Preparation, Characterization, and Anti-Adhesive Activity of Sulfate Polysaccharide from Caulerpa lentillifera against Helicobacter pylori
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Condition
2.2. Polysaccharide Extraction
2.3. Chemical Composition Analysis
2.4. Antimicrobial Activity Assays
2.5. Cell Line, Cell Culture, and Cell Viability Assay
2.6. Measurement of NO Production and iNOS Expression from RAW 264.7 Cells
2.7. Helicobacter pylori Adherence Assay
2.8. Evaluation of Inflammatory Responses in AGS Cells
2.9. Analysis of Gene Expression by Quantitative Reverse Transcription Polymerase Chain Reaction
2.10. Statistical Analysis
3. Results
3.1. Yields and Compositions of the Sulfate Polysaccharide from Caulerpa lentillifera
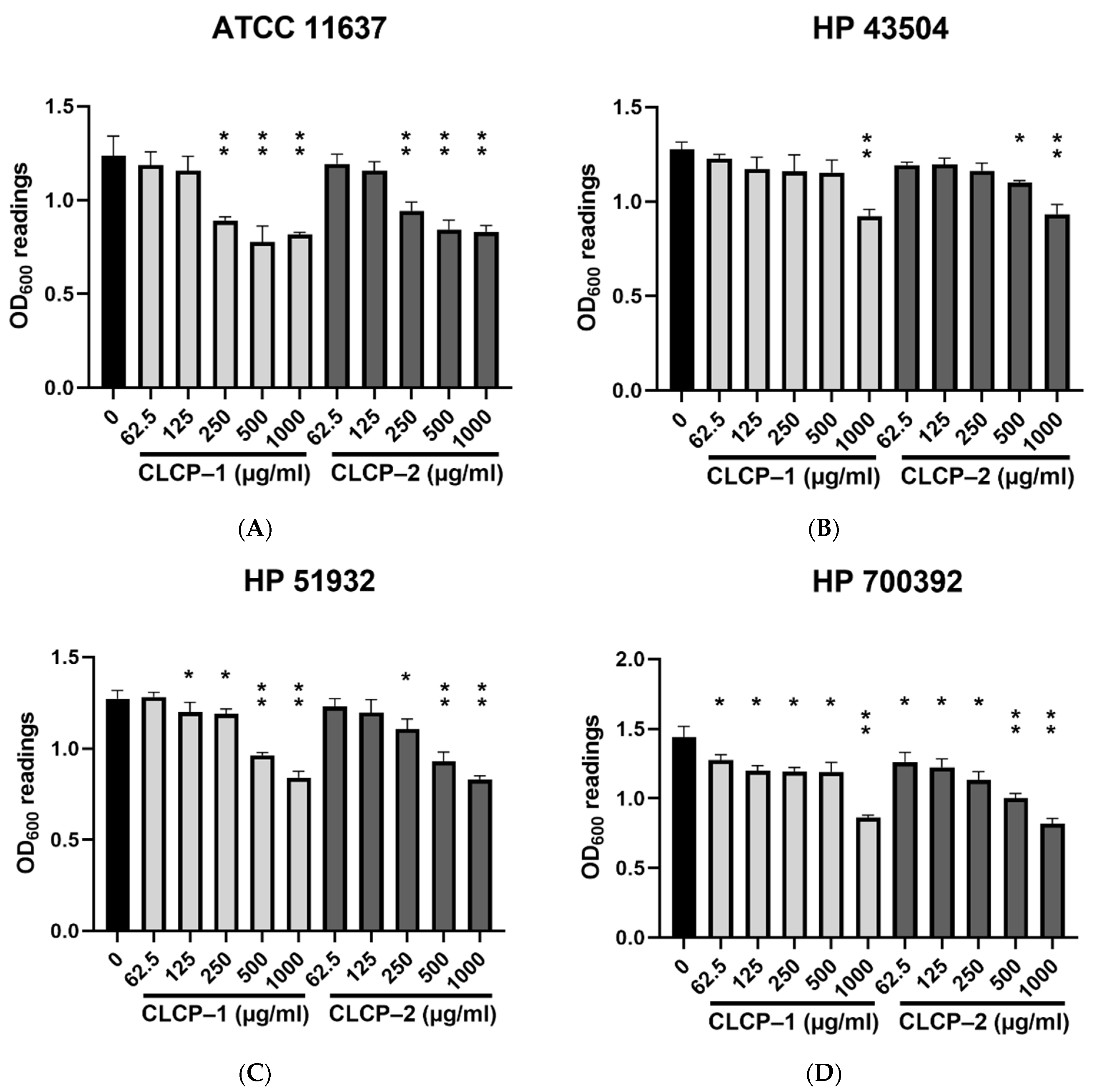
3.2. In Vitro Antimicrobial Potential of CLCP against H. pylori
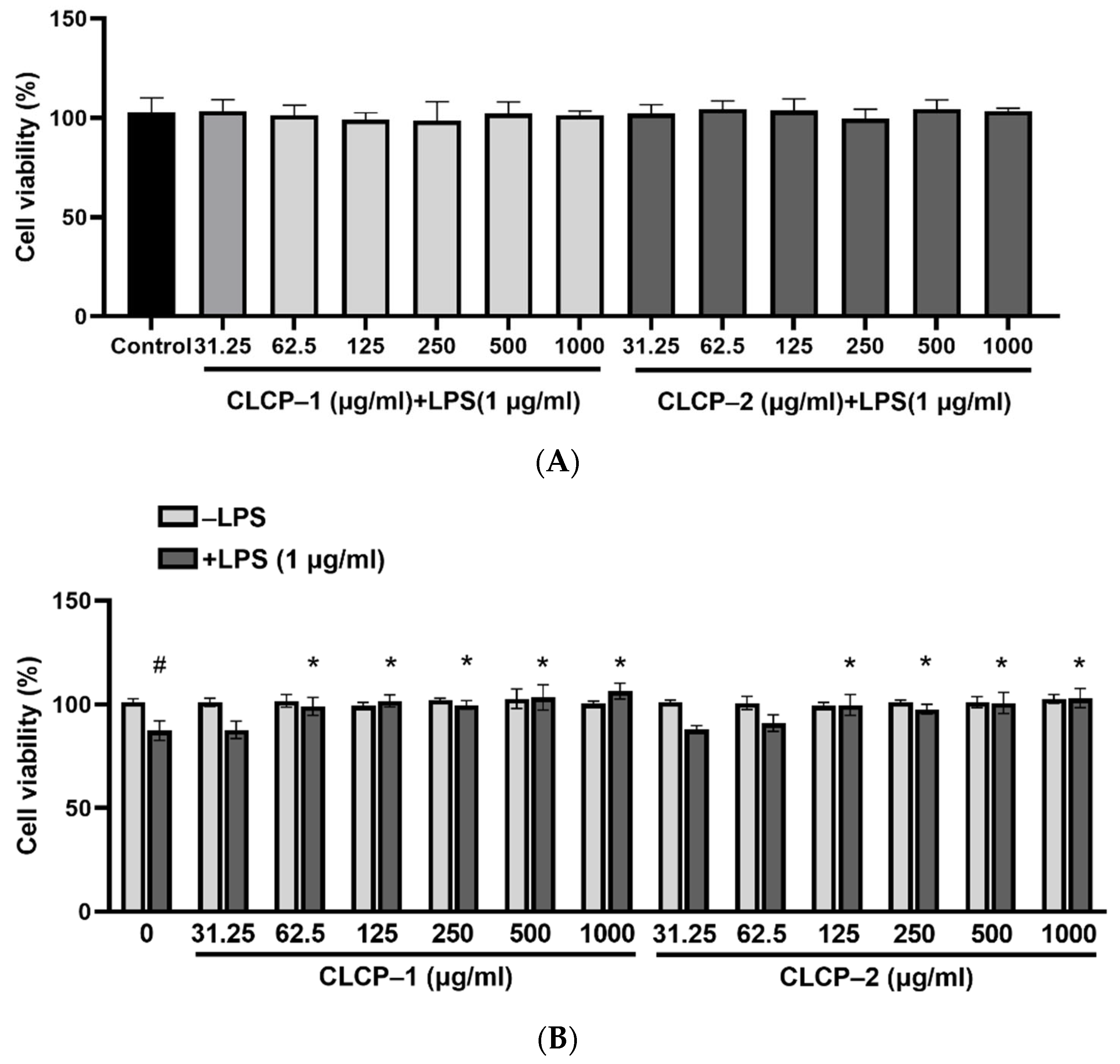
3.3. Effect of CLCP on Cell Viability
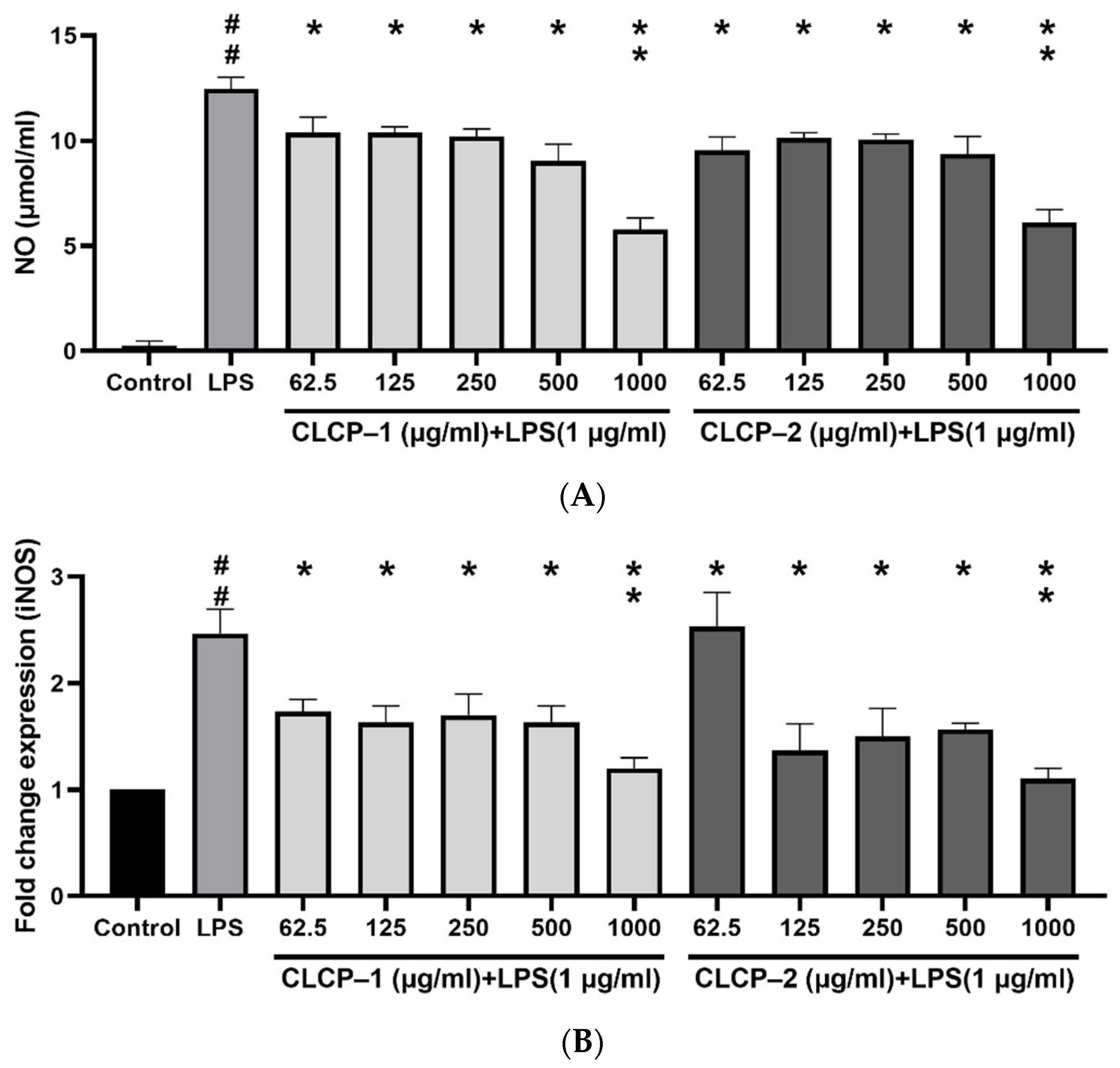
3.4. The Effect of CLCP on the Nitric Oxide Production in RAW 264.7 Cells
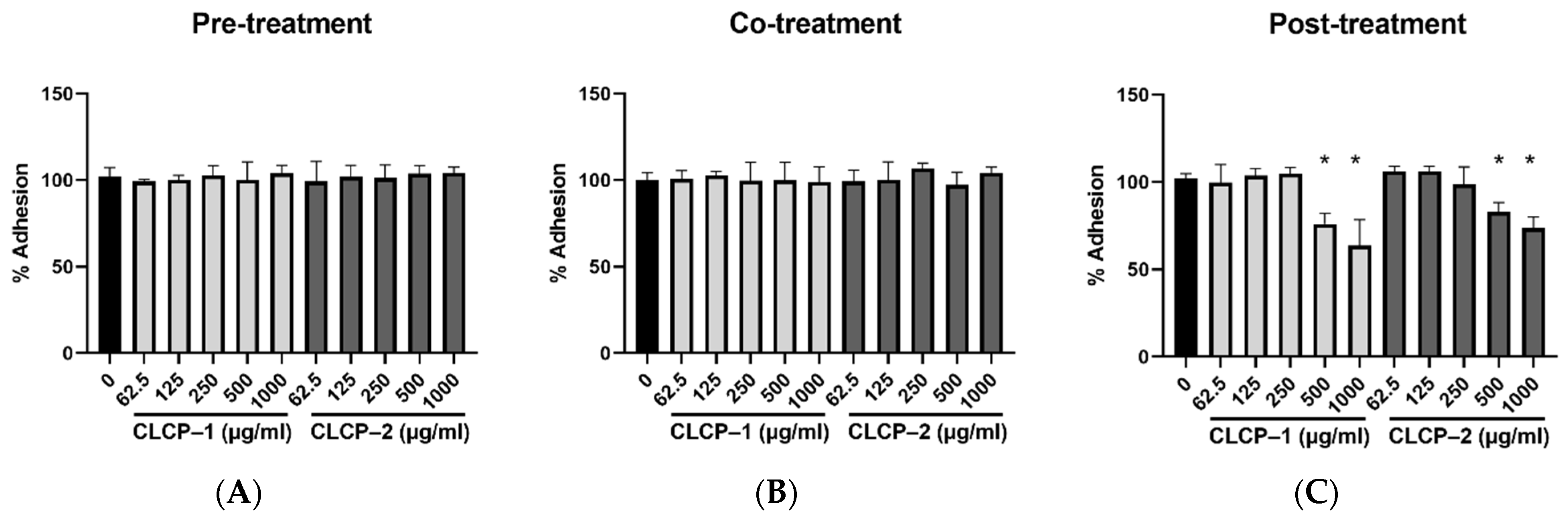
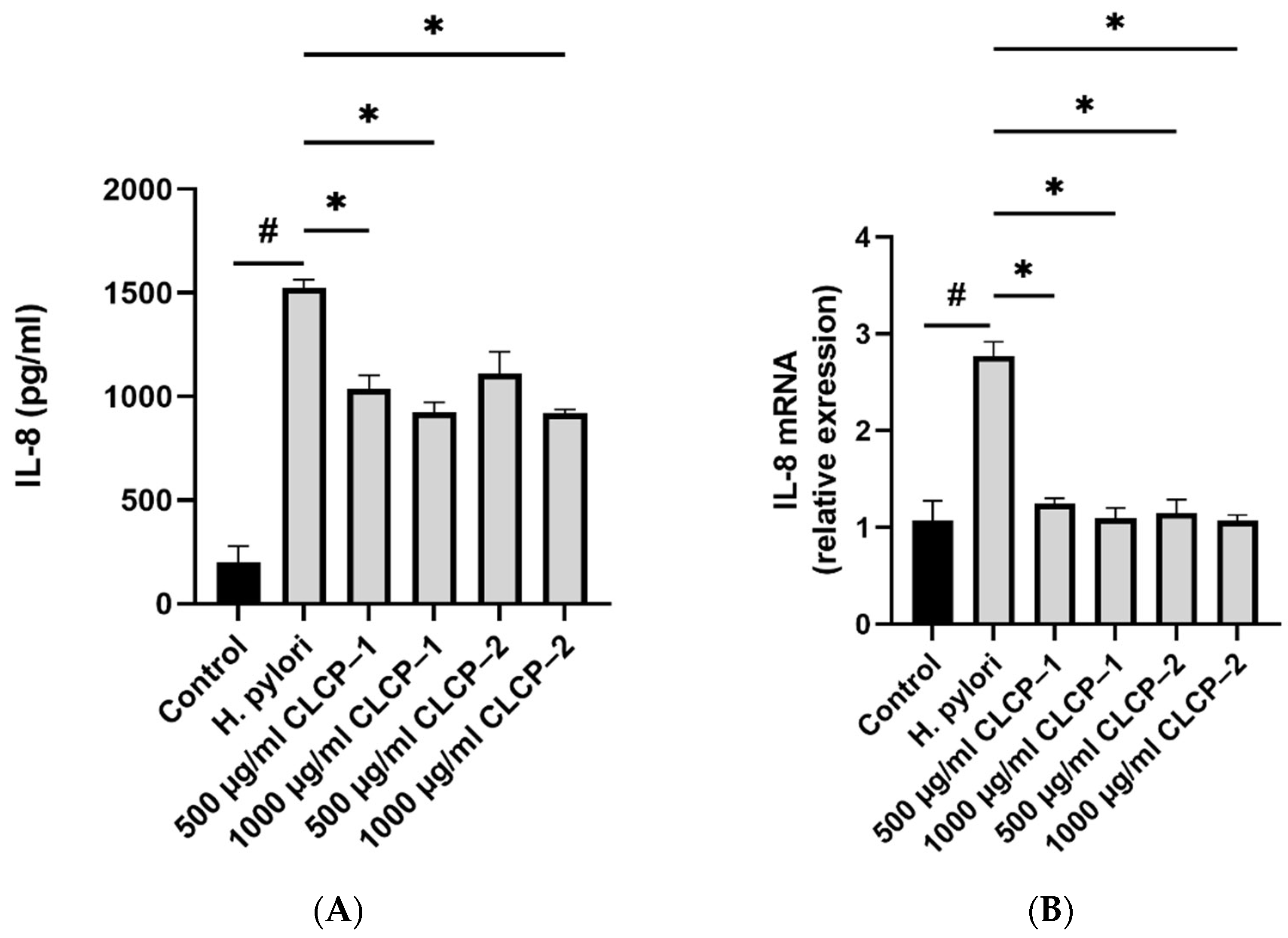
3.5. Inhibition of H. pylori Infection on AGS Cells by CLCPs
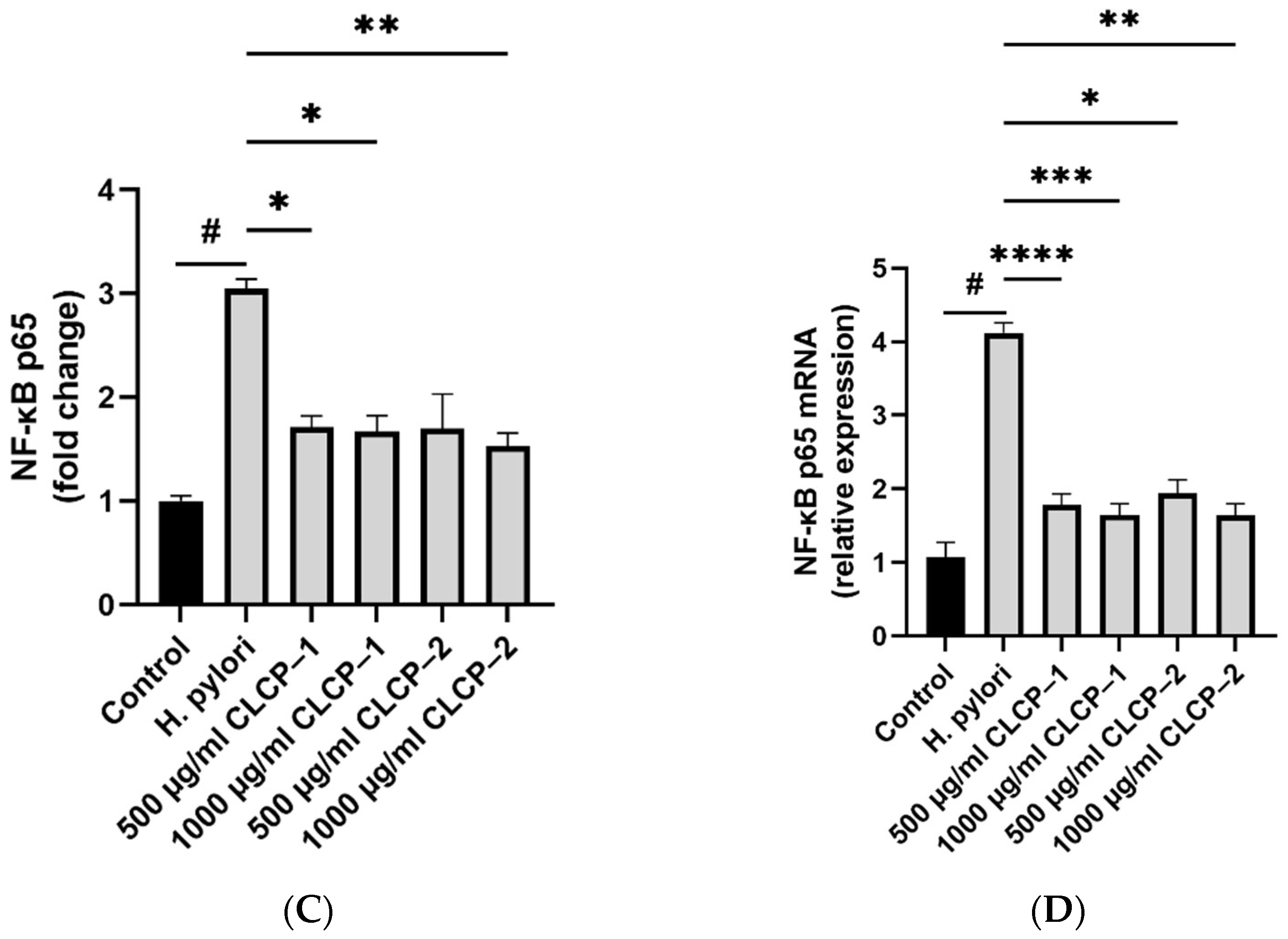
3.6. The Activity and Mechanism of CLCP in Reducing H. pylori Infection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gisbert, J.P.; Pajares, J.M. Helicobacter pylori infection and perforated peptic ulcer prevalence of the infection and role of antimicrobial treatment. Helicobacter 2003, 8, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-H.; Jin, S.-Y. Current guidelines for Helicobacter pylori treatment in East Asia 2022: Differences among China, Japan, and South Korea. World J. Clin. Cases 2022, 10, 6349. [Google Scholar] [CrossRef] [PubMed]
- Keikha, M.; Karbalaei, M. Prevalence of antibiotic heteroresistance associated with Helicobacter pylori infection: A systematic review and meta-analysis. Microb. Pathog. 2022, 170, 105720. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Kocsmár, É.; Buzás, G.M.; Szirtes, I.; Kocsmár, I.; Kramer, Z.; Szijártó, A.; Fadgyas-Freyler, P.; Szénás, K.; Rugge, M.; Fassan, M. Primary and secondary clarithromycin resistance in Helicobacter pylori and mathematical modeling of the role of macrolides. Nat. Commun. 2021, 12, 2255. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Shortridge, D.; Kibler, K.; Griffy, M.V.; Beyer, J.; Flamm, R.K.; Tanaka, S.K.; Graham, D.Y.; Go, M.F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 1996, 40, 477–480. [Google Scholar] [CrossRef]
- Rosenkranz, H.S.; Speck, W.T. Mutagenicity of metronidazole: Activation by mammalian liver microsomes. Biochem. Biophys. Res. Commun. 1975, 66, 520–525. [Google Scholar] [CrossRef]
- Goodwin, A. Identification of a Metronidazole-Resistance Gene(rdxA) in Helicobacter pylori; Dalhousie University: Halifax, NS, Canada, 1999. [Google Scholar]
- Thung, I.; Aramin, H.; Vavinskaya, V.; Gupta, S.; Park, J.; Crowe, S.; Valasek, M. the global emergence of Helicobacter pylori antibiotic resistance. Aliment. Pharmacol. Ther. 2016, 43, 514–533. [Google Scholar] [CrossRef]
- Sousa, C.; Ferreira, R.; Azevedo, N.F.; Oleastro, M.; Azeredo, J.; Figueiredo, C.; Melo, L.D. Helicobacter pylori infection: From standard to alternative treatment strategies. Crit. Rev. Microbiol. 2022, 48, 376–396. [Google Scholar] [CrossRef]
- Daelemans, S.; Deseck, V.; Levy, E.I.; Vandenplas, Y. Are pro-and/or synbiotics beneficial in Helicobacter pylori eradication therapy in children? A narrative review. Eur. J. Pediatr. 2022, 181, 3225–3234. [Google Scholar] [CrossRef]
- Sathianarayanan, S.; Aparna, V.; Biswas, R.; Anita, B.; Sukumaran, S.; Venkidasamy, B. A new approach against Helicobacter pylori using plants and its constituents: A review study. Microb. Pathog. 2022, 168, 105594. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.D.; Venmathi Maran, B.A.; Shaleh, S.R.M.; Zuldin, W.H.; Gnanaraj, C.; Yong, Y.S. Therapeutic potential and nutraceutical profiling of North Bornean seaweeds: A review. Mar. Drugs 2022, 20, 101. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, Y.; Liu, H.; Liu, S.; Qin, Y.; Li, P. Advances in cultivation, wastewater treatment application, bioactive components of Caulerpa lentillifera and their biotechnological applications. PeerJ 2019, 7, e6118. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Song, H.; Wang, L.; Peng, H.; Sun, Y.; Ai, C.; Wen, C.; Zhu, B.; Song, S. Structural characterization and SARS-CoV-2 inhibitory activity of a sulfated polysaccharide from Caulerpa lentillifera. Carbohydr. Polym. 2022, 280, 119006. [Google Scholar] [CrossRef]
- Tian, H.; Liu, H.; Song, W.; Zhu, L.; Yin, X. Polysaccharide from Caulerpa lentillifera: Extraction optimization with response surface methodology, structure and antioxidant activities. Nat. Prod. Res. 2021, 35, 3417–3425. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, M.; Qing, Y.; Luo, Y.; Xia, G.; Li, Y. Study on immunostimulatory activity and extraction process optimization of polysaccharides from Caulerpa lentillifera. Int. J. Biol. Macromol. 2020, 143, 677–684. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Z.; Song, S.; Zhu, B.; Zhao, L.; Jiang, J.; Liu, N.; Wang, J.; Chen, X. Anti-inflammatory activity and structural identification of a sulfated polysaccharide CLGP4 from Caulerpa lentillifera. Int. J. Biol. Macromol. 2020, 146, 931–938. [Google Scholar] [CrossRef]
- Khairuddin, K.; Sudirman, S.; Huang, L.; Kong, Z.-L. Caulerpa lentillifera polysaccharides-rich extract reduces oxidative stress and proinflammatory cytokines levels associated with male reproductive functions in diabetic mice. Appl. Sci. 2020, 10, 8768. [Google Scholar] [CrossRef]
- Maeda, R.; Ida, T.; Ihara, H.; Sakamoto, T. Induction of apoptosis in MCF-7 cells by β-1, 3-xylooligosaccharides prepared from Caulerpa lentillifera. Biosci. Biotechnol. Biochem. 2012, 76, 1032–1034. [Google Scholar] [CrossRef]
- Menchicchi, B.; Hensel, A.; Goycoolea, F.M. Polysaccharides as bacterial antiadhesive agents and “smart” constituents for improved drug delivery systems against Helicobacter pylori infection. Curr. Pharm. Des. 2015, 21, 4888–4906. [Google Scholar] [CrossRef]
- Sun, Y.; Gong, G.; Guo, Y.; Wang, Z.; Song, S.; Zhu, B.; Zhao, L.; Jiang, J. Purification, structural features and immunostimulatory activity of novel polysaccharides from Caulerpa lentillifera. Int. J. Biol. Macromol. 2018, 108, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, Q.; Zhang, H.; Wang, J.; Fu, Q.; Qiao, H.; Wang, Q. Insight into antibacterial mechanism of polysaccharides: A review. LWT 2021, 150, 111929. [Google Scholar] [CrossRef]
- Pham, T.N.A.; Le, B.; Yang, S.H. Anticancer activity of the potential Pyropia yezoensis galactan fractionated in human prostate cancer cells. Biotechnol. Bioprocess Eng. 2021, 26, 63–70. [Google Scholar] [CrossRef]
- Kochert, G. Carbohydrate determination by the phenol-sulfuric acid method. In Handbook of Phycological Methods, Phycological and Biochemical Methods; Springer: Berlin/Heidelberg, Germany, 1978; p. 95. [Google Scholar] [CrossRef]
- Dodgson, K.; Price, R. A note on the determination of the ester sulphate content of sulphated polysaccharides. Biochem. J. 1962, 84, 106. [Google Scholar] [CrossRef] [PubMed]
- Karamanos, N.; Hjerpe, A.; Tsegenidis, T.; Engfeldt, B.; Antonopoulos, C. Determination of iduronic acid and glucuronic acid in glycosaminoglycans after stoichiometric reduction and depolymerization using high-performance liquid chromatography and ultraviolet detection. Anal. Biochem. 1988, 172, 410–419. [Google Scholar] [CrossRef]
- Le, B.; Anh, P.-T.-N.; Yang, S.-H. Polysaccharide derived from Nelumbo nucifera lotus plumule shows potential prebiotic activity and ameliorates insulin resistance in HepG2 cells. Polymers 2021, 13, 1780. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Leardi, R.M.C.; Polotti, G. CAT (Chemometric Agile Software). Available online: http://gruppochemiometria.it/index.php/software (accessed on 1 November 2022).
- Zheng, Q.; Jia, R.-B.; Ou, Z.-R.; Li, Z.-R.; Zhao, M.; Luo, D.; Lin, L. Comparative study on the structural characterization and α-glucosidase inhibitory activity of polysaccharide fractions extracted from Sargassum fusiforme at different pH conditions. Int. J. Biol. Macromol. 2022, 194, 602–610. [Google Scholar] [CrossRef]
- Sanjeewa, K.K.A.; Fernando, I.P.S.; Kim, E.-A.; Ahn, G.; Jee, Y.; Jeon, Y.-J. Anti-inflammatory activity of a sulfated polysaccharide isolated from an enzymatic digest of brown seaweed Sargassum horneri in RAW 264.7 cells. Nutr. Res. Pract. 2017, 11, 3–10. [Google Scholar] [CrossRef]
- Souza, B.W.; Cerqueira, M.A.; Bourbon, A.I.; Pinheiro, A.C.; Martins, J.T.; Teixeira, J.A.; Coimbra, M.A.; Vicente, A.A. Chemical characterization and antioxidant activity of sulfated polysaccharide from the red seaweed Gracilaria birdiae. Food Hydrocoll. 2012, 27, 287–292. [Google Scholar] [CrossRef]
- Olasehinde, T.A.; Mabinya, L.V.; Olaniran, A.O.; Okoh, A.I. Chemical characterization, antioxidant properties, cholinesterase inhibitory and anti-amyloidogenic activities of sulfated polysaccharides from some seaweeds. Bioact. Carbohydr. Diet. Fibre 2019, 18, 100182. [Google Scholar] [CrossRef]
- Robic, A.; Bertrand, D.; Sassi, J.-F.; Lerat, Y.; Lahaye, M. Determination of the chemical composition of ulvan, a cell wall polysaccharide from Ulva spp.(Ulvales, Chlorophyta) by FT-IR and chemometrics. J. Appl. Phycol. 2009, 21, 451–456. [Google Scholar] [CrossRef]
- Sekkal, M.; Huvenne, J.-P.; Legrand, P.; Sombret, B.; Mollet, J.-C.; Mouradi-Givernaud, A.; Verdus, M.-C. Direct structural identification of polysaccharides from red algae by FTIR microspectrometry I: Localization of agar in Gracilaria verrucosa sections. Microchim. Acta 1993, 112, 11–18. [Google Scholar] [CrossRef]
- Hong, T.; Yin, J.-Y.; Nie, S.-P.; Xie, M.-Y. Applications of infrared spectroscopy in polysaccharide structural analysis: Progress, challenge and perspective. Food Chem. X 2021, 12, 100168. [Google Scholar] [CrossRef] [PubMed]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2017, 185, 317–335. [Google Scholar] [CrossRef]
- Mirzadeh, M.; Arianejad, M.R.; Khedmat, L. Antioxidant, antiradical, and antimicrobial activities of polysaccharides obtained by microwave-assisted extraction method: A review. Carbohydr. Polym. 2020, 229, 115421. [Google Scholar] [CrossRef]
- Besednova, N.N.; Zaporozhets, T.S.; Somova, L.M.; Kuznetsova, T.A. Prospects for the use of extracts and polysaccharides from marine algae to prevent and treat the diseases caused by Helicobacter pylori. Helicobacter 2015, 20, 89–97. [Google Scholar] [CrossRef]
- Chen, B.-R.; Li, W.-M.; Li, T.-L.; Chan, Y.-L.; Wu, C.-J. Fucoidan from Sargassum hemiphyllum inhibits infection and inflammation of Helicobacter pylori. Sci. Rep. 2022, 12, 429. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Herrmann, F.; König, S.; Goycoolea, F.; Hensel, A. Structural characterization of the carbohydrate and protein part of arabinogalactan protein from Basella alba stem and antiadhesive activity of polysaccharides from B. alba against Helicobacter pylori. Fitoterapia 2022, 157, 105132. [Google Scholar] [CrossRef]
- Wittschier, N.; Faller, G.; Hensel, A. Aqueous extracts and polysaccharides from liquorice roots (Glycyrrhiza glabra L.) inhibit adhesion of Helicobacter pylori to human gastric mucosa. J. Ethnopharmacol. 2009, 125, 218–223. [Google Scholar] [CrossRef]
- Piotrowski, I.; Kulcenty, K.; Suchorska, W. Interplay between inflammation and cancer. Rep. Pract. Oncol. Radiother. 2020, 25, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Butcher, L.D.; den Hartog, G.; Ernst, P.B.; Crowe, S.E. Oxidative stress resulting from Helicobacter pylori infection contributes to gastric carcinogenesis. Cell Mol. Gastroenterol. Hepatol. 2017, 3, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Je, J.-G.; Huang, C.; Oh, J.-Y.; Fu, X.; Wang, K.; Ahn, G.; Xu, J.; Gao, X.; Jeon, Y.-J. Anti-inflammatory effect of sulfated polysaccharides isolated from Codium fragile in vitro in RAW 264.7 macrophages and in vivo in zebrafish. Mar. Drugs 2022, 20, 391. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Wang, L.; Fu, X.; Duan, D.; Jeon, Y.-J.; Xu, J.; Gao, X. In vitro and in vivo anti-inflammatory activities of a fucose-rich fucoidan isolated from Saccharina japonica. Int. J. Biol. Macromol. 2020, 156, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-Z.; Goldberg, J.B.; Hatakeyama, M. Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 2010, 6, 851–862. [Google Scholar] [CrossRef]
- Maubach, G.; Vieth, M.; Boccellato, F.; Naumann, M. Helicobacter pylori-induced NF-κB: Trailblazer for gastric pathophysiology. Trends. Mol. Med. 2022, 28, 210–222. [Google Scholar] [CrossRef]
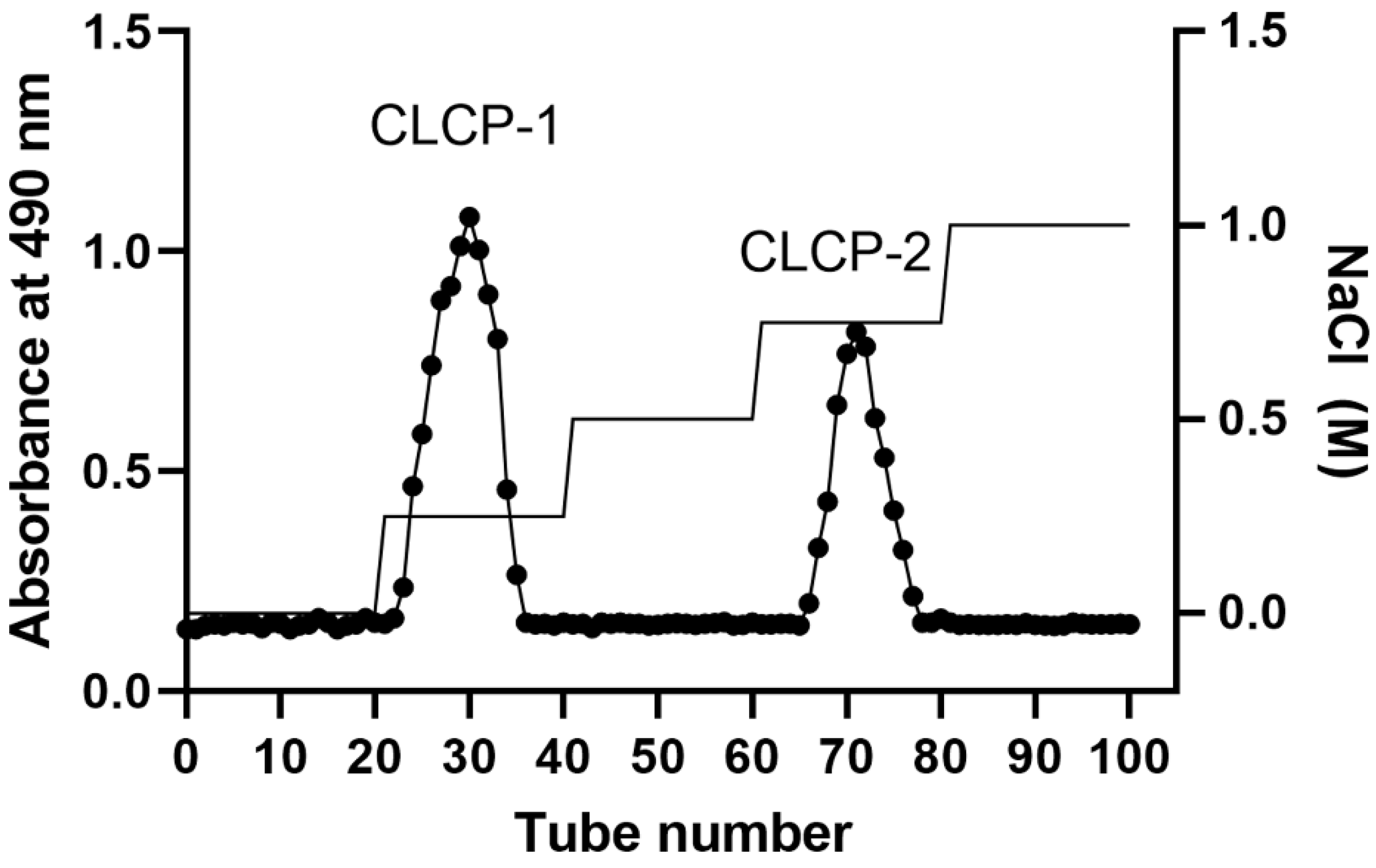
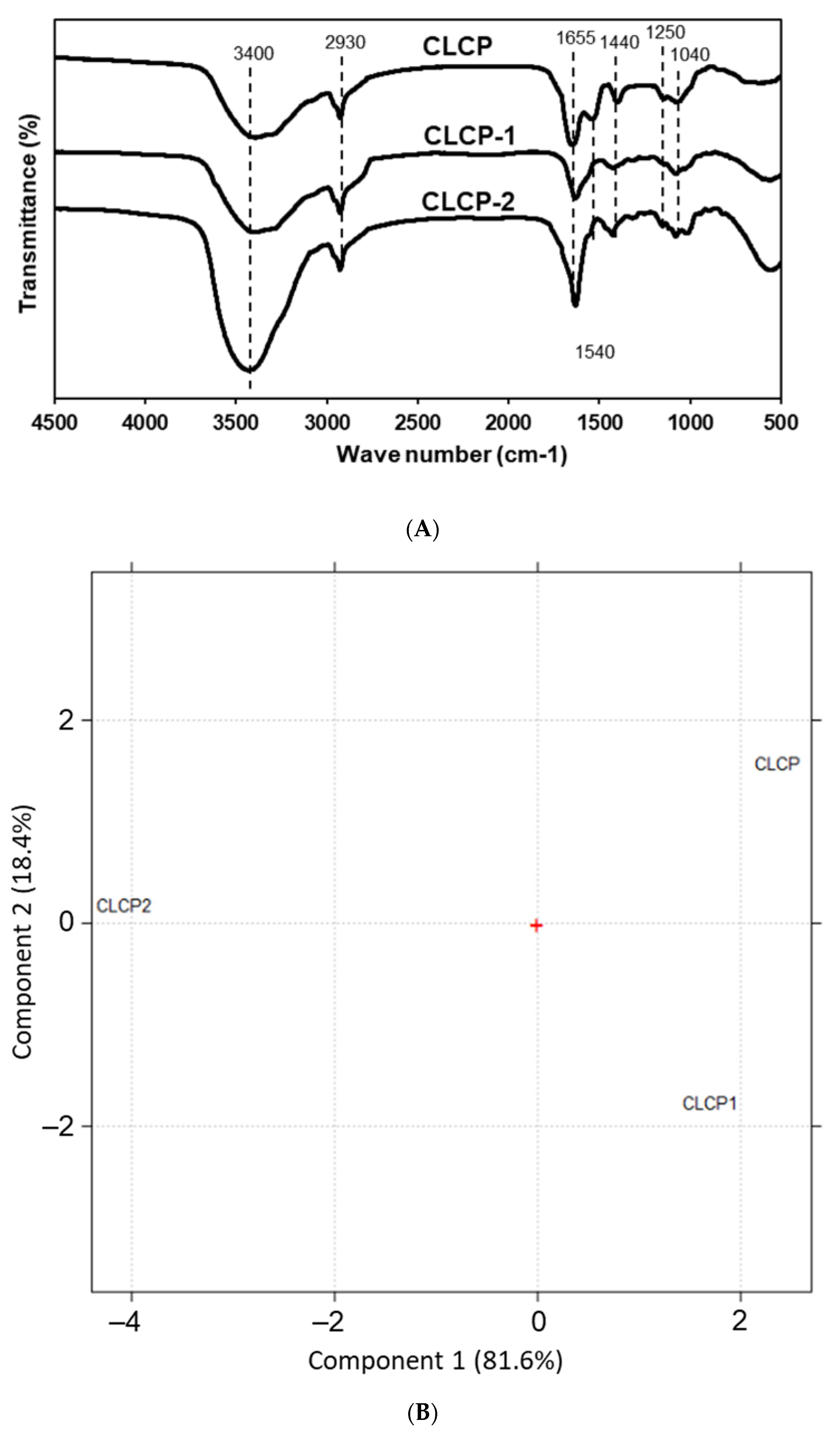
| CLCP | CLCP-1 | CLCP-2 | |
|---|---|---|---|
| Yield (%) | 8.93 ± 0.56 | 1.38 ± 0.17 | 1.03 ± 0.08 |
| Chemical characteristics (%) | |||
| Total sugar | 87.51 ± 2.13 | 72.19 ± 1.51 | 65.51 ± 3.04 |
| Protein | 2.94 ± 0.02 | 0.24 ± 0.02 | 0.35 ± 0.07 |
| Uronic acid | 12.48 ± 1.08 | 5.36 ± 0.48 | 2.81 ± 0.64 |
| Sulfate | 8.11 ± 0.24 | 16.21 ± 0.29 | 18.15 ± 1.95 |
| Mw (kDa) | NA 1 | 963.15 | 648.42 |
| Monosaccharide composition (%) | |||
| Man | 38.18 | 46.85 | 50.21 |
| Xyl | 11.23 | 20.81 | 23.21 |
| Gal | 29.31 | 22.78 | 24.86 |
| Glc | 3.51 | 2.18 | 1.85 |
| z-potential (mV) | −15.6 ± 0.2 | −22.61 ± 0.5 | −26.1 ± 0.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, B.; Do, D.T.; Nguyen, H.M.; Do, B.H.; Le, H.T. Preparation, Characterization, and Anti-Adhesive Activity of Sulfate Polysaccharide from Caulerpa lentillifera against Helicobacter pylori. Polymers 2022, 14, 4993. https://doi.org/10.3390/polym14224993
Le B, Do DT, Nguyen HM, Do BH, Le HT. Preparation, Characterization, and Anti-Adhesive Activity of Sulfate Polysaccharide from Caulerpa lentillifera against Helicobacter pylori. Polymers. 2022; 14(22):4993. https://doi.org/10.3390/polym14224993
Chicago/Turabian StyleLe, Bao, Duy Thanh Do, Hien Minh Nguyen, Bich Hang Do, and Huong Thuy Le. 2022. "Preparation, Characterization, and Anti-Adhesive Activity of Sulfate Polysaccharide from Caulerpa lentillifera against Helicobacter pylori" Polymers 14, no. 22: 4993. https://doi.org/10.3390/polym14224993
APA StyleLe, B., Do, D. T., Nguyen, H. M., Do, B. H., & Le, H. T. (2022). Preparation, Characterization, and Anti-Adhesive Activity of Sulfate Polysaccharide from Caulerpa lentillifera against Helicobacter pylori. Polymers, 14(22), 4993. https://doi.org/10.3390/polym14224993







