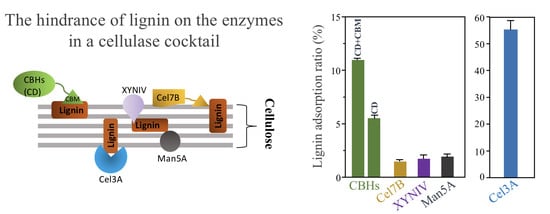
Comparative Evaluation of Adsorption of Major Enzymes in a Cellulase Cocktail Obtained from Trichoderma reesei onto Different Types of Lignin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enzyme Preparation
2.2. Lignin Isolation
2.3. Lignin Adsorption
2.4. Adsorption of Cellobiohydrolases (CBHs) onto Lignin
2.5. Adsorption of Endoglucanases (EGs) onto Lignin
2.6. Lignin Adsorption on the β-Glucosidases from Different Fungi
2.7. Lignin Adsorption on the Xylanases under Different Enzyme Conditions
2.8. Adsorption of Mannanase in the Cellulase Cocktail Obtained from T. reesei
3. Result and Discussion
3.1. Adsorption of Extracellular Enzymes of T. reesei onto Lignin
| Biomass | Chemical Composition (%) | Ref. | |||
|---|---|---|---|---|---|
| Guaiacyl Unit (G) | p-hydroxyphenyl Unit (H) | Syringyl Unit (S) | S/G Ratio | ||
| Hardwood | 25–50 | 0–8 | 46–75 | 1.50–1.84 | [28] |
| Populus tremuloides | 37.8 | 0.3 | 61.9 | 1.64 | [23] |
| Quercus suber | 44 | 1 | 55 | 1.2 | [31] |
| Eucalyptus globulus | 10 | 1 | 39 | 3.8 | [31] |
| Softwood | >95 | <5 | 0 | - | [28] |
| Pinus daeda | 98.3 | 1.7 | 0–1.3 | - | [23] |
| Herbaceous plants | |||||
| Rice straw | 71 | 5 | 22 | 0.31 | [29] |
| Kenaf | 40.9 | 1.0 | 58.1 | 1.40 | [30] |
| Corn stove | 42.2 | 44.9 | 12.9 | 0.31 | [32] |
| Arabidopsis | 77.1 | 2.8 | 20.1 | 0.26 | [23] |
3.2. Lignin Adsorption on Cellobiohydrolases (CBHs) and Individual Extracellular Enzyme of T. reesei
3.3. Adsorption of Endoglucanases onto Lignin
3.4. Adsorption on β-Glucosidases onto Lignin
3.5. Xylanase Adsorption by the Lignins
3.6. Adsorption of Mannanase onto Lignin
3.7. Summary of Lignin Adsorption of Major Enzymes in the Cellulase Cocktail of T. reesei
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anwar, Z.; Gulfraz, M.; Irshad, M. Agro-industrial lignocellulosic biomass a key to unlock the future bio-energy: A brief review. J. Radiation Res. App. Sci. 2014, 168–173. [Google Scholar] [CrossRef]
- Kahar, P. Synergistic effects of pretreatment process on enzymatic digestion of rice straw for efficient ethanol fermentation. Environ. Biotechnol. New Approaches Prospect. Appl. 2013, 73–77. [Google Scholar] [CrossRef] [Green Version]
- Himmel, H.E.; Ding, S.Y.; Johnson, D.K.; Adney, W.S.; Nimlos, M.R.; Brady, J.W.; Foust, T.D. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science 2007, 315, 804–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.S.; Lee, Y.G.; Song, Y.; Cho, E.J.; Bae, H.J. Hydrolysis patterns of xylem tissues of hardwood pretreated with acetic acid and hydrogen peroxide. Front. Energy Res. 2020, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Sun, J.; Leu, S.Y.; Chen, S.C. Toward a fundamental understanding of cellulase-lignin interactions in the whole slurry enzymatic saccharification process. Biofuels Bioprod. Bioref. 2016, 10, 648–663. [Google Scholar] [CrossRef]
- Valjamae, P.; Sild, V.; Pettersson, G.; Johansson, G. The initial kinetics of hydrolysis by cellobiohydrolases I and II is consistent with a cellulose surface- erosion model. Eur. J. Biochem. 1998, 253, 469–475. [Google Scholar] [CrossRef] [Green Version]
- Igarashi, K.; Uchihashi, T.; Koivula, A.; Mad, M.; Kumura, S.; Okamoto, T.; Penttila, M.; Ando, T.; Samejima, M. Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 2011, 333, 1279–1282. [Google Scholar] [CrossRef] [Green Version]
- Vermaas, J.V.; Petridis, L.; Qi, X.; Schulz, R.; Lindner, B.; Smith, J.C. Mechanism of lignin inhibition of enzymatic biomass deconstruction. Biotechnol. Biofuels 2015, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Sammond, D.W.; Yarbrough, J.M.; Mansfield, E.; Bomble, Y.J.; Hobdey, S.E.; Decker, S.R.; Taylor, L.E.; Resch, M.G.; Bozell, J.J.; Himmel, M.E.; et al. Predicting enzyme adsorption to lignin films by calculating enzyme surface hydrophobicity. J. Biol. Chem. 2014, 289, 20960–20969. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Wyman, C.E. Access of cellulase to cellulose and lignin for poplar solids produced by leading pretreatment technologies. Biotechnol. Prog. 2009, 25, 807–819. [Google Scholar] [CrossRef]
- Yoo, C.G.; Meng, X.; Pu, Y.; Ragauskas, A.J. The critical role of lignin in lignocellulosic biomass conversion and recent pretreatment strategies: A comprehensive review. Bioresour. Technol. 2020, 301, 122784. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Pan, X. Correlation between lignin physicochemical properties and inhibition to enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 2016, 113, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Berlin, A.; Balakshin, M.; Gilkes, N.; Kadla, J.; Maximenko, V.; Kubo, S.; Saddler, J. Inhibition of cellulase, xylanase and β-glucosidase activities by softwood lignin preparations. J. Biotechnol. 2006, 125, 198–209. [Google Scholar] [CrossRef]
- Lu, X.; Zheng, X.; Li, X.; Zhao, J. Adsorption and mechanism of cellulase enzymes onto lignin isolated from corn stover pretreated with liquid hot water. Biotechnol. Biofuels 2016, 9, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pareek, N.; Gillgren, T.; Jonsson, L.J. Adsorption of proteins involved in hydrolysis of lignocellulose on lignins and hemicelluloses. Bioresour. Technol. 2013, 148, 70–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, D.; Chundawat, S.P.S.; Uppugundla, N.; Balan, V.; Dale, B.E. Binding characteristics of Trichoderma reesei cellulases on untreated, ammonia fiber expansion (AFEX), and dilute-acid pretreated lignocellulosic biomass. Biotehcnol. Bioeng. 2011, 108, 1788–1800. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Shi, Z.; Yang, H.; Xu, G.; Zheng, Z.; Yang, J. Effect of alkaline lignin modification on cellulase-lignin interactions and enzymatic saccharification yield. Biotechnol. Biofuels 2018, 11, 1–13. [Google Scholar] [CrossRef]
- Guo, F.; Shi, W.; Sun, W.; Li, X.; Wang, F.; Zhao, J.; Qu, Y. Differences in the adsorption of enzymes onto lignins from diverse types of lignocellulosic biomass and the underlying mechanism. Biotechnol. Biofuels 2014, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wi, S.G.; Chung, B.Y.; Lee, Y.G.; Yang, D.J.; Bae, H.J. Enhanced enzymatic hydrolysis of rapeseed straw by popping pretreatment for bioethanol production. Bioresour. Technol. 2011, 5788–5793. [Google Scholar] [CrossRef]
- Wi, S.G.; Choi, I.S.; Kim, K.H.; Kim, H.M.; Bae, H.J. Bioethanol production from rice straw by popping pretreatment. Biotechnol. Biofules 2013, 6, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Luhtala, N.; Parker, R. LSM1 over-expression in Saccharomyces cerevisiae depletes U6 snRNA levels. Nucleic Acids Res. 2009, 37, 5529–5536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant. Biol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [Green Version]
- Mansfiled, S.D.; Kim, H.; Lu, F.; Ralph, J. Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 2012, 7, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Sewalt, V.J.H.; Glasser, W.G.; Beauchemin, K.A. Lignin impact on fiber degradation. 3. Reversal of inhibition of enzymatic hydrolysis by chemical modification of lignin and by addictives. J. Agric. Food. Chem. 1997, 45, 1823–1828. [Google Scholar] [CrossRef]
- Li, X.; Zheng, Y. Lignin-enzyme interaction: Mechanism, mitigation approach, modeling, and research prospects. Biotechnol. Adv. 2017, 35, 466–489. [Google Scholar] [CrossRef] [PubMed]
- Hage, R.E.; Brosse, N.; Chrusciel, L.; Sanchez, C.; Sannigrahi, P.; Ragauskas, A. Characterization of milled wood lignin and ethanol organosolv lignin from miscanthus. Polym. Degrad. Stab. 2009, 94, 1632–1638. [Google Scholar] [CrossRef]
- Tayyab, M.; Noman, A.; Islam, W.; Waheed, S.; Arafat, Y.; Ali, F.; Zaynab, M.; Lin, S.; Zhang, H.; Lin, W. Bioethanol production from lignocellulosic biomass by environment-friendly pretreatment methods: A review. Appl. Ecol. Environ. Res. 2018, 16, 225–249. [Google Scholar] [CrossRef]
- Liu, X.; Bouxin, F.; Fan, J.; Budarin, V.L.; Hu, C.; Clark, J.H. Recent advances in the catalytic depolymerization of lignin towards phenolic chemicals: A review. ChemSusChem. 2020, 13, 4296–4317. [Google Scholar] [CrossRef]
- Rosado, M.J.; Rencoret, J.; Marques, G.; Gutierrez, A.; Rio, J.C. Structural characteristics of the guaiacyl-rich lignins from rice (Oryza sativa L.) husks and straw. Front. Plant Sci. 2021, 12, 195. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Izumi, A.; Mazumder, B.B.; Ohtani, Y.; Sameshima, K. Characterization of kenaf (Hibiscus cannabinus) lignin by pyrolysis-gas chromatography-mass spectrometry in the presence of tetramethylammonium hydroxide. J. Anal. Appl. Pyrolysis 2002, 64, 453–463. [Google Scholar] [CrossRef]
- Yarbrough, J.M.; Mittal, A.; Mansfield, E.; Taylor, L.E.; Hobdey, S.E.; Sammond, D.W.; Blomble, Y.J.; Crowley, M.F.; Decker, S.R.; Himmel, M.E.; et al. New perspective on glycoside hydrolase binding to lignin from pretreated corn stover. Biotechnol. Biofuels 2015, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Adav, S.; Ravindran, L.T.; Chao, L.; Tan, S.; Singh, S.K. Proteomic analysis of pH and strains dependent protein secretion of Trichoderma reesei. J. Proteome Res. 2011, 10, 4579–4596. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, A.; Pereira, H. Chapter 3: Compositional variability of lignin in biomass. In Lignin-Trends and Applications; IntechOpen: London, UK, 2017; pp. 65–98. [Google Scholar]
- Mouthier, T.; Aledoorm, M.M.; Pel, H.; Schols, H.A.; Gruppen, H.; Kabel, M.A. Corn stover lignin is modified differently by acetic acid compared to sulfuric acid. Industrial Crops Products 2018, 121, 160–168. [Google Scholar] [CrossRef]
- Biely, P.; Puchart, V.; Stringer, M.A.; Morkeberg Krogh, K.B.R. Trichoderma reesei XYN VI- a novel appendage-dependent eukaryotic glucuronoxylan hydrolase. FEBS J. 2014, 281, 3894–3903. [Google Scholar] [CrossRef] [PubMed]
- Herpoel-Gimbert, I.; Margeot, A.; Dolla, A.; Jan, G.; Molle, D.; Lignon, S.; Mathis, H.; Sigoillot, J.C.; Monot, F.; Asther, M. Comparative secretome analyses of two Trichoderma reesei RUT-30 and CL847 hypersecretory strains. Biotechnol. Biofuels 2008, 18. [Google Scholar]
- Ramoni, J.; Darchetti-Deschman, M.; Seidl-Seiboth, V. Trichoderma reesei xylanase 5 is defective in the reference strain QM6a but functional alleles are present in other wild-type strains. App. Microbiol. Biotechnol. 2017, 101, 4139–4149. [Google Scholar] [CrossRef] [Green Version]
- Palonene, H.; Tjerneld, F.; Zacchi, G.; Tenkanen, M. Adsorption of Trichoderma reesei CBH1 and EG II and their catalytic domains on steam pretreated softwood and isolated lignin. J. Biotechnol. 2004, 107, 65–72. [Google Scholar] [CrossRef]
- Rahikainen, J.L.; Martin-Sampeder, R.; Hikkinen, H.; Rovo, S.; Marjamaa, K.; Tamminen, T.; Rojas, O.J.; Kruus, K. Inhibitory effect of lignin during cellulose bioconversion: The effect of lignin chemistry on non-productive enzyme adsorption. Bioresour. Technol. 2013, 133, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, Y.; Nagata, T.; Suetomi, T.; Oshiro, S.; Kondo, K.; Katahira, M.; Watanabe, T. NMR analysis on molecular interaction of lignin with amino acid residues of carbohydrate-binding module from Trichoderma reesei Cel7A. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.K.; Ximenes, E.; Kim, Y.; Laisch, M.R. Adsorption of enzyme onto lignin of liquid hot water pretreated hardwoods. Biotechnol. Bioeng. 2015, 112, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Singh, S.; Singh, O.V. Bioconversion of lignocellulosic biomass: Biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 2008, 35, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Carli, S.; Carneiro, L.A.B.D.C.; Ward, R.J.; Meleiro, L.P. Immobilization of a β-glucosidase and an endoglucanase in ferromagnetic nanoparticles: A study of synergistic effects. Protein Expr. Purif. 2019, 160, 28–35. [Google Scholar] [CrossRef]
- Guo, B.; Sato, N.; Biely, P.; Amano, Y.; Nozaki, K. Comparison of catalytic properties of multiple β-glucosidases of Trichoderma reesei. Appl. Microbiol. Biotechnol. 2016, 100, 4959–4968. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.L.; Chang, C.K.; Jeng, W.Y.; Wang, A.H.J.; Kiang, P.H. Mutations in the substrate entrance region of β-glucosidase from Trichoderma reesei improve enzyme activity and thermostability. Protein Eng. Des. Sel. 2012, 25, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Murphy, L.; Bohlin, C.; Baumann, M.J.; Olsen, S.N.; Sorensen, T.H.; Anderson, L.; Borch, K.; Westh, P. Product inhibition of five Hypocrea jecorina cellulases. Enzyme Microb. Technol. 2013, 52, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Yeo, S.L.; Shazilah, K.; Suhaila, S.; Abu, B.F.D.; Murad, A.M.A. In-silico analysis of Aspergillus niger beta-glucosidases. AIP Conf. Proc. 2014, 1614, 537–5543. [Google Scholar] [CrossRef] [Green Version]
- Souza, W.R.D.; Gouvea, P.F.D.; Saboldi, M.S.; Malavazi, I.; Bernardes, L.A.D.S.; Goldman, M.H.S.; Vries, R.P.D.; Oliveiar, J.V.D.C.; Goldman, G.H. Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnol. Biofuels. 2011, 4, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Seidle, H.F.; Marten, I.; Shoseyov, O.; Huber, R.E. Physical and kinetic properties of the family 3 β-glucosidase from Aspergillus niger which is important for cellulose breakdown. Protein. J. 2004, 23, 11–23. [Google Scholar] [CrossRef]
- Junior, A.B.; Borges, D.G.; Tardioli, P.W.; Farinas, C.S. Characterization of glucosidase produced by Aspergillus niger under solid-state fermentation and partially purified using MANAE-Agarose. Biotechnol. Res. Inter. 2014. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Shi, D.; Yang, S.; Lin, H.; Chen, H. Identification of an intracellular β-glucosidase in Aspergillus niger with transglycosylation activity. App. Microbiol. Biotechnol. 2020, 104, 8367–8380. [Google Scholar] [CrossRef]
- Maki-Arvela, P.; Salmi, T.; Holmbom, B.; Willfor, S.; Murzin, D.Y. Synthesis of sugars by hydrolysis of hemicelluloses-A review. Chem. Rev. 2011, 111, 5638–5666. [Google Scholar] [CrossRef]
- Quyang, J.; Yan, M.; Kong, D.; Xu, L. A complete protein pattern of cellulase and hemicellulose genes in the filamentous fungus Trichoderma reesei. Biotechnol. J. 2006, 1, 1266–1274. [Google Scholar]
- Qing, Q.; Yang, B.; Wyman, C.E. Xylooligomers are strong inhibitors of cellobiose hydrolysis by enzymes. Bioresour. Technol. 2010, 101, 9624–9630. [Google Scholar] [CrossRef]
- Shivastava, S.; Shukla, P.; Mukhopadhyay, K. Purification and preliminary characterization of a xylanase from Thermomyces lanuginosus strain SS-8. 3 Biotech. 2011, 1, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Winger, A.M.; Heazlewood, J.L.; Chan, L.J.G.; Petzold, C.J.; Permaul, K.; Singh, S. Secretome analysis of the thermophilic xylanase hyper-producer Thermomyces lanuginosus SSBP cultivated on corn cobs. J. Ind. Microbiol. Biotechnol. 2014, 41, 1687–1696. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.J.; Feng, Y.L.; Cao, Q.L.; Huang, M.Y.; Feng, J.X. Identification of a novel family of carbohydrate-binding modules with broad ligand specificity. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, P.S.; Puri, N.; Sharma, P.L.; Gupta, N. Mannanases: Microbial sources, production, properties and potential biotechnological applications. Appl. Microbiol. Biotehcnol. 2012, 93, 1817–1830. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.H.; Davidson, K.; Rixon, J.E.; Halstead, J.R.; Fransen, M.P.; Gilbert, H.J.; Hazlewood, G.P. A comparison of enzyme-aided bleaching of softwood paper pulp using combinations of xylanase, mannanase and α-galactosidase. Appl. Microbiol. Biotechnol. 2000, 53, 661–667. [Google Scholar] [CrossRef] [PubMed]
- von Freiesleben, P.; Spodsberg, N.; Stenbak, A.; Stalbrand, H.; Krogh, K.B.R.M.; Meyer, A.S. Boosting of enzymatic softwood saccharification by fungal GH5 and GH26 endomannanases. Biotechnol. Biofuels. 2018, 11, 194. [Google Scholar] [CrossRef]
- Varnai, A.; Huikko, L.; Pere, J.; Siika-aho, M.; Viikari, L. Synergistic action of xylanase and mannanase improves the total hydrolysis of softwood. Bioresour. Technol. 2011, 102, 9096–9104. [Google Scholar] [CrossRef]
- Hagglund, P.; Eriksson, T.; Collen, A.; Nerinckx, W.; Claeyssens, M.; Stalbrand, H. A cellulose-binding module of the Trichoderma reesei β-mannanase Man5A increases the mannan-hydrolysis of complex substrates. J. Biotechnol. 2003, 101, 37–48. [Google Scholar] [CrossRef]






| Molecular Weight (kDa) | Lignin Adsorption Rate (%) | ||||
|---|---|---|---|---|---|
| Oak | Pine | Rice Straw | Kenaf | ||
| Cellobiohydrolase | |||||
| CD 1 + CBM 2 | 55 kDa | 14.61 ± 1.94 | 7.40 ± 1.63 | 8.74 ± 3.27 | 12.52 ± 2.69 |
| CD | 48 kDa | 4.00 ± 0.84 | 8.77 ± 0.42 | 7.73 ± 1.51 | 1.63 ± 0.17 |
| CD/CD+CBM | 0.27 | 1.19 | 0.88 | 0.14 | |
| Endoglucanase | |||||
| Cel7B | 55 kDa | 1.21 ± 0.14 | 1.02 ± 0.05 | 1.96 ± 0.34 | 2.09 ± 0.37 |
| ꞵ-glucosidase | |||||
| Cel3A | 81 kDa | 51.66 ± 8.66 | 56.71 ± 2.08 | 94.68 ± 5.91 | 19.24 ± 5.03 |
| Xylanase | |||||
| XYNIV | 55 kDa | 2.10 ± 0.26 | 2.03 ± 0.21 | 1.50 ± 0.17 | 1.01 ± 0.06 |
| Mannanase | |||||
| Man5A | 53 kDa | 1.74 ± 0.71 | 1.21 ± 0.80 | 1.83 ± 0.36 | 2.50 ± 0.57 |
| Sum 3 | 9.09 | 6.67 | 7.61 | 7.12 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.-S.; Song, Y.; Lee, Y.-G.; Bae, H.-J. Comparative Evaluation of Adsorption of Major Enzymes in a Cellulase Cocktail Obtained from Trichoderma reesei onto Different Types of Lignin. Polymers 2022, 14, 167. https://doi.org/10.3390/polym14010167
Lee D-S, Song Y, Lee Y-G, Bae H-J. Comparative Evaluation of Adsorption of Major Enzymes in a Cellulase Cocktail Obtained from Trichoderma reesei onto Different Types of Lignin. Polymers. 2022; 14(1):167. https://doi.org/10.3390/polym14010167
Chicago/Turabian StyleLee, Dae-Seok, Younho Song, Yoon-Gyo Lee, and Hyeun-Jong Bae. 2022. "Comparative Evaluation of Adsorption of Major Enzymes in a Cellulase Cocktail Obtained from Trichoderma reesei onto Different Types of Lignin" Polymers 14, no. 1: 167. https://doi.org/10.3390/polym14010167
APA StyleLee, D. -S., Song, Y., Lee, Y. -G., & Bae, H. -J. (2022). Comparative Evaluation of Adsorption of Major Enzymes in a Cellulase Cocktail Obtained from Trichoderma reesei onto Different Types of Lignin. Polymers, 14(1), 167. https://doi.org/10.3390/polym14010167