Marked Difference in the Conformational Transition of DNA Caused by Propanol Isomer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Preparation of Samples
2.2. Observation of the Higher-Order Structure of DNA by Fluorescence Microscopy
2.3. Circular Dichronism (CD) Measurements
3. Results
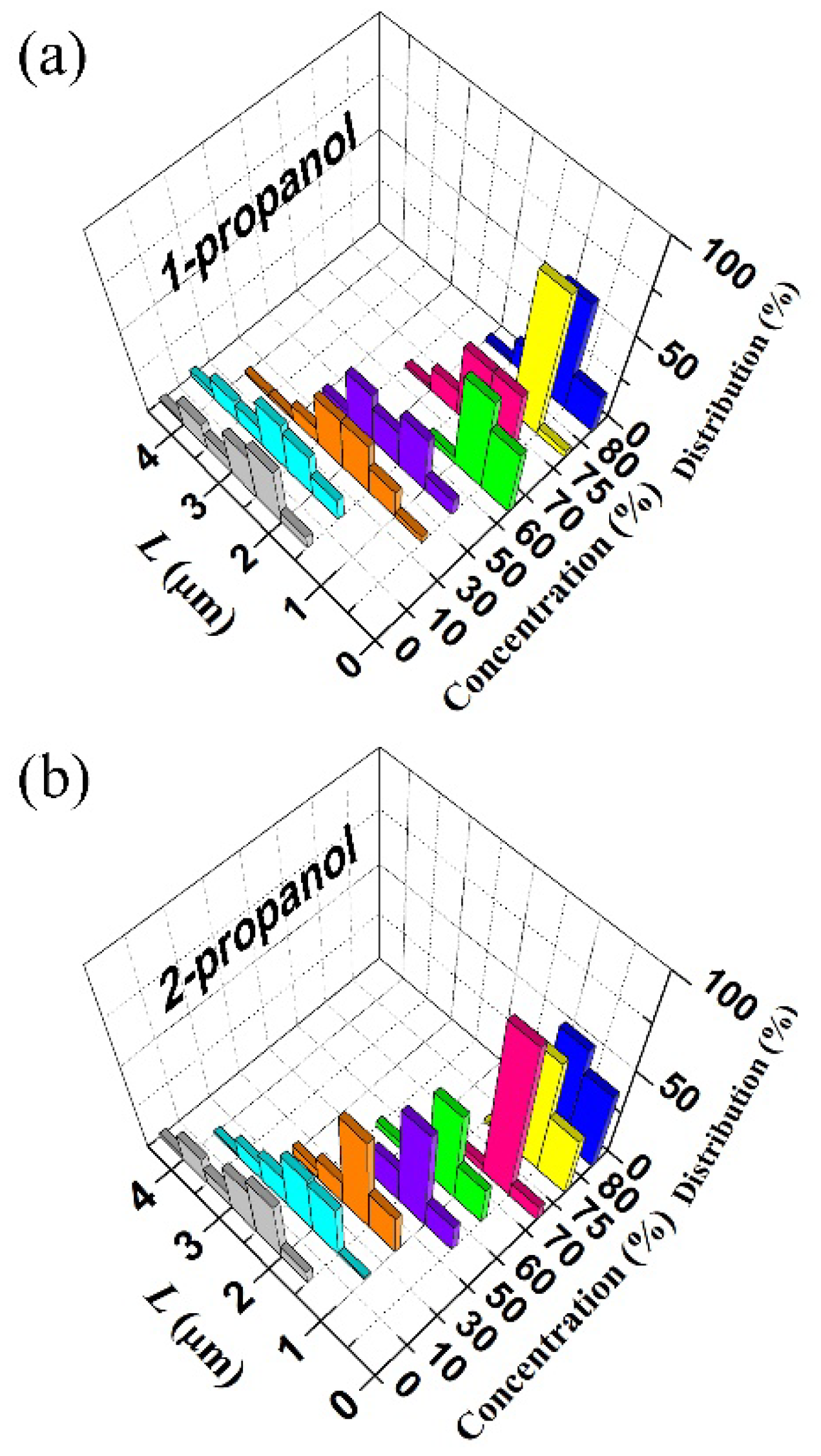
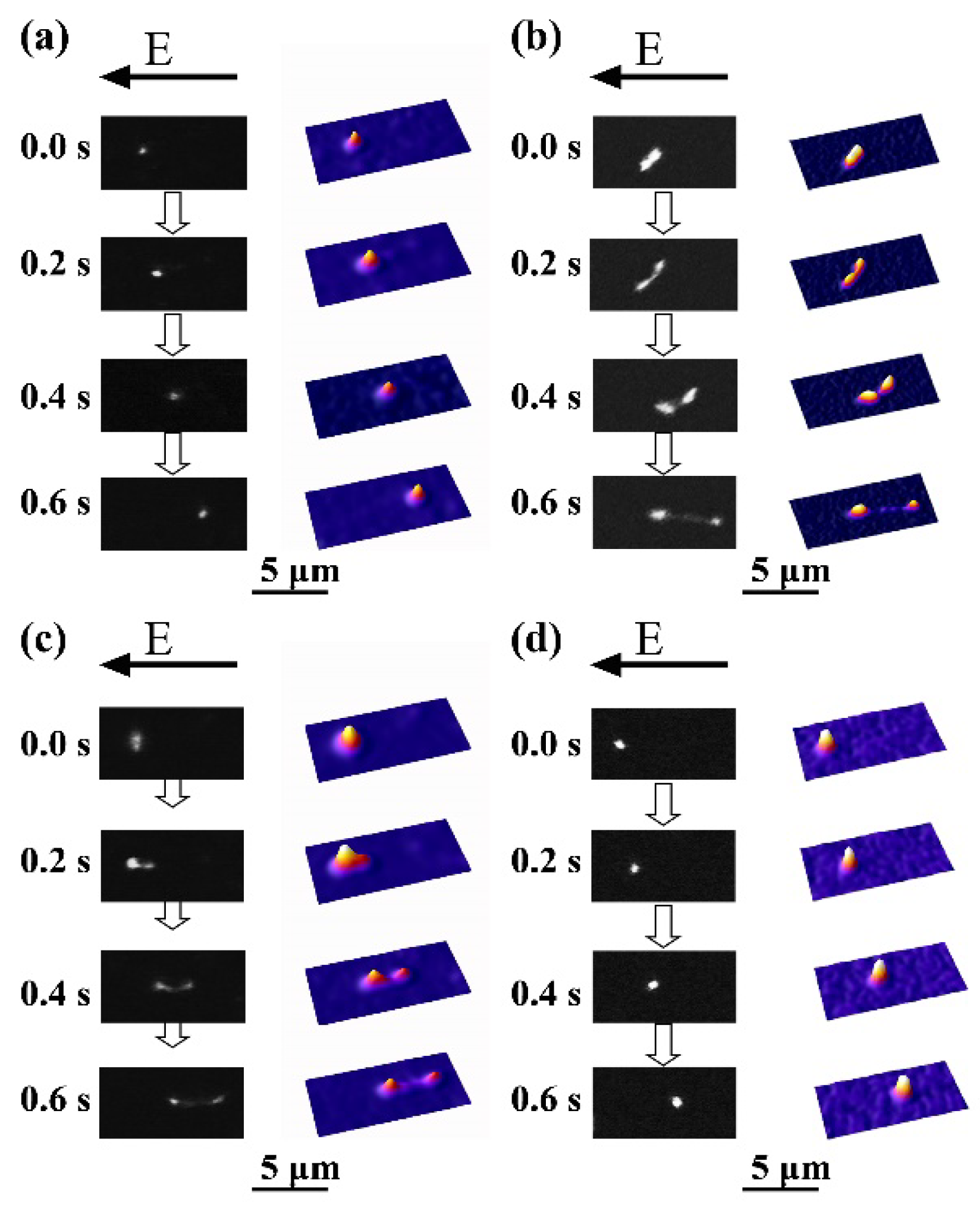
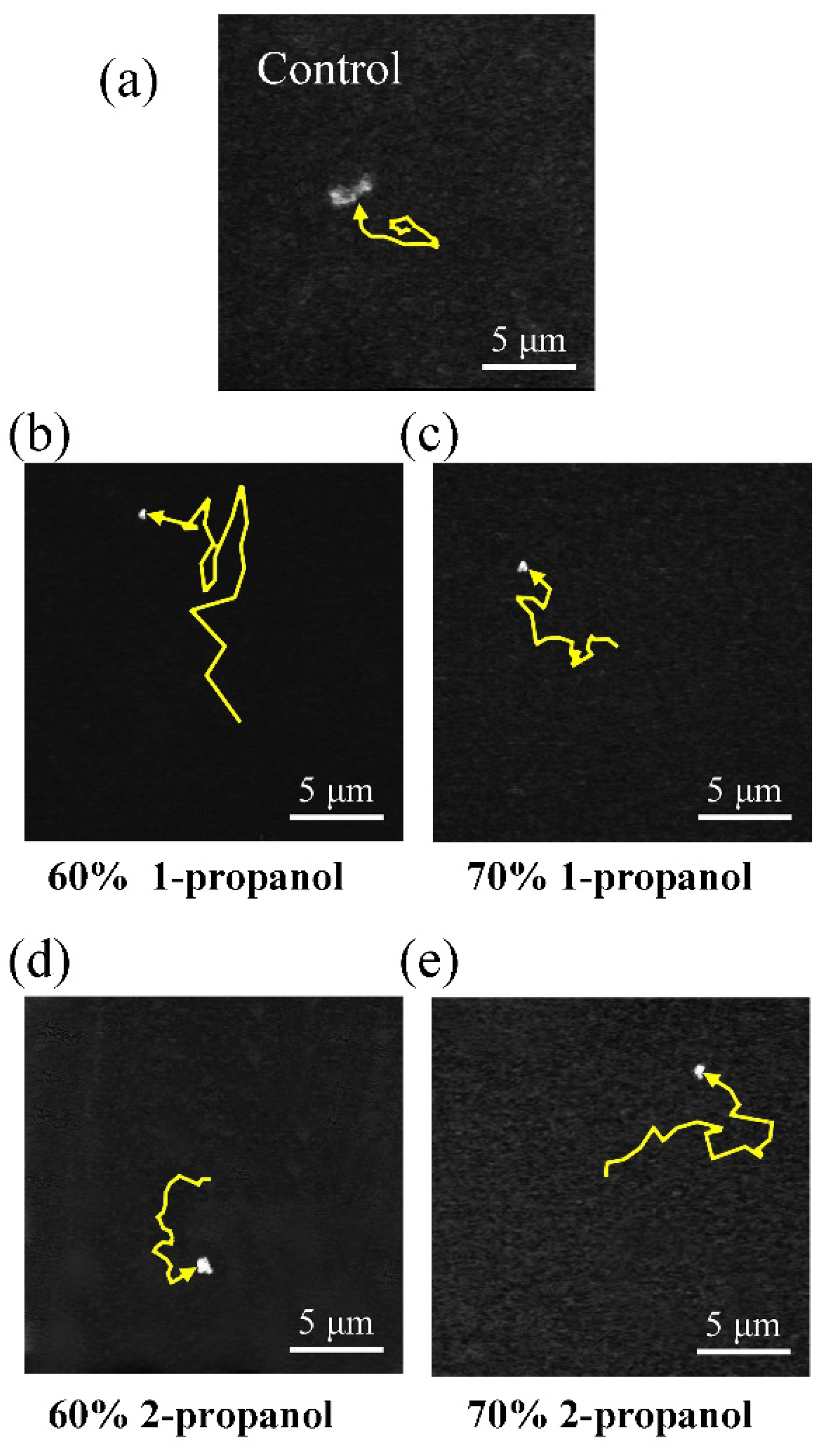
3.1. Higher-Order Structural Change of DNA Molecules with Alcohol
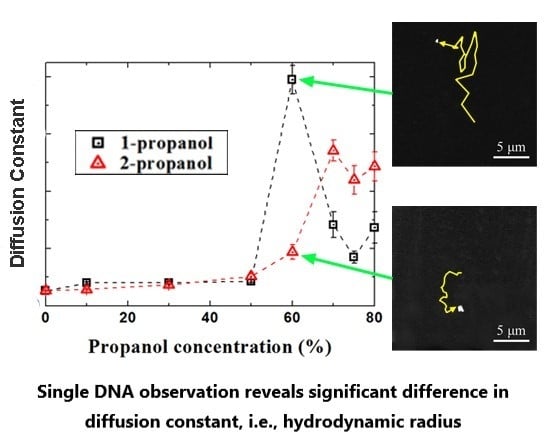
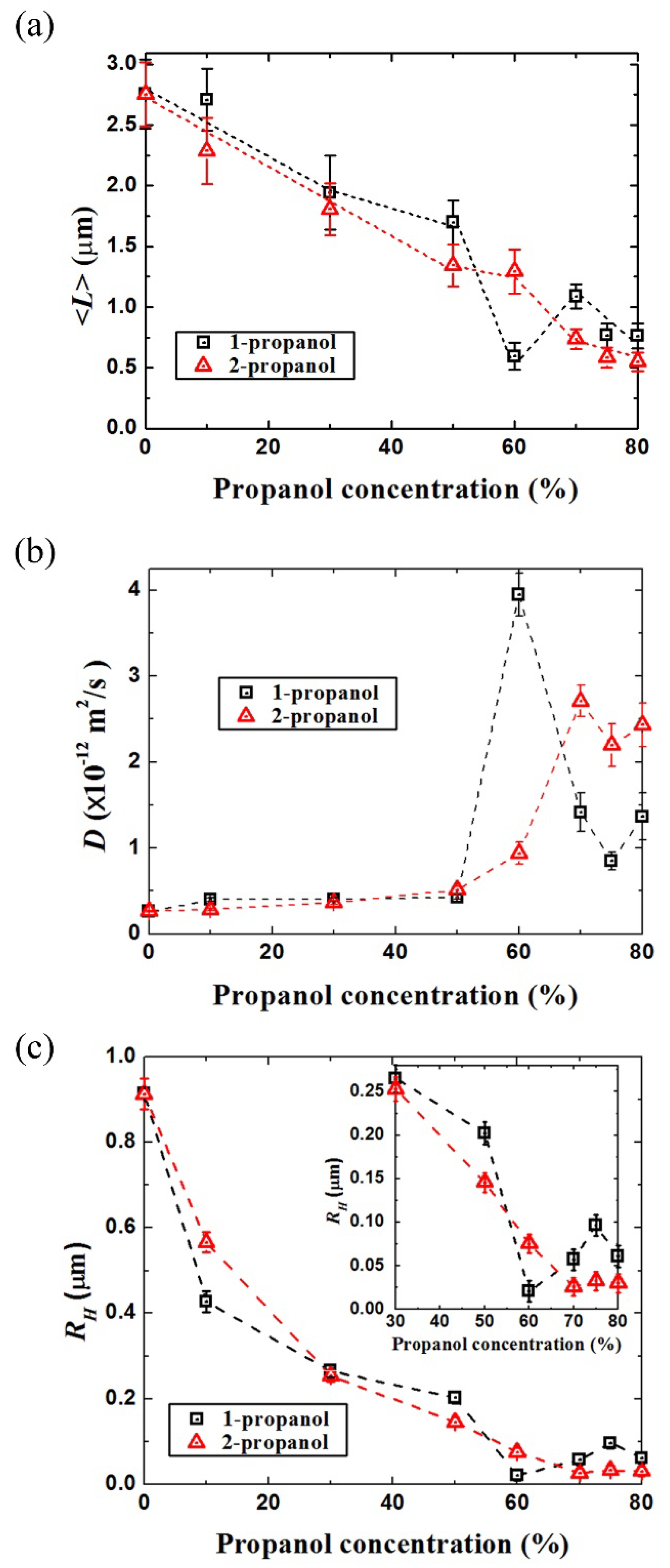
3.2. Analysis of the Brownian Motion of Single DNA
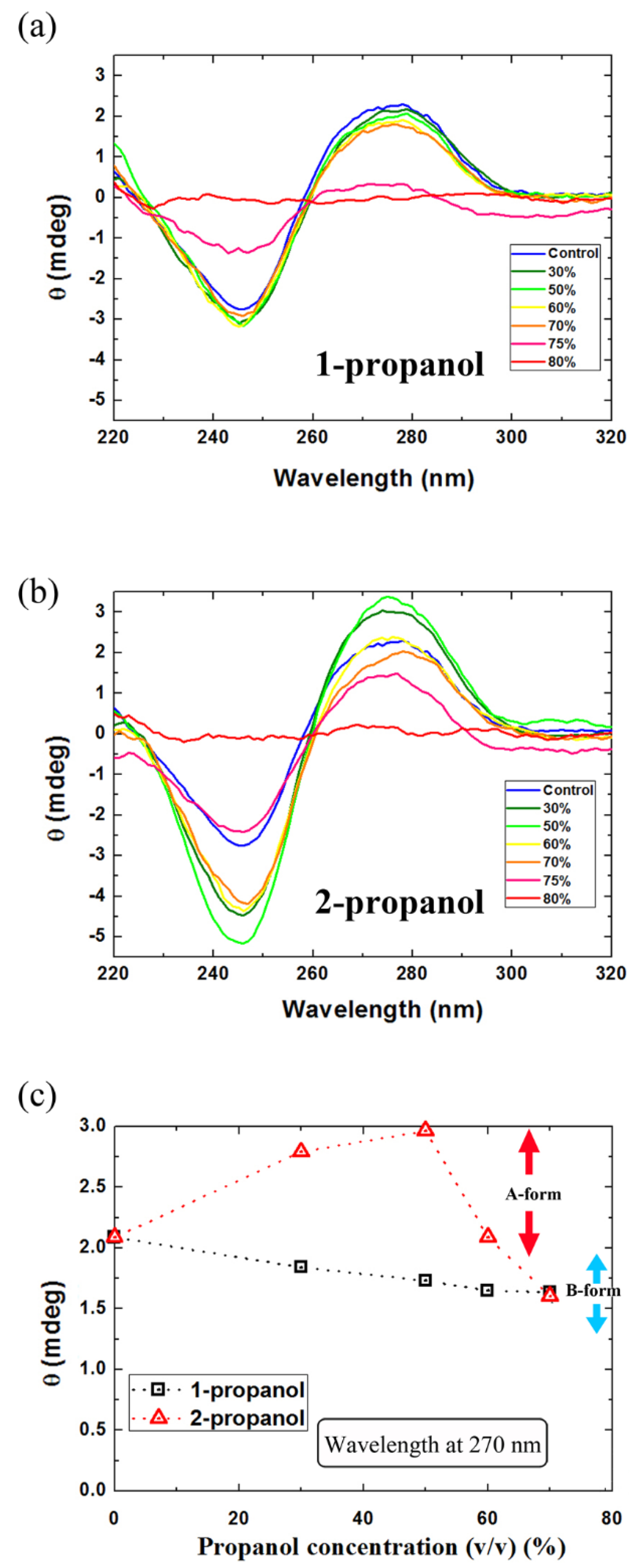
3.3. Secondary Structure of DNA Molecules in Alcohol Solutions
4. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Crouse, J.; Amorese, D. Ethanol precipitation: Ammonium acetate as an alternative to sodium acetate. Focus 1987, 9, 3–5. [Google Scholar]
- Gaillard, C.; Strauss, F.O. Ethanol precipitation of DNA with linear polyacrylamide as carrier. Nucleic Acids Res. 1990, 18, 378. [Google Scholar] [CrossRef] [Green Version]
- Horn, N.A.; Meek, J.A.; Budahazi, G.; Marquet, M. Cancer gene therapy using plasmid DNA: Purification of DNA for human clinical trials. Hum. Gene Ther. 1995, 6, 565–573. [Google Scholar] [CrossRef]
- Michaels, S.; John, M.; Amasino, R. Removal of polysaccharides from plant DNA by ethanol precipitation. Biotechniques 1994, 17, 274–276. [Google Scholar] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paithankar, K.; Prasad, K. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 1991, 19, 1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paterson, A.H.; Brubaker, C.L.; Wendel, J.F. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. 1993, 11, 122–127. [Google Scholar] [CrossRef]
- Piškur, J.; Rupprecht, A. Aggregated DNA in ethanol solution. FEBS Lett. 1995, 375, 174–178. [Google Scholar] [CrossRef] [Green Version]
- Raeder, U.; Broda, P. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1985, 1, 17–20. [Google Scholar] [CrossRef]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 241–245. [Google Scholar] [CrossRef]
- Zeugin, J.A.; Hartley, J.L. Ethanol precipitation of DNA. Focus 1985, 7, 1–2. [Google Scholar]
- Djurhuus, A.; Port, J.; Closek, C.J.; Yamahara, K.M.; Romero-Maraccini, O.; Walz, K.R.; Goldsmith, D.B.; Michisaki, R.; Breitbart, M.; Boehm, A.B. Evaluation of filtration and DNA extraction methods for environmental DNA biodiversity assessments across multiple trophic levels. Front. Mar. Sci. 2017, 4, 314. [Google Scholar] [CrossRef] [Green Version]
- Gallyamov, M.; Pyshkina, O.; Sergeev, V.; Yaminskii, I. DNA T4 condensation in water-alcohol media. Surf. Invest. 2001, 16, 1177–1182. [Google Scholar]
- Hinlo, R.; Gleeson, D.; Lintermans, M.; Furlan, E. Methods to maximise recovery of environmental DNA from water samples. PLoS ONE 2017, 12, e0179251. [Google Scholar] [CrossRef]
- Matzeu, M.; Onori, G.; Santucci, A. Condensation of DNA by monohydric alcohols. Colloids Surf. B 1999, 13, 157–163. [Google Scholar] [CrossRef]
- Ruiz, R.; Hoyuelos, F.J.; Navarro, A.M.; Leal, J.M.; García, B. Unequal effect of ethanol-water on the stability of ct-DNA, poly [(dA–dT)] 2 and poly (rA) poly (rU). Thermophysical properties. Phys. Chem. Chem. Phys. 2015, 17, 2025–2033. [Google Scholar] [CrossRef]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: The past and the present. Biomed. Res. Int. 2009, 2009, 574398/1–10. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ran, S.; Man, B.; Yang, G. Ethanol induces condensation of single DNA molecules. Soft Matter 2011, 7, 4425–4434. [Google Scholar] [CrossRef]
- Ali, S.H.; Tarakmah, A.; Merchant, S.Q.; Al-Sahhaf, T. Synthesis of esters: Development of the rate expression for the Dowex 50 Wx8-400 catalyzed esterification of propionic acid with 1-propanol. Chem. Eng. Sci. 2007, 62, 3197–3217. [Google Scholar] [CrossRef]
- Miyazawa, T.; Koso, S.; Kunimori, K.; Tomishige, K. Development of a Ru/C catalyst for glycerol hydrogenolysis in combination with an ion-exchange resin. Appl. Catal. A Gen. 2007, 318, 244–251. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Precipitation of DNA with isopropanol. Cold Spring Harb. Protoc. 2017, 2017, pdb–prot093385. [Google Scholar] [CrossRef]
- Ishizawa, M.; Kobayashi, Y.; Miyamura, T.; Matsuura, S. Simple procedure of DNA isolation from human serum. Nucleic Acids Res. 1991, 19, 5792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bednar, J.; Horowitz, R.A.; Grigoryev, S.A.; Carruthers, L.M.; Hansen, J.C.; Koster, A.J.; Woodcock, C.L. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA 1998, 95, 14173–14178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloomfield, V.A. DNA condensation by multivalent cations. Biopolymers 1997, 44, 269–282. [Google Scholar] [CrossRef]
- Daban, J.-R. Physical constraints in the condensation of eukaryotic chromosomes. Local concentration of DNA versus linear packing ratio in higher order chromatin structures. Biochemistry 2000, 39, 3861–3866. [Google Scholar] [CrossRef]
- Dias, R.S.; Innerlohinger, J.; Glatter, O.; Miguel, M.G.; Lindman, B. Coil-globule transition of DNA molecules induced by cationic surfactants: A dynamic light scattering study. J. Phys. Chem. B 2005, 109, 10458–10463. [Google Scholar] [CrossRef] [Green Version]
- Lyubchenko, Y.; Oden, P.; Lampner, D.; Lindsay, S.; Dunker, K. Atomic force microscopy of DNA and bacteriophage in air, water and propanol: The role of adhesion forces. Nucleic Acids Res. 1993, 21, 1117–1123. [Google Scholar] [CrossRef] [Green Version]
- Oda, Y.; Sadakane, K.; Yoshikawa, Y.; Imanaka, T.; Takiguchi, K.; Hayashi, M.; Kenmotsu, T.; Yoshikawa, K. Highly Concentrated Ethanol Solutions: Good Solvents for DNA as Revealed by Single-Molecule Observation. Chem. Phys. Chem. 2016, 17, 471–473. [Google Scholar] [CrossRef] [Green Version]
- Schaper, A.; Starink, J.P.; Jovin, T.M. The scanning force microscopy of DNA in air and in n-propanol using new spreading agents. FEBS Lett. 1994, 355, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Simpson, R.T.; Thoma, F.; Brubaker, J.M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: A model system for study of higher order structure. Cell 1985, 42, 799–808. [Google Scholar] [CrossRef]
- Tang, M.; Szoka, F. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther. 1997, 4, 823–832. [Google Scholar] [CrossRef] [Green Version]
- Ueda, M.; Yoshikawa, K. Phase transition and phase segregation in a single double-stranded DNA molecule. Phys. Rev. Lett. 1996, 77, 2133–2136. [Google Scholar] [CrossRef]
- Woodcock, C.L.; Ghosh, R.P. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect Biol. 2010, 2, a000596. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, Y.; Yoshikawa, K. Higher order structure of DNA controlled by the redox state of Fe2+/Fe3+. J. Am. Chem. Soc. 1997, 119, 10573–10578. [Google Scholar] [CrossRef]
- Yoshikawa, K. Controlling the higher-order structure of giant DNA molecules. Adv. Drug Deliv. Rev. 2001, 52, 235–244. [Google Scholar] [CrossRef]
- Schiessel, H. The physics of chromatin. J. Phys. Condens. Matter 2003, 15, R699. [Google Scholar] [CrossRef]
- Everaers, R.; Schiessel, H. The physics of chromatin. J. Phys. Condens. Matter 2015, 27, 060301. [Google Scholar] [CrossRef] [PubMed]
- Zinchenko, A.; Berezhnoy, N.V.; Wang, S.; Rosencrans, W.M.; Korolev, N.; van der Maarel, J.R.C.; Nordenskiöld, L. Single-molecule compaction of megabase-long chromatin molecules by multivalent cations. Nucleic Acids Res. 2018, 46, 635–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, Y.; Velichko, Y.S.; Ichiba, Y.; Yoshikawa, K. Self-assembled pearling structure of long duplex DNA with histone H1. Eur. J. Biochem. 2001, 268, 2593–2599. [Google Scholar] [CrossRef]
- Arnott, S.; Hukins, D. Optimised parameters for A-DNA and B-DNA. Biochem. Biophys. Res. Commun. 1972, 47, 1504–1509. [Google Scholar] [CrossRef]
- Arnott, S.; Selsing, E. The conformation of C-DNA. J. Mol. Biol. 1975, 98, 265–269. [Google Scholar] [CrossRef]
- Baase, W.A.; Johnson, W.C., Jr. Circular dichroism and DNA secondary structure. Nucleic Acids Res. 1979, 6, 797–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brahms, J.; Mommaerts, W. A study of conformation of nucleic acids in solution by means of circular dichroism. J. Mol. Biol. 1964, 10, 73–88. [Google Scholar] [CrossRef]
- Tunis-Schneider, M.J.B.; Maestre, M.F. Circular dichroism spectra of oriented and unoriented deoxyribonucleic acid films—A preliminary study. J. Mol. Biol. 1970, 52, 521–541. [Google Scholar] [CrossRef]
- Zimmer, C. Effects of the antibiotics netropsin and distamycin A on the structure and function of nucleic acids. Prog. Nucleic Acid Res. Mol. Biol. 1975, 15, 285–318. [Google Scholar]
- Matsumoto, M.; Sakaguchi, T.; Kimura, H.; Doi, M.; Minagawa, K.; Matsuzawa, Y.; Yoshikawa, K. Direct observation of brownian motion of macromolecules by fluorescence microscope. J. Polym. Sci. Part B Polym. Phys. 1992, 30, 779–783. [Google Scholar] [CrossRef]
- Araki, S.; Nakai, T.; Hizume, K.; Takeyasu, K.; Yoshikawa, K. Hydrodynamic radius of circular DNA is larger than that of linear DNA. Chem. Phys. Lett. 2006, 418, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, Y.; Teramoto, Y.; Yoshikawa, K. Disappearance of the Negative Charge in Giant DNA witha Folding Transition. Biophys. J. 2001, 80, 2823–2832. [Google Scholar] [CrossRef] [Green Version]
- Dünweg, B.; Reith, D.; Steinhauser, M.; Kremer, K. Corrections to scaling in the hydrodynamic properties of dilute polymer solutions. J. Chem. Phys. 2002, 117, 914–924. [Google Scholar] [CrossRef] [Green Version]
- Ladd, A.J.; Frenkel, D. Computer simulation studies of static and dynamical scaling in dilute solutions of excluded-volume polymers. Macromolecules 1992, 25, 3435–3438. [Google Scholar] [CrossRef] [Green Version]
- Pang, F.M.; Seng, C.E.; Teng, T.T.; Ibrahim, M.H. Densities and viscosities of aqueous solutions of 1-propanol and 2-propanol at temperatures from 293.15 K to 333.15 K. J. Mol. Liq. 2007, 136, 71–78. [Google Scholar] [CrossRef]
- Dixit, S.; Crain, J.; Poon, W.; Finney, J.; Soper, A. Molecular segregation observed in a concentrated alcohol-water solution. Nature 2002, 416, 829–832. [Google Scholar] [CrossRef]
- Großmann, G.H.; Ebert, K.H. Formation of Clusters in 1-Propanol/Water-Mixtures. Ber. Bunsenges. Phys. Chem. 1981, 85, 1026–1029. [Google Scholar] [CrossRef]
- Koga, Y. Fluctuations in aqueous methanol, ethanol, and propan-1-ol: Amplitude and wavelength of fluctuation. Can. J. Chem. 1999, 77, 2039–2045. [Google Scholar] [CrossRef]
- Matsumoto, M.; Nishi, N.; Furusawa, T.; Saita, M.; Takamuku, T.; Yamagami, M.; Yamaguchi, T. Structure of clusters in ethanol-water binary solutions studied by mass spectrometry and X-ray diffraction. Bull. Chem. Soc. Jpn. 1995, 68, 1775–1783. [Google Scholar] [CrossRef]
- Nishi, N.; Takahashi, S.; Matsumoto, M.; Tanaka, A.; Muraya, K.; Takamuku, T.; Yamaguchi, T. Hydrogen-bonded cluster formation and hydrophobic solute association in aqueous solutions of ethanol. J. Phys. Chem. 1995, 99, 462–468. [Google Scholar] [CrossRef]
- Turner, J.; Soper, A. The effect of apolar solutes on water structure: Alcohols and tetraalkylammonium ions. J. Chem. Phys. 1994, 101, 6116–6125. [Google Scholar] [CrossRef]
- Mijaković, M.; Kežić, B.; Zoranić, L.; Sokolić, F.; Asenbaum, A.; Pruner, C.; Wilhelm, E.; Perera, A. Ethanol-water mixtures: Ultrasonics, Brillouin scattering and molecular dynamics. J. Mol. Liq. 2011, 164, 66–73. [Google Scholar] [CrossRef]
- Riobóo, R.J.; Philipp, M.; Ramos, M.; Krüger, J.-K. Concentration and temperature dependence of the refractive index of ethanol-water mixtures: Influence of intermolecular interactions. Eur. Phys. J. E 2009, 30, 19. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Protoc. Mol. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Yoshikawa, Y.; Oana, H.; Yoshikawa, K. Marked Difference in the Conformational Transition of DNA Caused by Propanol Isomer. Polymers 2020, 12, 1607. https://doi.org/10.3390/polym12071607
Ma Y, Yoshikawa Y, Oana H, Yoshikawa K. Marked Difference in the Conformational Transition of DNA Caused by Propanol Isomer. Polymers. 2020; 12(7):1607. https://doi.org/10.3390/polym12071607
Chicago/Turabian StyleMa, Yue, Yuko Yoshikawa, Hidehiro Oana, and Kenichi Yoshikawa. 2020. "Marked Difference in the Conformational Transition of DNA Caused by Propanol Isomer" Polymers 12, no. 7: 1607. https://doi.org/10.3390/polym12071607
APA StyleMa, Y., Yoshikawa, Y., Oana, H., & Yoshikawa, K. (2020). Marked Difference in the Conformational Transition of DNA Caused by Propanol Isomer. Polymers, 12(7), 1607. https://doi.org/10.3390/polym12071607