Architecture and Composition Dictate Viscoelastic Properties of Organ-Derived Extracellular Matrix Hydrogels
Abstract
1. Introduction
2. Materials and Methods
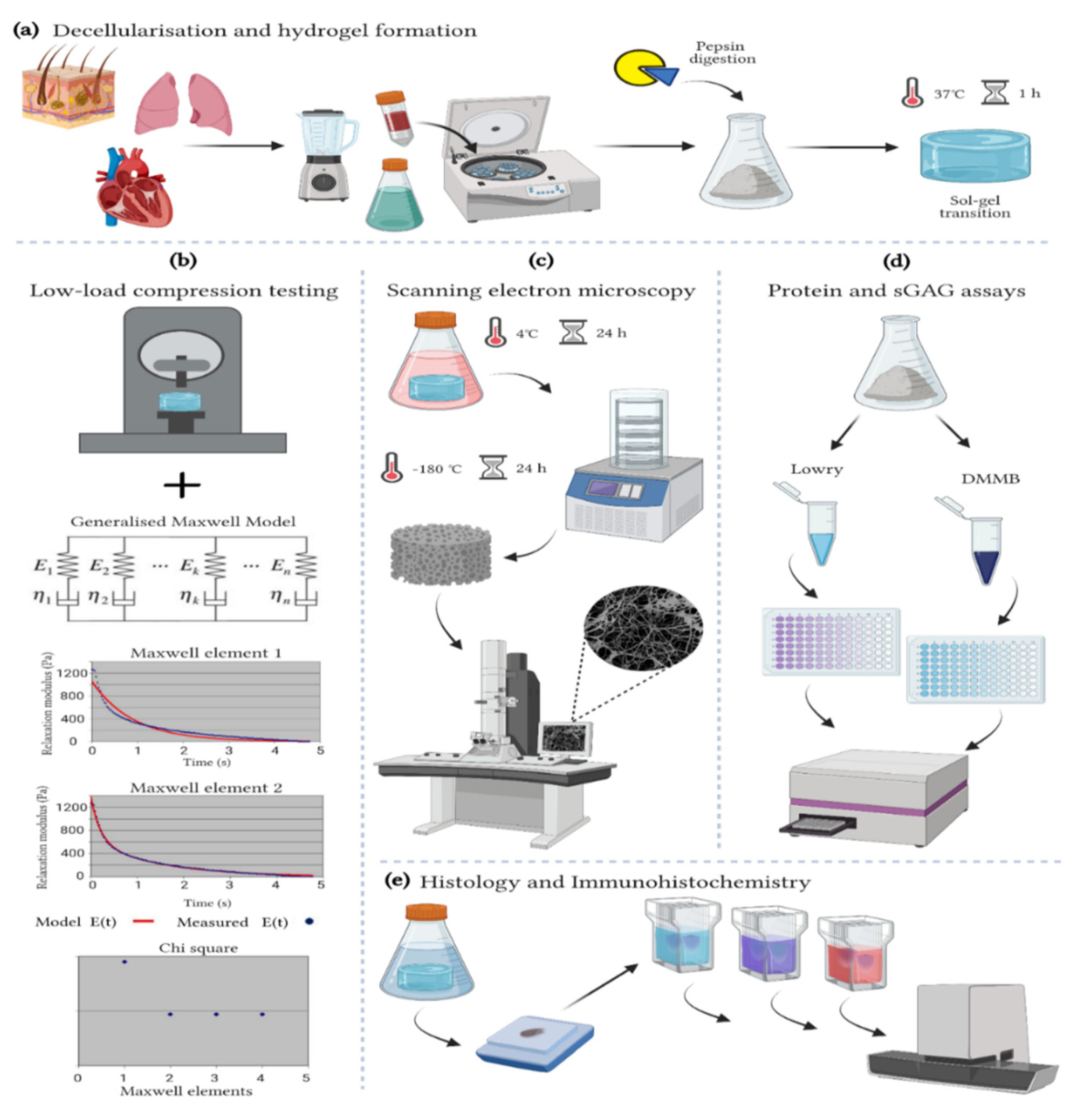
2.1. Decellularization
2.2. Hydrogel Preparation
2.3. Turbidity Assay
2.4. Viscoelastic Properties
2.5. Protein Quantification
2.6. Sulphated Glycosaminoglycans (sGAGs) Quantification
2.7. Histological Characterisation
2.8. Immunohistochemistry
2.9. Imaging and Image Analysis
2.10. Hydrogel Ultrastructure
2.11. Statistical Analysis
3. Results
3.1. Turbidity
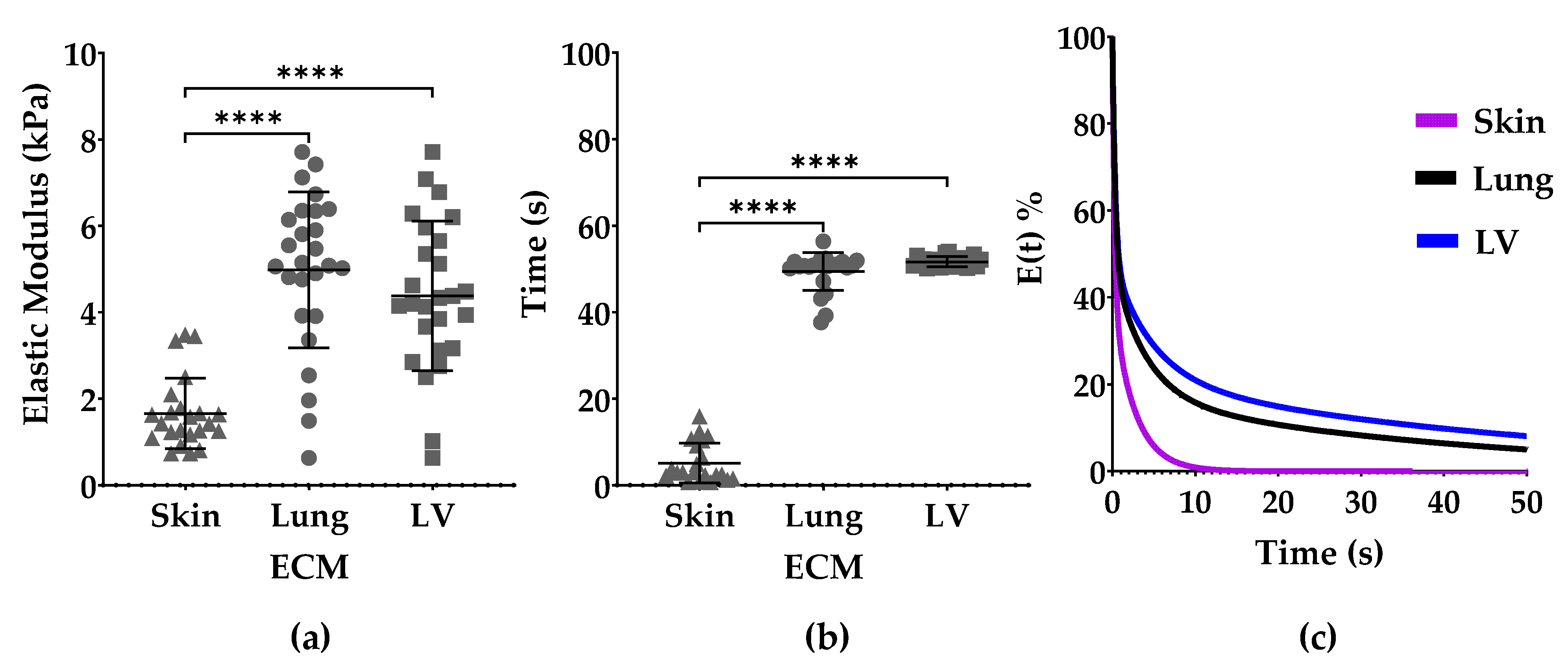
3.2. Elastic Modulus and Stress Relaxation
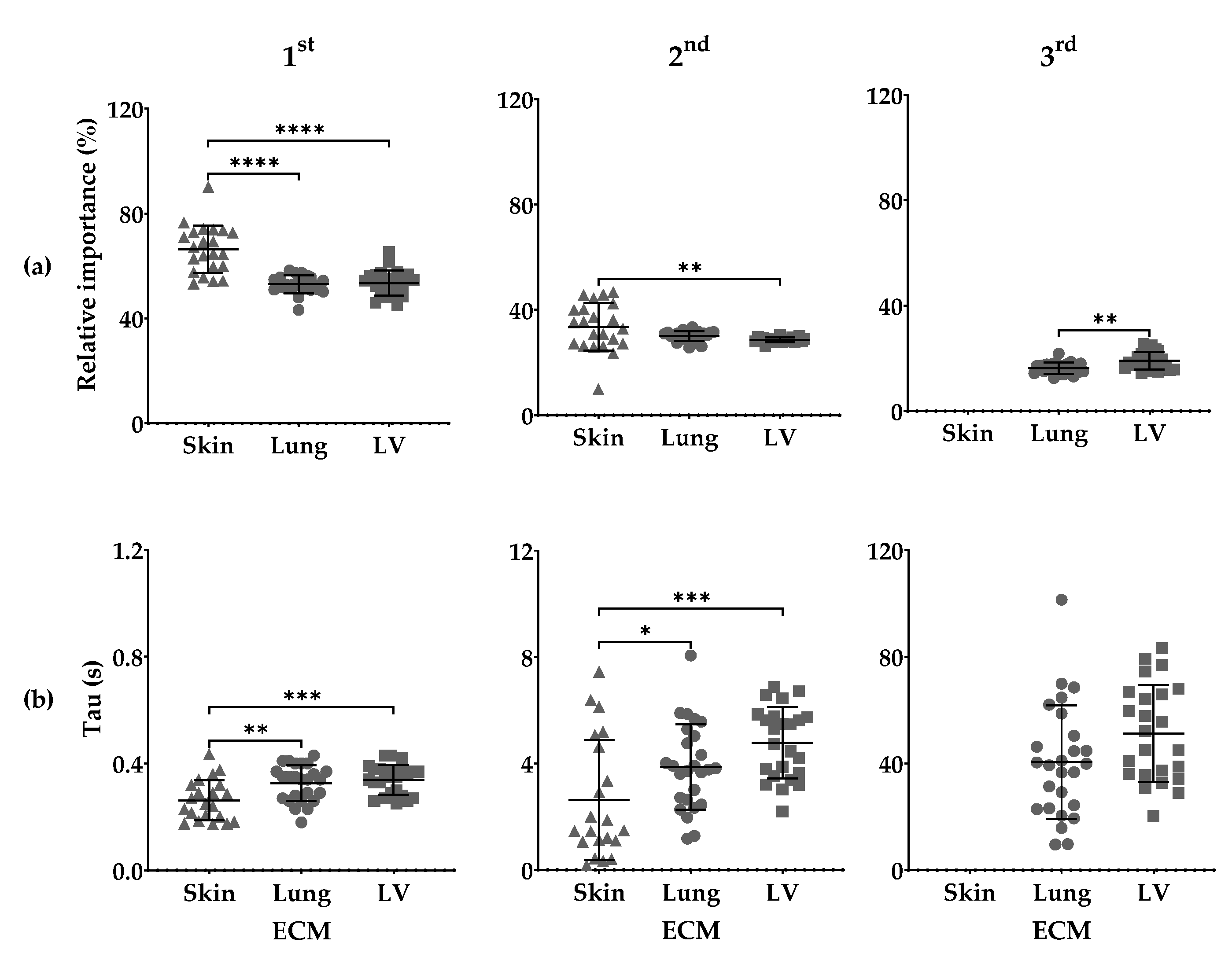
3.3. Maxwell Analysis
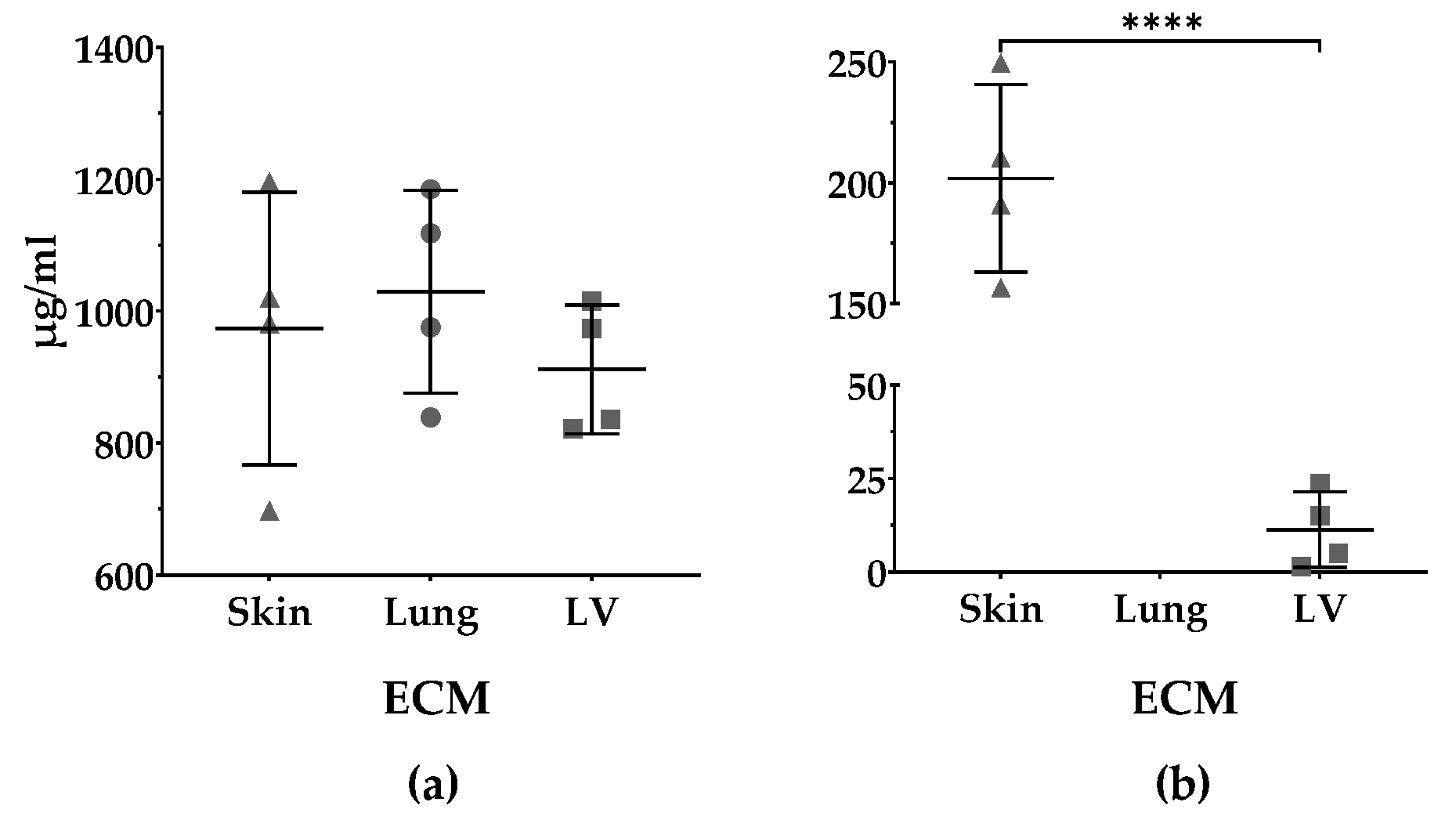
3.4. Protein and sGAGs Content
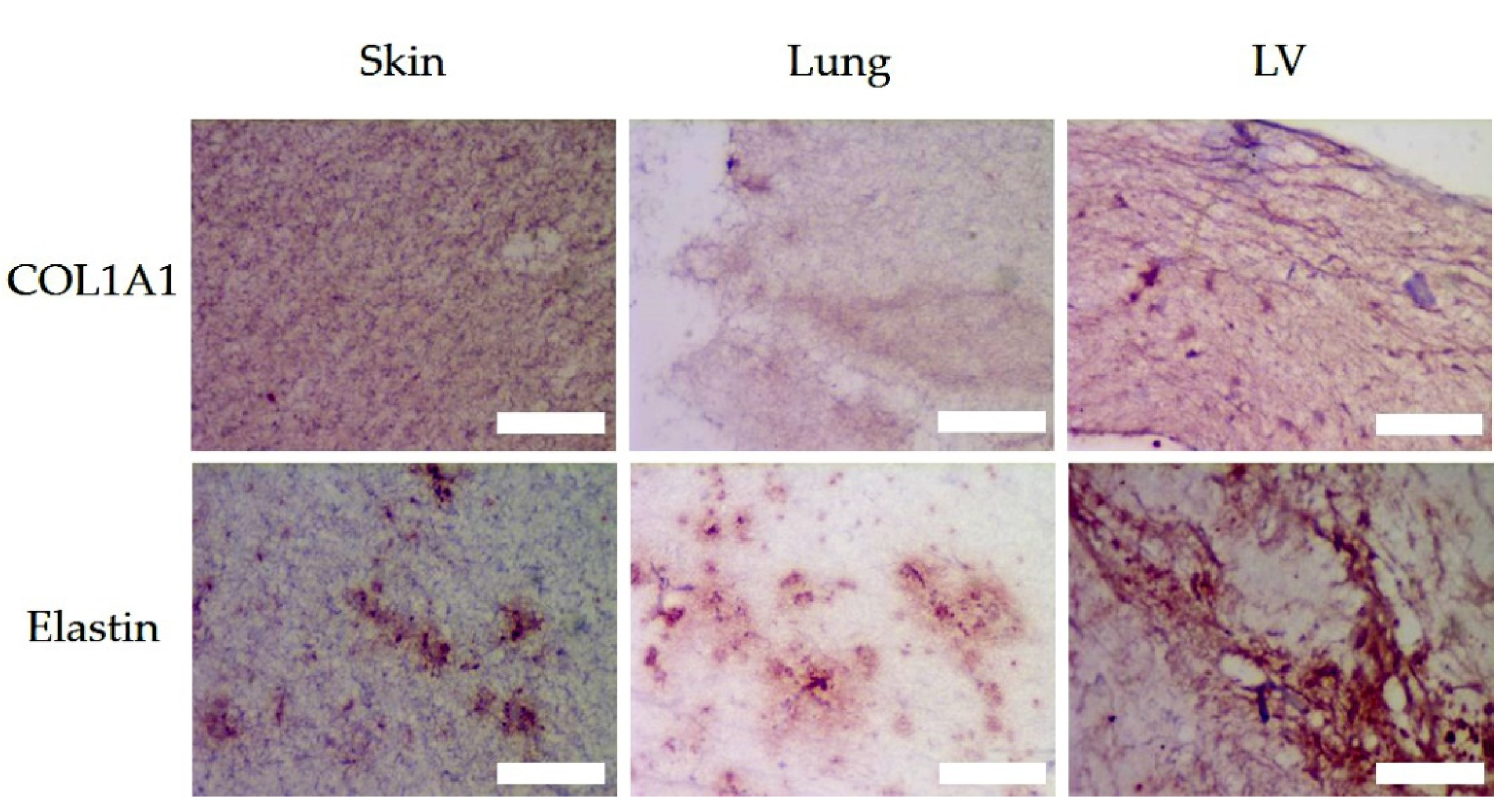
3.5. Histologic Assessment
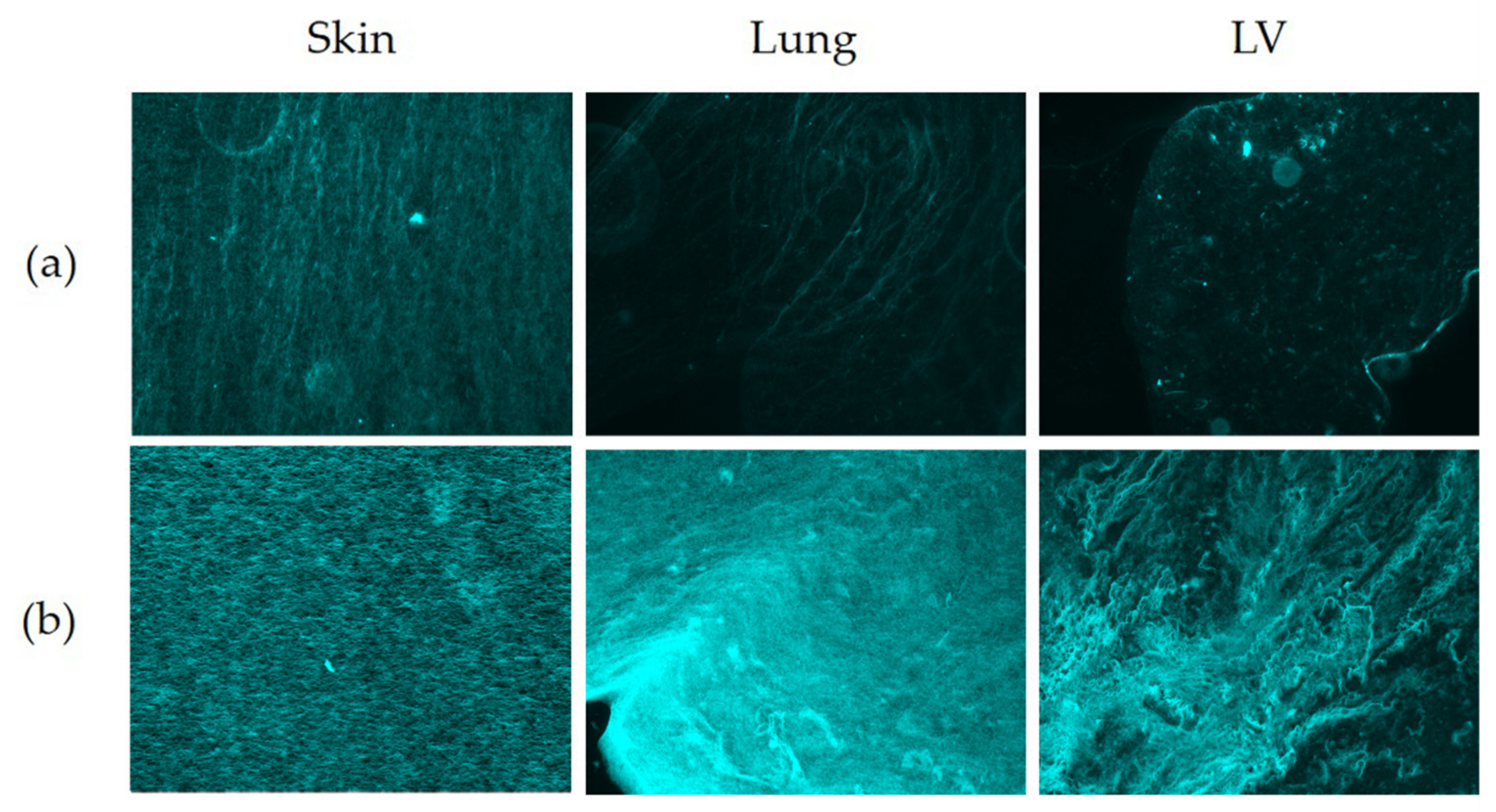
3.6. Matrix Organisation
3.6.1. Number of Fibers and Length (Mean and Total)
3.6.2. Branch Points and End Points
3.6.3. Lacunarity and High-Density Matrix (HDM)
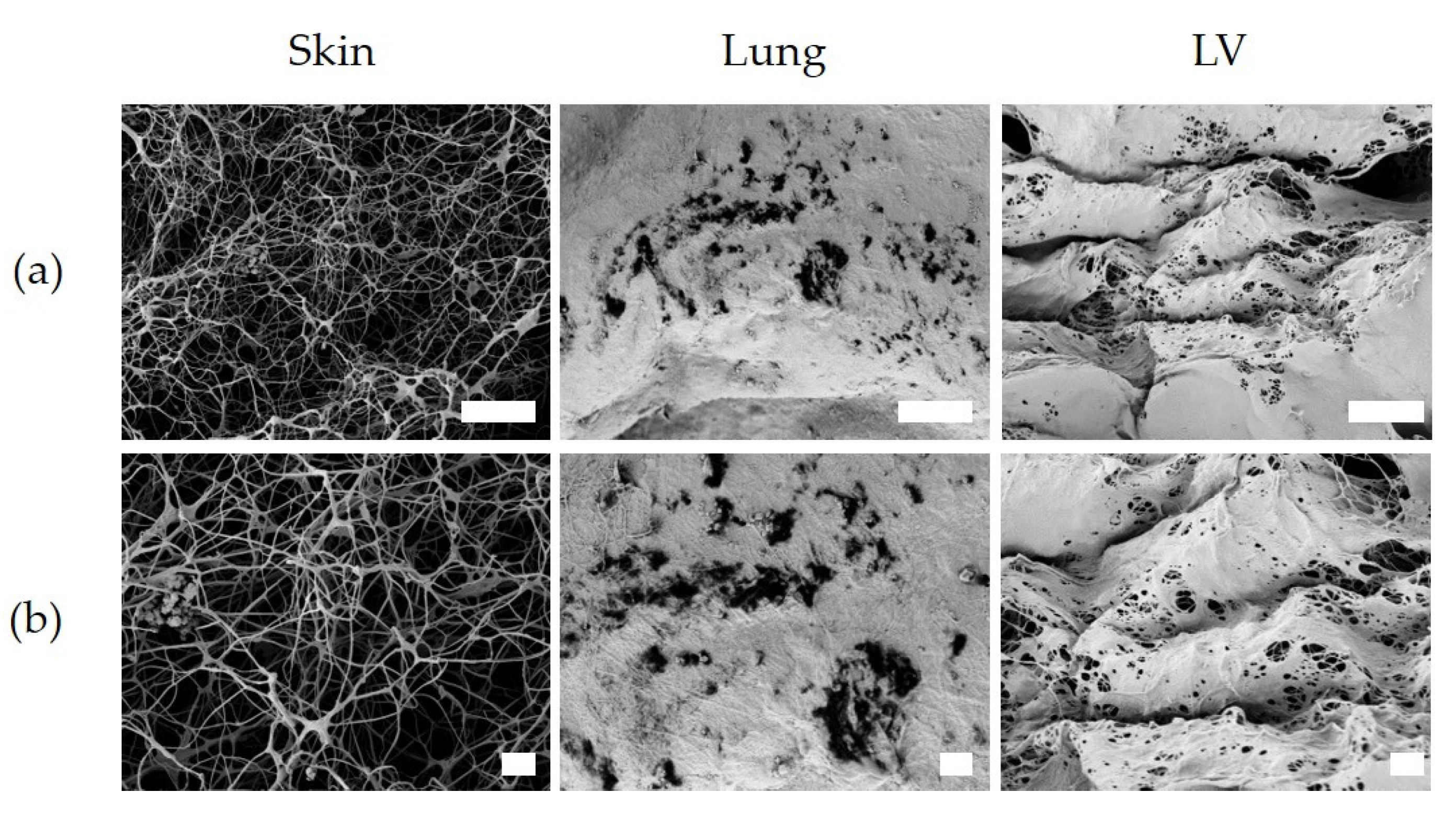
3.7. Ultrastrucure
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular Matrix Structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the Extracellular Matrix in Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Jana, S.; Hu, M.; Shen, M.; Kassiri, Z. Extracellular Matrix, Regional Heterogeneity of the Aorta, and Aortic Aneurysm. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef]
- Burgess, J.K.; Mauad, T.; Tjin, G.; Karlsson, J.C.; Westergren-Thorsson, G. The Extracellular Matrix—The Under-recognized Element in Lung Disease? J. Pathol. 2016, 240, 397–409. [Google Scholar] [CrossRef]
- Hastings, J.F.; Skhinas, J.N.; Fey, D.; Croucher, D.R.; Cox, T.R. The Extracellular Matrix as a Key Regulator of Intracellular Signalling Networks: ECM Regulation of Intracellular Signalling Networks. Br. J. Pharmacol. 2019, 176, 82–92. [Google Scholar] [CrossRef]
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Model. Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef]
- Elosegui-Artola, A. The extracellular matrix viscoelasticity as a regulator of cell and tissue dynamics. Curr. Opin. Cell Biol. 2021, 72, 10–18. [Google Scholar] [CrossRef]
- Wang, K. Die Swell of Complex Polymeric Systems. In Viscoelasticity—From Theory to Biological Applications; De Vicente, J., Ed.; InTech: Rijeka, Croatia, 2012; ISBN 978-953-51-0841-2. [Google Scholar]
- Guimarães, C.F.; Gasperini, L.; Marques, A.P.; Reis, R.L. The stiffness of living tissues and its implications for tissue engineering. Nat. Rev. Mater. 2020, 5, 351–370. [Google Scholar] [CrossRef]
- Chaudhuri, O.; Cooper-White, J.; Janmey, P.A.; Mooney, D.J.; Shenoy, V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 2020, 584, 535–546. [Google Scholar] [CrossRef]
- Chaudhuri, O.; Gu, L.; Klumpers, D.; Darnell, M.; Bencherif, S.A.; Weaver, J.C.; Huebsch, N.; Lee, H.-P.; Lippens, E.; Duda, G.N.; et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2015, 15, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Harjanto, D.; Zaman, M.H. Modeling Extracellular Matrix Reorganization in 3D Environments. PLoS ONE 2013, 8, e52509. [Google Scholar] [CrossRef][Green Version]
- Ahmed, E.M. Hydrogel: Preparation, characterization, and applications: A review. J. Adv. Res. 2013, 6, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Kim, H.-W. Emerging properties of hydrogels in tissue engineering. J. Tissue Eng. 2018, 9, 2041731418768285. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Mooney, D. Hydrogels for Tissue Engineering. Chem. Rev. 2001, 101, 1869–1880. [Google Scholar] [CrossRef]
- Laronda, M.M.; Rutz, A.; Xiao, S.; Whelan, K.A.; Duncan, F.E.; Roth, E.W.; Woodruff, T.K.; Shah, R.N. A bioprosthetic ovary created using 3D printed microporous scaffolds restores ovarian function in sterilized mice. Nat. Commun. 2017, 8, 15261. [Google Scholar] [CrossRef]
- Habib, A.; Sathish, V.; Mallik, S.; Khoda, B. 3D Printability of Alginate-Carboxymethyl Cellulose Hydrogel. Materials 2018, 11, 454. [Google Scholar] [CrossRef]
- Doyle, A.D.; Carvajal, N.; Jin, A.; Matsumoto, K.; Yamada, K.M. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat. Commun. 2015, 6, 8720. [Google Scholar] [CrossRef] [PubMed]
- Sapudom, J.; Rubner, S.; Martin, S.; Kurth, T.; Riedel, S.; Mierke, C.T.; Pompe, T. The phenotype of cancer cell invasion controlled by fibril diameter and pore size of 3D collagen networks. Biomaterials 2015, 52, 367–375. [Google Scholar] [CrossRef]
- Ivankova, E.; Dobrovolskaya, I.P.; Popryadukhin, P.; Kryukov, A.; Yudin, V.E.; Morganti, P. In-situ cryo-SEM investigation of porous structure formation of chitosan sponges. Polym. Test. 2016, 52, 41–45. [Google Scholar] [CrossRef]
- Cantini, M.; Donnelly, H.; Dalby, M.; Salmeron-Sanchez, M. The Plot Thickens: The Emerging Role of Matrix Viscosity in Cell Mechanotransduction. Adv. Health Mater. 2019, 9, e1901259. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-N.; Park, J.-C.; Kim, H.O.; Song, M.J.; Suh, H. Characterization of porous collagen/hyaluronic acid scaffold modified by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide cross-linking. Biomaterials 2001, 23, 1205–1212. [Google Scholar] [CrossRef]
- Annabi, N.; Mithieux, S.; Boughton, E.A.; Ruys, A.J.; Weiss, A.; Dehghani, F. Synthesis of highly porous crosslinked elastin hydrogels and their interaction with fibroblasts in vitro. Biomaterials 2009, 30, 4550–4557. [Google Scholar] [CrossRef]
- Mattson, J.M.; Turcotte, R.; Zhang, Y. Glycosaminoglycans contribute to extracellular matrix fiber recruitment and arterial wall mechanics. Biomech. Model. Mechanobiol. 2016, 16, 213–225. [Google Scholar] [CrossRef]
- Choudhury, D.; Tun, H.W.; Wang, T.; Naing, M.W. Organ-Derived Decellularized Extracellular Matrix: A Game Changer for Bioink Manufacturing? Trends Biotechnol. 2018, 36, 787–805. [Google Scholar] [CrossRef]
- Liguori, G.R.; Liguori, T.T.A.; De Moraes, S.R.; Sinkunas, V.; Terlizzi, V.; Van Dongen, J.A.; Sharma, P.; Moreira, L.F.P.; Harmsen, M.C. Molecular and Biomechanical Clues from Cardiac Tissue Decellularized Extracellular Matrix Drive Stromal Cell Plasticity. Front. Bioeng. Biotechnol. 2020, 8, 520. [Google Scholar] [CrossRef]
- Pouliot, R.A.; Link, P.; Mikhaiel, N.S.; Schneck, M.B.; Valentine, M.S.; Gninzeko, F.J.K.; Herbert, J.A.; Sakagami, M.; Heise, R.L. Development and characterization of a naturally derived lung extracellular matrix hydrogel. J. Biomed. Mater. Res. Part A 2016, 104, 1922–1935. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pérez, J.; Ahearne, M. The impact of decellularization methods on extracellular matrix derived hydrogels. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Pouliot, R.A.; Young, B.; Link, P.; Park, H.E.; Kahn, A.R.; Shankar, K.; Schneck, M.B.; Weiss, D.J.; Heise, R.L. Porcine Lung-Derived Extracellular Matrix Hydrogel Properties Are Dependent on Pepsin Digestion Time. Tissue Eng. Part C: Methods 2020, 26, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Kreger, S.T.; Bell, B.J.; Bailey, J.; Stites, E.; Kuske, J.; Waisner, B.; Voytik-Harbin, S.L. Polymerization and matrix physical properties as important design considerations for soluble collagen formulations. Biopolymers 2010, 93, 690–707. [Google Scholar] [CrossRef] [PubMed]
- De Hilster, R.H.J.; Sharma, P.; Jonker, M.R.; White, E.S.; A Gercama, E.; Roobeek, M.; Timens, W.; Harmsen, M.; Hylkema, M.N.; Burgess, J.K. Human lung extracellular matrix hydrogels resemble the stiffness and viscoelasticity of native lung tissue. Am. J. Physiol. Cell. Mol. Physiol. 2020, 318, L698–L704. [Google Scholar] [CrossRef]
- Van Dongen, J.A.; Getova, V.; Brouwer, L.A.; Liguori, G.R.; Sharma, P.; Stevens, H.P.; Van Der Lei, B.; Harmsen, M.C. Adipose tissue-derived extracellular matrix hydrogels as a release platform for secreted paracrine factors. J. Tissue Eng. Regen. Med. 2019, 13, 973–985. [Google Scholar] [CrossRef]
- Coulson-Thomas, V.; Gesteira, T. Dimethylmethylene Blue Assay (DMMB). Bio-Protocol 2014, 4, e1236. [Google Scholar] [CrossRef]
- Farndale, R.W.; Sayers, C.A.; Barrett, A. A Direct Spectrophotometric Microassay for Sulfated Glycosaminoglycans in Cartilage Cultures. Connect. Tissue Res. 1982, 9, 247–248. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; Lü, B. Tissue Preparation for Microscopy and Histology. In Comprehensive Sampling and Sample Preparation; Elsevier: Amsterdam, The Netherlands, 2012; pp. 53–93. ISBN 978-0-12-381374-9. [Google Scholar]
- Junqueira, L.C.U.; Bignolas, G.; Brentani, R.R. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. J. Mol. Histol. 1979, 11, 447–455. [Google Scholar] [CrossRef]
- Foot, N.C. The Masson Trichrome Staining Methods in Routine Laboratory Use. Stain. Technol. 1933, 8, 101–110. [Google Scholar] [CrossRef]
- Vogel, B.; Siebert, H.; Hofmann, U.; Frantz, S. Determination of collagen content within picrosirius red stained paraffin-embedded tissue sections using fluorescence microscopy. MethodsX 2015, 2, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Chem. Biol. 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Wershof, E.; Park, D.; Barry, D.J.; Jenkins, R.P.; Rullan, A.; Wilkins, A.; Schlegelmilch, K.; Roxanis, I.; I Anderson, K.; Bates, A.P.; et al. A FIJI macro for quantifying pattern in extracellular matrix. Life Sci. Alliance 2021, 4, e202000880. [Google Scholar] [CrossRef] [PubMed]
- Landini, G. Quantitative analysis of the epithelial lining architecture in radicular cysts and odontogenic keratocysts. Head Face Med. 2006, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Motulsky, H.J.; E Brown, R. Detecting outliers when fitting data with nonlinear regression–a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123–220. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, A.; Zahediasl, S. Normality Tests for Statistical Analysis: A Guide for Non-Statisticians. Int. J. Endocrinol. Metab. 2012, 10, 486–489. [Google Scholar] [CrossRef]
- Das, K.R. A Brief Review of Tests for Normality. Am. J. Theor. Appl. Stat. 2016, 5, 5–12. [Google Scholar] [CrossRef]
- Saldin, L.T.; Cramer, M.C.; Velankar, S.S.; White, L.J.; Badylak, S.F. Extracellular matrix hydrogels from decellularized tissues: Structure and function. Acta Biomater. 2016, 49, 1–15. [Google Scholar] [CrossRef]
- Birk, D.; Fitch, J.; Babiarz, J.; Doane, K.; Linsenmayer, T. Collagen fibrillogenesis in vitro: Interaction of types I and V collagen regulates fibril diameter. J. Cell Sci. 1990, 95, 649–657. [Google Scholar] [CrossRef]
- Brightman, A.O.; Rajwa, B.P.; Sturgis, J.E.; McCallister, M.E.; Robinson, J.P.; Voytik-Harbin, S.L. Time-Lapse Confocal Reflection Microscopy of Collagen Fibrillogenesis and Extracellular Matrix Assembly in Vitro. Biopolymers 2000, 54, 222–234. [Google Scholar] [CrossRef]
- Gelman, R.; Poppke, D.; Piez, K. Collagen fibril formation in vitro. The role of the nonhelical terminal regions. J. Biol. Chem. 1979, 254, 11741–11745. [Google Scholar] [CrossRef]
- Peterson, B.; van der Mei, H.C.; Sjollema, J.; Busscher, H.J.; Sharma, P.K. A Distinguishable Role of eDNA in the Viscoelastic Relaxation of Biofilms. mBio 2013, 4, e00497-13. [Google Scholar] [CrossRef]
- Colliec-Jouault, S. Skin tissue engineering using functional marine biomaterials. In Functional Marine Biomaterials; Elsevier: Amsterdam, The Netherlands, 2015; pp. 69–90. ISBN 978-1-78242-086-6. [Google Scholar]
- Al Jamal, R.; Roughley, P.J.; Ludwig, M.S. Effect of glycosaminoglycan degradation on lung tissue viscoelasticity. Am. J. Physiol. Cell. Mol. Physiol. 2001, 280, L306–L315. [Google Scholar] [CrossRef]
- Papakonstantinou, E.; Karakiulakis, G. The ‘sweet’ and ‘bitter’ involvement of glycosaminoglycans in lung diseases: Pharmacotherapeutic relevance. Br. J. Pharmacol. 2009, 157, 1111–1127. [Google Scholar] [CrossRef]
- Uhl, F.E.; Zhang, F.; Pouliot, R.A.; Uriarte, J.J.; Enes, S.R.; Han, X.; Ouyang, Y.; Xia, K.; Westergren-Thorsson, G.; Malmström, A.; et al. Functional role of glycosaminoglycans in decellularized lung extracellular matrix. Acta Biomater. 2019, 102, 231–246. [Google Scholar] [CrossRef]
- Karageorgiou, V.; Kaplan, D. Porosity of 3D biomaterial scaffolds and osteogenesis. Biomaterials 2005, 26, 5474–5491. [Google Scholar] [CrossRef]
- León, C.A.L.Y. New perspectives in mercury porosimetry. Adv. Colloid Interface Sci. 1998, 76–77, 341–372. [Google Scholar] [CrossRef]
- Roosa, S.M.M.; Kemppainen, J.M.; Moffitt, E.N.; Krebsbach, P.H.; Hollister, S.J. The pore size of polycaprolactone scaffolds has limited influence on bone regeneration in anin vivomodel. J. Biomed. Mater. Res. Part A 2010, 92, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Włodarczyk-Biegun, M.K. Faithful scanning electron microscopic (SEM) visualization of 3D printed alginate-based scaffolds. Bioprinting 2020, 20, e00098. [Google Scholar] [CrossRef]
- McKinlay, K.J.; Allison, F.J.; Scotchford, C.; Grant, D.; Oliver, J.M.; King, J.R.; Wood, J.V.; Brown, P. Comparison of environmental scanning electron microscopy with high vacuum scanning electron microscopy as applied to the assessment of cell morphology. J. Biomed. Mater. Res. 2004, 69, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Gungor-Ozkerim, P.S.; Zhang, Y.S.; Yue, K.; Zhu, K.; Liu, W.; Pi, Q.; Byambaa, B.; Dokmeci, M.R.; Shin, S.R.; et al. Direct 3D bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials 2016, 106, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Doyle, A.D.; Yamada, K.M. Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp. Cell Res. 2015, 343, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.; Hayn, A.; Mierke, C.T. Fast and reliable advanced two-step pore-size analysis of biomimetic 3D extracellular matrix scaffolds. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]


| Organ ECM | Amin | Amax | Tlag (min) | T1/2 (min) | Tend (min) | S (%/ min) |
|---|---|---|---|---|---|---|
| Skin | 2.3 ± 0.2 ****b | 3.5 ± 0.0 ****b | 13.1 ± 2.9 | 12.2 ± 2.9 ****b | 16.9 ± 3.2 ****b | 5.3 ± 0.8 ****b |
| Lung | 1.0 ± 0.1 ****c | 1.4 ± 0.2 ****c | 10.4 ± 4.7 | 43.2 ± 16.8 ****c | 94.1 ± 11.7 *c | 1.2 ± 0.3 |
| LV | 1.5 ± 0.1 ****a | 1.8 ± 0.2 ****a | 10.9 ± 6.5 | 20.9 ± 4.3 **a | 75.7 ± 17.5 ****a | 1.5 ± 0.6 ****a |
| COL1A1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Organ ECM | Number of Fibers | 1 Mean Fiber Length | 1 Total Fiber Length | Fiber Alignment | Number of Branch Points | Number of End Points | Lacunarity | 2 HDM^1000% |
| Skin | 15.3 ± 1.6 | 25.3 ± 0.5 | 8504 ± 2064 | 0.19 ± 0.04 | 121.3 ± 81.8 | 552 ± 95 | 14.7 ± 0.8 | 82.7 ± 44.0 |
| Lung | 14.9 ± 0.7 | 25.4 ± 1.2 | 28,316 ± 17,989 | 0.11 ± 0.07 | 350.7 ± 254.0 | 1902 ± 1179 | 16.7 ± 3.1 | 0.9 ± 0.1 *a |
| LV | 13.9 ± 1.6 | 24.3 ± 1.2 | 15,337 ± 785 | 0.11 ± 0.01 | 158.7 ± 39.0 | 1106 ± 45 | 21.6 ± 4.7 | 4.2 ± 2.7 *a |
| Elastin | ||||||||
| Skin | 10.7 ± 1.7 *c | 19.3 ± 2.3 *c | 2852 ± 270 **c | 0.04 ± 0.01 | 27.0 ± 7.8 *c | 273 ± 55c ***c | 95.2 ± 15.6 ***c | 9.1 ± 6.1 **c |
| Lung | 11.7 ± 0.7 | 26.0 ± 2.7 | 9349 ± 1330 *c | 0.04 ± 0.04 | 95.7 ± 23.2 *c | 808 ± 153 **c | 98.5 ± 16.0 ***c | 11.2 ± 0.9 **c |
| LV | 16.09 ± 2.60 | 26.0 ± 2.7 *a | 17,869 ± 5099 | 0.09 ± 0.12 | 259.0 ± 109.7 | 1101 ± 169 | 18.5 ± 1.7 | 89.5 ± 33.0 |
| Picrosirius Red (Fluorescent) | ||||||||
| Skin | 14.19 ± 1.08 *b | 27.8 ± 1.9 *b | 33,481 ± 20,453 *b | 0.07 ± 0.01 | 54.0 ± 41.7 | 2322 ± 1382 **b | 95.4 ± 32.4 ****b | 933.5 ± 35.1 |
| Lung | 16.71 ± 2.61 | 32.5 ± 5.0 | 18,818 ± 2972 | 0.07 ± 0.04 | 35.0 ± 16.7 | 18,818 ± 2972 *c | 530.0 ± 129.0 | 31.1 ± 7.0 ****a |
| LV | 12.66 ± 1.11 **b | 24.7 ± 2.1 **b | 30,346 ± 11,839 **b | 0.07 ± 0.02 | 62.5 ± 26.8 | 30,346 ± 11,839 ****a | 136.4 ± 29.0 ****b | 59.3 ± 35.7 ****a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Garcia, F.D.; de Hilster, R.H.J.; Sharma, P.K.; Borghuis, T.; Hylkema, M.N.; Burgess, J.K.; Harmsen, M.C. Architecture and Composition Dictate Viscoelastic Properties of Organ-Derived Extracellular Matrix Hydrogels. Polymers 2021, 13, 3113. https://doi.org/10.3390/polym13183113
Martinez-Garcia FD, de Hilster RHJ, Sharma PK, Borghuis T, Hylkema MN, Burgess JK, Harmsen MC. Architecture and Composition Dictate Viscoelastic Properties of Organ-Derived Extracellular Matrix Hydrogels. Polymers. 2021; 13(18):3113. https://doi.org/10.3390/polym13183113
Chicago/Turabian StyleMartinez-Garcia, Francisco Drusso, Roderick Harold Jan de Hilster, Prashant Kumar Sharma, Theo Borghuis, Machteld Nelly Hylkema, Janette Kay Burgess, and Martin Conrad Harmsen. 2021. "Architecture and Composition Dictate Viscoelastic Properties of Organ-Derived Extracellular Matrix Hydrogels" Polymers 13, no. 18: 3113. https://doi.org/10.3390/polym13183113
APA StyleMartinez-Garcia, F. D., de Hilster, R. H. J., Sharma, P. K., Borghuis, T., Hylkema, M. N., Burgess, J. K., & Harmsen, M. C. (2021). Architecture and Composition Dictate Viscoelastic Properties of Organ-Derived Extracellular Matrix Hydrogels. Polymers, 13(18), 3113. https://doi.org/10.3390/polym13183113






