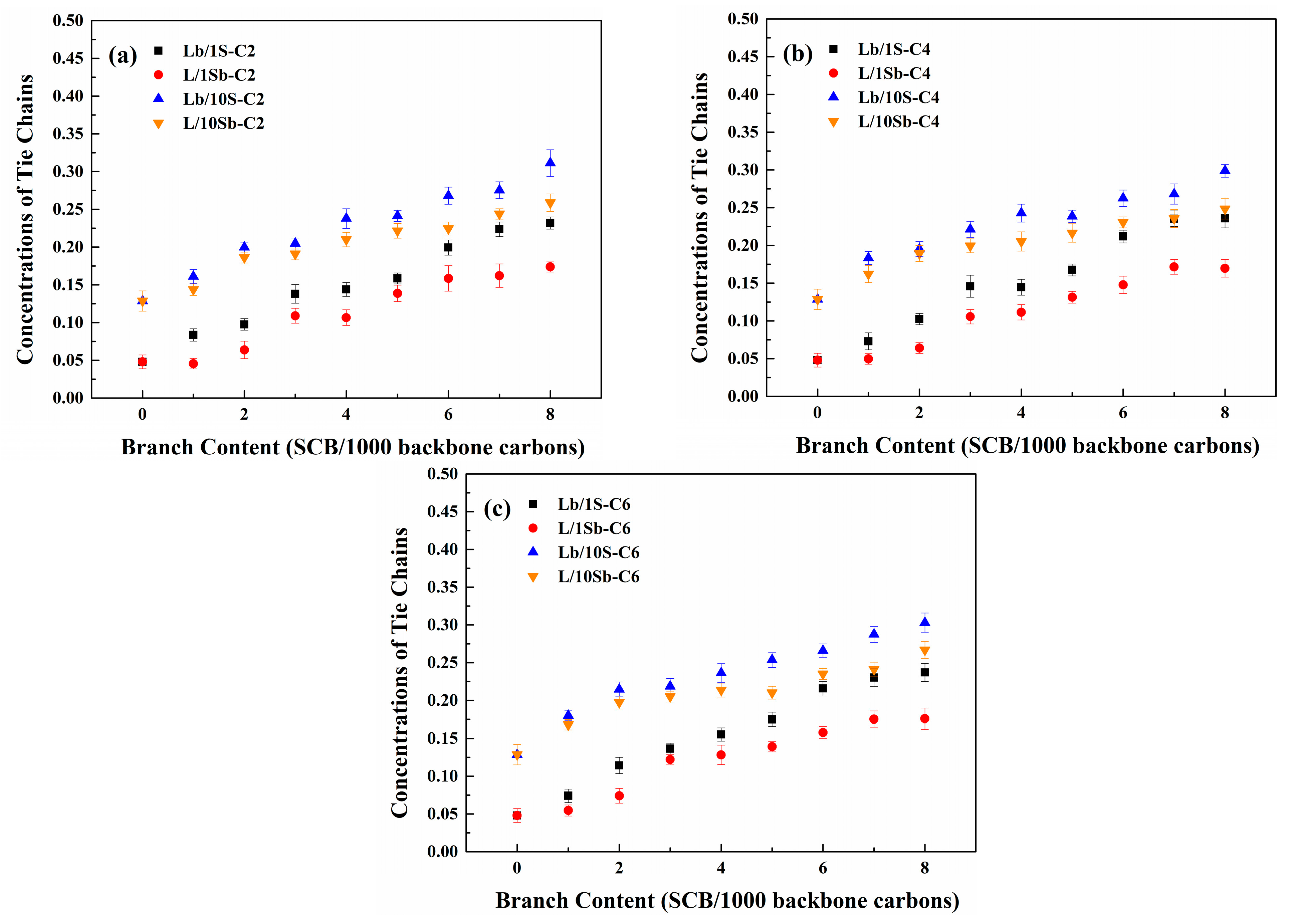
3.4. Tie Chains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SCB | Short-chain branching |
| MSL | Methylene sequence length |
| SCBD | Short-chain branching distribution |
| BPE | Bimodal polyethylene |
| PE | Polyethylene |
| MWD | Molecular weight distribution |
| MD | Molecular dynamics |
| ADMET | Acyclic diene metathesis |
| HDPE | High-density polyethylene |
| SOP | Site order parameter |
| Xc | Crystallinity |
| ICF | Interchain contact fraction |
| tp* | Induction time |
| LCP | Long-chain proportion |
| tn* | Formation time of the first stable nucleus |
References
- Hubert, L.; David, L.; Seguela, R.; Vigier, G.; Degoulet, C.; Germain, Y. Physical and mechanical properties of polyethylene for pipes in relation to molecular architecture. I. Microstructure and crystallisation kinetics. Polymer 2001, 42, 8425–8434. [Google Scholar] [CrossRef]
- Shan, C.L.P.; Soares, J.B.P.; Penlidis, A. HDPE/LLDPE reactor blends with bimodal microstructures—Part II: Rheological properties. Polymer 2003, 44, 177–185. [Google Scholar] [CrossRef]
- Gupta, S.; Olson, J.D. Industrial needs in physical properties. Ind. Eng. Chem. Res. 2003, 42, 6359–6374. [Google Scholar] [CrossRef]
- Domínguez, C.; García, R.; Aroca, M.; Carrero, A. Study of the PENT test conditions for reducing failure times in high-resistance polyethylene resins for pipe applications. Mech. Time Depend. Mater. 2012, 16, 105–115. [Google Scholar] [CrossRef]
- Chen, K.; Tian, Z.; Luo, N.; Liu, B. Modeling and simulation of Borstar bimodal polyethylene process based on a rigorous PC-SAFT equation of state model. Ind. Eng. Chem. Res. 2014, 53, 19905–19915. [Google Scholar] [CrossRef]
- Tian, Z.; Chen, K.; Liu, B.; Luo, N.; Du, W.; Qian, F. Short-chain branching distribution oriented model development for Borstar bimodal polyethylene process and its correlation with product performance of slow crack growth. Chem. Eng. Sci. 2015, 130, 41–55. [Google Scholar] [CrossRef]
- Huang, Y.F.; Kao, H.L.; Ruan, J.; Su, A.C. Effects of solution status on single-crystal growth habit of Poly(l-lactide). Macromolecules 2010, 43, 7222–7227. [Google Scholar] [CrossRef]
- Lan, Y.K.; Su, A.C. Nucleation of polymer crystals: The “δ Mystery”. Macromolecules 2010, 43, 7908–7912. [Google Scholar] [CrossRef]
- Hoffman, J.D.; Miller, R.L. Kinetic of crystallization from the melt and chain folding in polyethylene fractions revisited: Theory and experiment. Polymer 1997, 38, 3151–3212. [Google Scholar] [CrossRef]
- Waheed, N.; Lavine, M.S.; Rutledge, G.C. Molecular simulation of crystal growth in n-eicosane. J. Chem. Phys. 2002, 116, 2301–2309. [Google Scholar] [CrossRef]
- Hu, W.; Frenkel, D.; Mathot, V.B.F. Simulation of shish-kebab crystallite induced by a single prealigned macromolecule. Macromolecules 2002, 35, 7172–7174. [Google Scholar] [CrossRef]
- Tashiro, K.; Sasaki, S. Structural changes in the ordering process of polymers as studied by an organized combination of the various measurement techniques. Prog. Polym. Sci. 2003, 28, 451–519. [Google Scholar] [CrossRef]
- Yamamoto, T. Molecular dynamics modeling of polymer crystallization from the melt. Polymer 2004, 45, 1357–1364. [Google Scholar] [CrossRef]
- Strobl, G. Crystallization and melting of bulk polymers: New observations, conclusions and a thermodynamic scheme. Prog. Polym. Sci. 2006, 31, 398–442. [Google Scholar] [CrossRef]
- Zhang, J.; Muthukumar, M. Monte Carlo simulations of single crystals from polymer solutions. J. Chem. Phys. 2007, 126, 234904. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, J.K.; Farrance, O.E.; Kailas, L. How atomic force microscopy has contributed to our understanding of polymer crystallization. Polymer 2009, 50, 4281–4292. [Google Scholar] [CrossRef] [Green Version]
- Cormia, R.L.; Price, F.P.; Turnbull, D. Kinetics of crystal nucleation in polyethylene. J. Chem. Phys. 1962, 37, 1333–1340. [Google Scholar] [CrossRef]
- Yamamoto, T. Computer modeling of polymer crystallization—Toward computer-assisted materials’ design. Polymer 2009, 50, 1975–1985. [Google Scholar] [CrossRef]
- Anwar, M.; Schilling, T. Crystallization of polyethylene: A molecular dynamics simulation study of the nucleation and growth mechanisms. Polymer 2015, 76, 307–312. [Google Scholar] [CrossRef] [Green Version]
- Zerze, H.; Mittal, J.; McHugh, A.J. Ab initio crystallization of alkanes: Structure and kinetics of nuclei formation. Macromolecules 2013, 46, 9151–9157. [Google Scholar] [CrossRef]
- Yi, P.; Rutledge, G.C. Molecular simulation of crystal nucleation in n-octane melts. J. Chem. Phys. 2009, 131, 134902. [Google Scholar] [PubMed] [Green Version]
- Yi, P.; Locker, C.R.; Rutledge, G.C. Molecular dynamics simulation of homogeneous crystal nucleation in polyethylene. Macromolecules 2013, 46, 4723–4733. [Google Scholar] [CrossRef]
- Lan, Y.K.; Su, A.C. Primary nucleation of polyethylene: Embryogenesis from a semidilute solution. Polymer 2014, 55, 3087–3092. [Google Scholar] [CrossRef]
- Sanmartín, S.; Ramos, J.; Vega, J.F.; Martínez-Salazar, J. Strong influence of branching on the early stage of nucleation and crystal formation of fast cooled ultralong n-alkanes as revealed by computer simulation. Eur. Polym. J. 2014, 50, 190–199. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; He, X.; Shao, Y.; Hu, Y.; Zhang, H.; Liu, Z.; Liu, B. Effects of branch content and branch length on polyethylene crystallization: Molecular dynamics simulation. Macromol. Theory Simul. 2016, 25, 303–311. [Google Scholar] [CrossRef]
- Gao, R.; He, X.; Zhang, H.; Shao, Y.; Liu, Z.; Liu, B. Molecular dynamics study of the isothermal crystallization mechanism of polyethylene chain: The combined effects of chain length with temperature. J. Mol. Model. 2016, 22, 67. [Google Scholar] [CrossRef]
- Hu, Y.; Shao, Y.; Liu, Z.; He, X.; Liu, B. Effect of short-chain branching on the tie chains and dynamics of bimodal polyethylene: Molecular dynamics simulation. Eur. Polym. J. 2018, 103, 312–321. [Google Scholar] [CrossRef]
- Alamo, R.G.; Mandelkern, L. Thermodynamic and structural properties of ethylene copolymers. Macromolecules 1989, 22, 1273–1277. [Google Scholar] [CrossRef]
- Alamo, R.G.; Viers, B.D.; Mandelkern, L. Phase structure of random ethylene copolymers: A study of counit content and molecular weight as independent variables. Macromolecules 1993, 26, 5740–5747. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, F.; He, T. Effect of branch content on the transition of crystalline structure and morphology of metallocene-catalyzed branched polyethylene. J. Mater. Sci. 2001, 36, 5345–5349. [Google Scholar] [CrossRef]
- Smith, J.A.; Brzezinska, K.R.; Valenti, D.J.; Wagener, K.B. Precisely controlled methyl branching in polyethylene via acyclic diene metathesis (ADMET) polymerization. Macromolecules 2000, 33, 3781–3794. [Google Scholar] [CrossRef]
- Sworen, J.C.; Smith, J.A.; Wagener, K.B.; Baugh, L.S.; Rucker, S.P. Modeling random methyl branching in ethylene/propylene copolymers using metathesis chemistry: Synthesis and thermal behavior. J. Am. Chem. Soc. 2003, 125, 2228–2240. [Google Scholar] [CrossRef] [PubMed]
- Sworen, J.C.; Wagener, K.B. Linear low-density polyethylene containing precisely placed hexyl branches. Macromolecules 2007, 40, 4414–4423. [Google Scholar] [CrossRef]
- Rojas, G.; Inci, B.; Wei, Y.; Wagener, K.B. Precision polyethylene: Changes in morphology as a function of alkyl branch size. J. Am. Chem. Soc. 2009, 131, 17376–17386. [Google Scholar] [CrossRef] [PubMed]
- Inci, B.; Wagener, K.B. Decreasing the alkyl branch frequency in precision polyethylene: Pushing the limits toward longer run lengths. J. Am. Chem. Soc. 2011, 133, 11872–11875. [Google Scholar] [CrossRef]
- Nozue, Y.; Kawashima, Y.; Seno, S.; Nagamatsu, T.; Hosoda, S.; Berda, E.B.; Rojas, G.; Baughman, T.W.; Wagener, K.B. Unusual crystallization behavior of polyethylene having precisely spaced branches. Macromolecules 2011, 44, 4030–4034. [Google Scholar] [CrossRef]
- Inci, B.; Lieberwirth, I.; Steffen, W.; Mezger, M.; Graf, R.; Landfester, K.; Wagener, K.B. Decreasing the alkyl branch frequency in precision polyethylene: Effect of alkyl branch size on nanoscale morphology. Macromolecules 2012, 45, 3367–3376. [Google Scholar] [CrossRef]
- Silva, L.C.D.; Graf, R.; Bowers, C.R.; Wagener, K.B. Branch-Induced heterogeneous chain motion in precision polyolefins. Macromolecules 2015, 48, 8858–8866. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Z.; Lu, Z.; Sun, C. Molecular dynamics simulation of the linear low-density polyethylene crystallization. J. Chem. Phys. 2001, 115, 3916–3922. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Z.; Lu, Z.; Sun, C. The crystallization of low-density polyethylene: A molecular dynamics simulation. Polymer 2002, 43, 3223–3227. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Z.; Lu, Z.; Sun, C. Roles of branch content and branch length in copolyethylene crystallization: Molecular dynamics simulations. Macromolecules 2002, 35, 106–111. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Z.; Yang, H.; Sun, C. Molecular dynamics simulations on crystallization of polyethylene copolymer with precisely controlled branching. Macromolecules 2004, 37, 7393–7400. [Google Scholar] [CrossRef]
- Ramos, J.; Vega, J.F.; Sanmartín, S.; Martínez-Salazar, J. Coarse-grained simulations on the crystallization, melting and annealing processes of short chain branched polyolefins. Eur. Polym. J. 2016, 85, 478–488. [Google Scholar] [CrossRef]
- Zhang, M.; Yuen, F.; Choi, P. Differences in the solid-state structures of single-site and Ziegler−Natta linear low-density polyethylenes as revealed by molecular dynamics simulation. Macromolecules 2006, 39, 8517–8525. [Google Scholar] [CrossRef]
- Li, C.; Choi, P. Molecular dynamics study of the solid-state structures of linear-low-density polyethylenes with blocky branches. Macromolecules 2008, 41, 7109–7114. [Google Scholar] [CrossRef]
- Choi, P.; Wang, Q.; Vignola, E. Molecular dynamics study of the conformation and dynamics of precisely branched polyethylene. Polymer 2014, 55, 5734–5738. [Google Scholar] [CrossRef]
- Kumar, V.; Locker, C.R.; in’t Veld, P.J.; Rutledge, G.C. Effect of short chain branching on the interlamellar structure of semicrystalline polyethylene. Macromolecules 2017, 50, 1206–1214. [Google Scholar] [CrossRef]
- Krishnaswamy, R.K.; Yang, Q.; Fernandez-Ballester, L.; Kornfield, J.A. Effect of the distribution of short-chain branches on crystallization kinetics and mechanical properties of high-density polyethylene. Macromolecules 2008, 41, 1693–1704. [Google Scholar] [CrossRef]
- Kavassalis, T.A.; Sundararajan, P.R. A molecular dynamics study of polyethylene crystallization. Macromolecules 1993, 26, 4144–4150. [Google Scholar] [CrossRef]
- Lindahl, E.; Hess, B.; van der Spoel, D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 2001, 7, 306–317. [Google Scholar] [CrossRef]
- Mayo, S.L.; Olafson, B.D.; Goddard, W.A., III. DREIDING: A generic force field for molecular simulations. J. Phys. Chem. 1990, 94, 8897–8909. [Google Scholar] [CrossRef]
- Yu, X.; Kong, B.; Yang, X. Molecular dynamics study on the crystallization of a cluster of polymer chains depending on the initial entanglement structure. Macromolecules 2008, 41, 6733–6740. [Google Scholar] [CrossRef]
- Wang, S.; Kong, B.; Nies, E.; Li, X.; Yang, X. Two mechanisms of polymer chain crystallization within nanoglobule. Polymer 2013, 54, 4030–4036. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, X. The effect of chain interpenetration on an ordering process in the early stage of polymer crystal nucleation. Polymer 2006, 47, 5213–5219. [Google Scholar] [CrossRef]
- Mao, W.; Kong, B.; Yang, X.; Nies, E. Nascent crystallization of a growing chain on a catalyst surface: A nonequilibrium molecular dynamics simulation study. J. Phys. Chem. B 2008, 112, 6753–6761. [Google Scholar] [CrossRef]
- Li, C.; Choi, P.; Sundararajan, P.R. Simulation of chain folding in polyethylene: A comparison of united atom and explicit hydrogen atom models. Polymer 2010, 51, 2803–2808. [Google Scholar] [CrossRef]
- Ungar, G.; Stejny, J.; Keller, A.; Bidd, I.; Whiting, M.C. The crystallization of ultralong normal paraffins: The onset of chain folding. Science 1985, 229, 386–389. [Google Scholar] [CrossRef]
- Flory, P.J. Theory of crystallization in copolymers. Trans. Faraday Soc. 1955, 51, 848–857. [Google Scholar] [CrossRef]
- Alamo, R.G.; Chan, E.K.M.; Mandelkern, L.; Voigt-Martin, I.G. Influence of molecular weight on the melting and phase structure of random copolymers of ethylene. Macromolecules 1992, 25, 6381–6394. [Google Scholar] [CrossRef]
- Wignall, G.D.; Alamo, R.G.; Ritchson, E.J.; Mandelkern, L.; Schwahn, D. SANS studies of liquid−liquid phase separation in heterogeneous and metallocene-based linear low-density polyethylenes. Macromolecules 2001, 34, 8160–8165. [Google Scholar] [CrossRef]
- Moyassari, A.; Mostafavi, H.; Gkourmpis, T.; Hedenqvist, M.S.; Gedde, U.W.; Nilsson, F. Simulation of semi-crystalline polyethylene: Effect of short-chain branching on tie chains and trapped entanglements. Polymer 2015, 72, 177–184. [Google Scholar] [CrossRef]
- Huang, Y.L.; Brown, N. Dependence of slow crack growth in polyethylene on butyl branch density: Morphology and theory. J. Polym. Sci. Part B Polym. Phys. 1991, 29, 129–137. [Google Scholar] [CrossRef]







| Complex BPE Model | Branch | Branch Content (SCB/1000 Backbone Carbons) | Methylene Sequence Length (CH2) | Total Number of CH2 |
|---|---|---|---|---|
| HDPE | None | 0 | - | 20,000 |
| Lb/10S-C2-1 | Ethyl | 1 | 476 | 20,036 |
| L/10Sb-C2-1 | Ethyl | 1 | 334 | 20,060 |
| Lb/10S-C2-2 | Ethyl | 2 | 244 | 20,084 |
| L/10Sb-C2-2 | Ethyl | 2 | 200 | 20,080 |
| Lb/10S-C2-3 | Ethyl | 3 | 164 | 20,124 |
| L/10Sb-C2-3 | Ethyl | 3 | 142 | 20,060 |
| Lb/10S-C2-4 | Ethyl | 4 | 124 | 20,204 |
| L/10Sb-C2-4 | Ethyl | 4 | 112 | 20,240 |
| Lb/10S-C2-5 | Ethyl | 5 | 100 | 20,300 |
| L/10Sb-C2-5 | Ethyl | 5 | 92 | 20,320 |
| Lb/10S-C2-6 | Ethyl | 6 | 82 | 20,162 |
| L/10Sb-C2-6 | Ethyl | 6 | 76 | 20,120 |
| Lb/10S-C2-7 | Ethyl | 7 | 70 | 20,150 |
| L/10Sb-C2-7 | Ethyl | 7 | 66 | 20,180 |
| Lb/10S-C2-8 | Ethyl | 8 | 62 | 20,302 |
| L/10Sb-C2-8 | Ethyl | 8 | 58 | 20,180 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Shao, Y.; Liu, Z.; He, X.; Liu, B. Dominant Effects of Short-Chain Branching on the Initial Stage of Nucleation and Formation of Tie Chains for Bimodal Polyethylene as Revealed by Molecular Dynamics Simulation. Polymers 2019, 11, 1840. https://doi.org/10.3390/polym11111840
Hu Y, Shao Y, Liu Z, He X, Liu B. Dominant Effects of Short-Chain Branching on the Initial Stage of Nucleation and Formation of Tie Chains for Bimodal Polyethylene as Revealed by Molecular Dynamics Simulation. Polymers. 2019; 11(11):1840. https://doi.org/10.3390/polym11111840
Chicago/Turabian StyleHu, Yanling, Yunqi Shao, Zhen Liu, Xuelian He, and Boping Liu. 2019. "Dominant Effects of Short-Chain Branching on the Initial Stage of Nucleation and Formation of Tie Chains for Bimodal Polyethylene as Revealed by Molecular Dynamics Simulation" Polymers 11, no. 11: 1840. https://doi.org/10.3390/polym11111840
APA StyleHu, Y., Shao, Y., Liu, Z., He, X., & Liu, B. (2019). Dominant Effects of Short-Chain Branching on the Initial Stage of Nucleation and Formation of Tie Chains for Bimodal Polyethylene as Revealed by Molecular Dynamics Simulation. Polymers, 11(11), 1840. https://doi.org/10.3390/polym11111840







