Solid State NMR a Powerful Technique for Investigating Sustainable/Renewable Cellulose-Based Materials
Abstract
1. Introduction
- bacterial cellulose [16], preparation method: Gluconacetobater hansenii bacteria using corn steep liquor as nutrient,
- cellulose ester (e.g., cellulose acetate) [17], preparation method: reaction of cellulose with acetic anhydride and acetic acid in the presence of sulfuric acid,
2. Cellulose Structure Investigated via Solid State NMR Spectroscopy
2.1. Pure Cellulose with 13C CP MAS and Different Crystal Phases
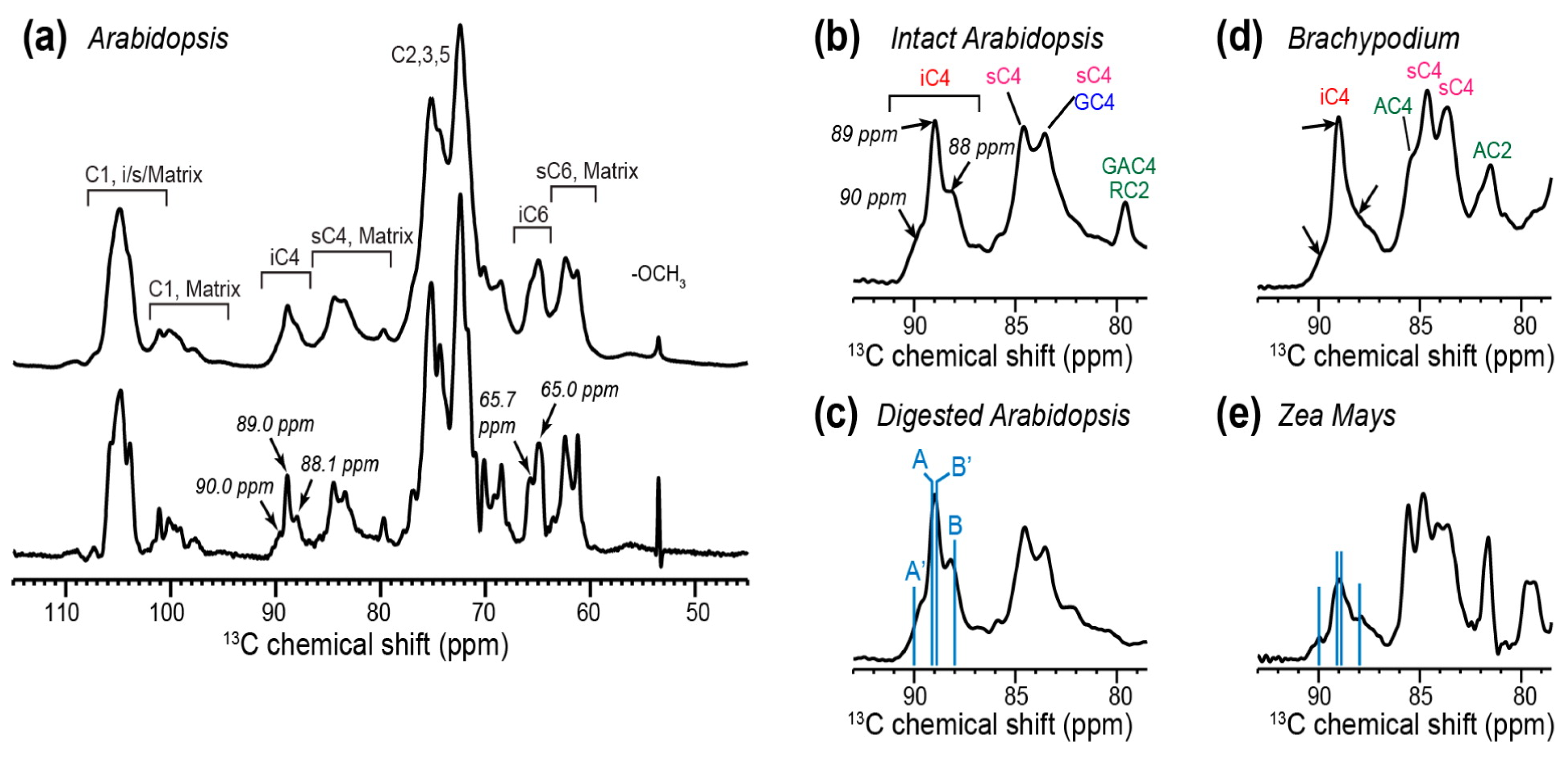
2.2. Cellulose Polymorphism in Plant Primary Cell Walls
3. Lignocellulosic Biomass Structure Interpretation
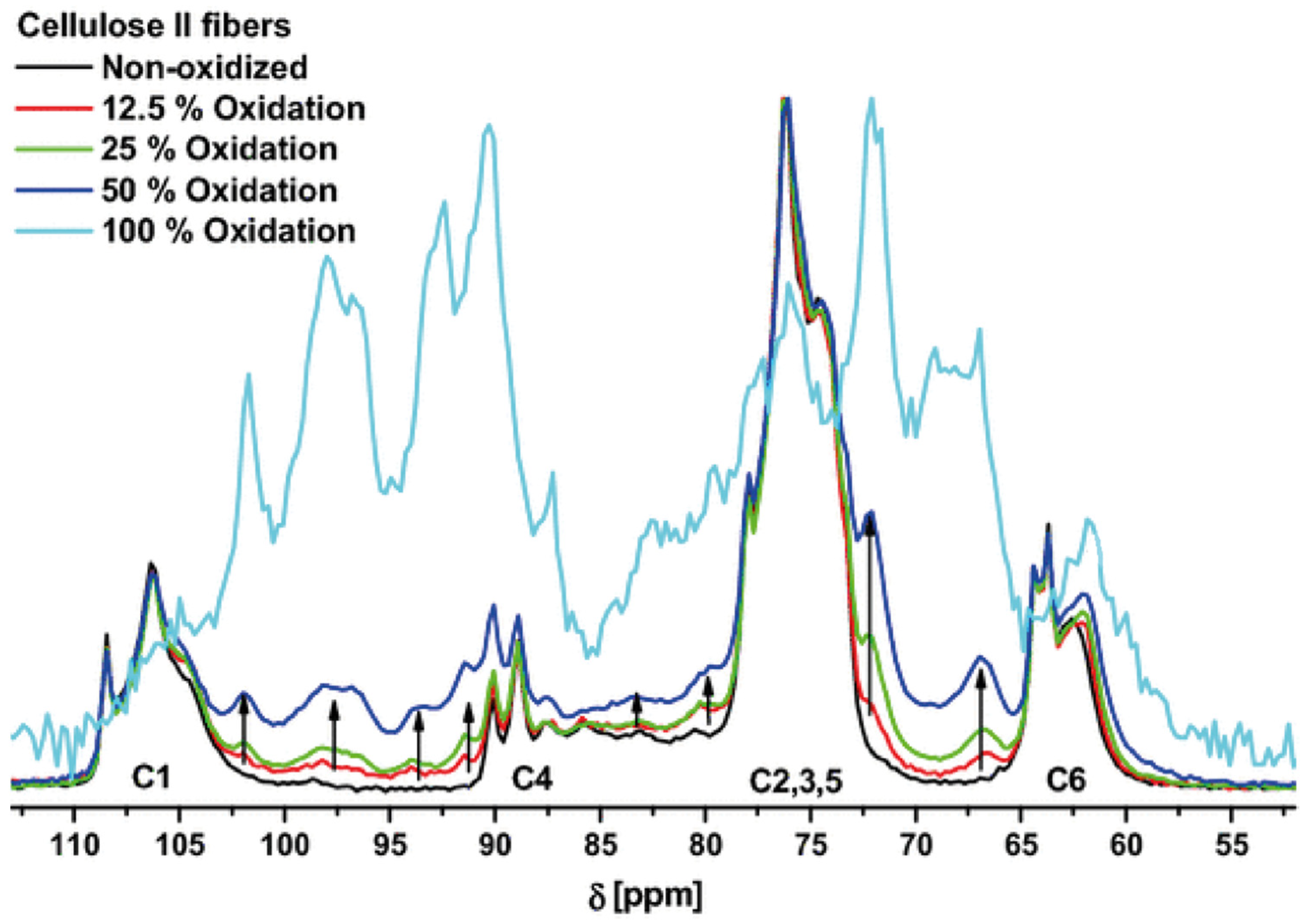
4. Effect of Oxidation on Pulp and Viscous Cellulose
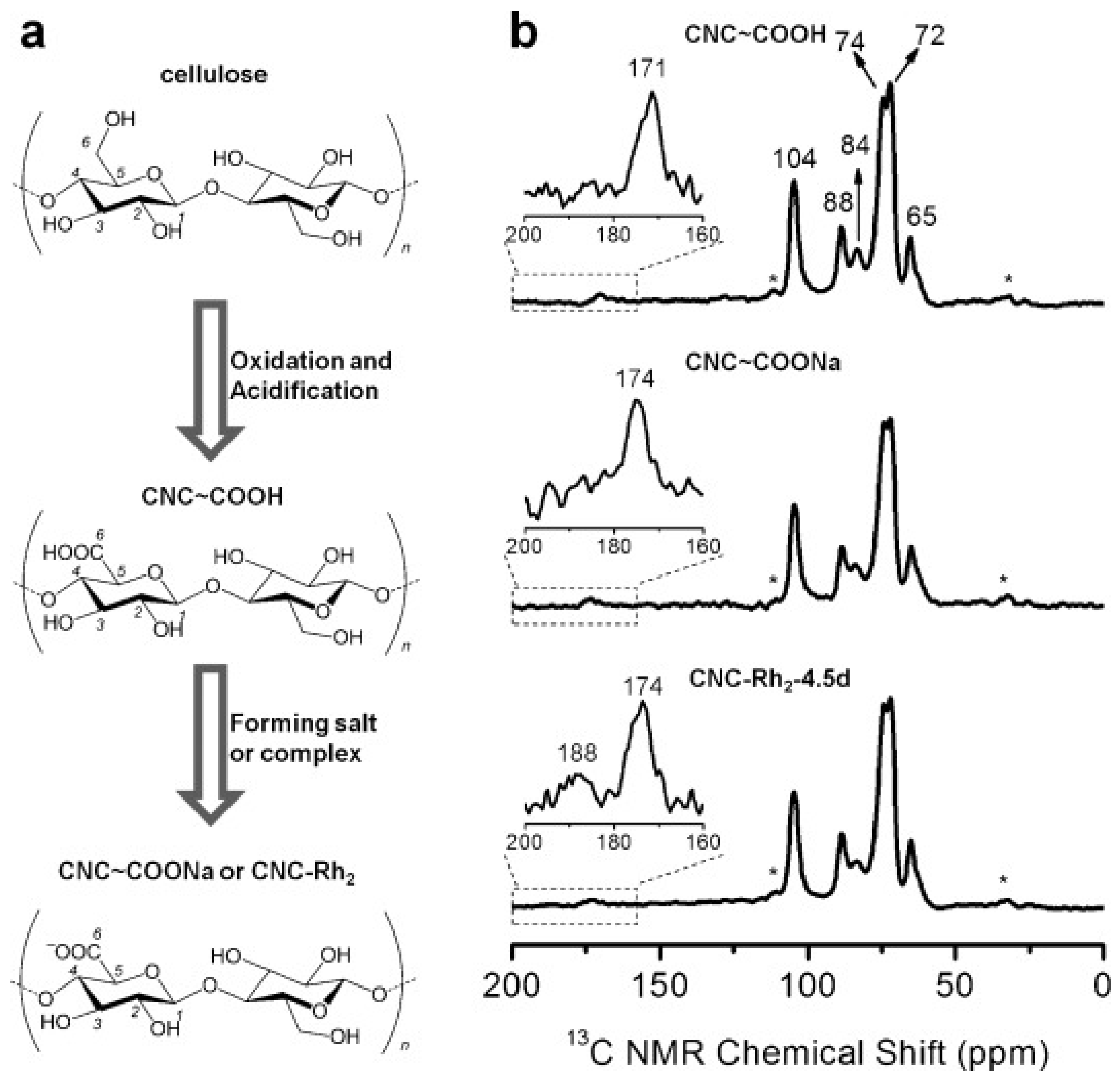
5. Production of Crystalline Nanocelluloses via Oxidation of Microcrystalline Cellulose
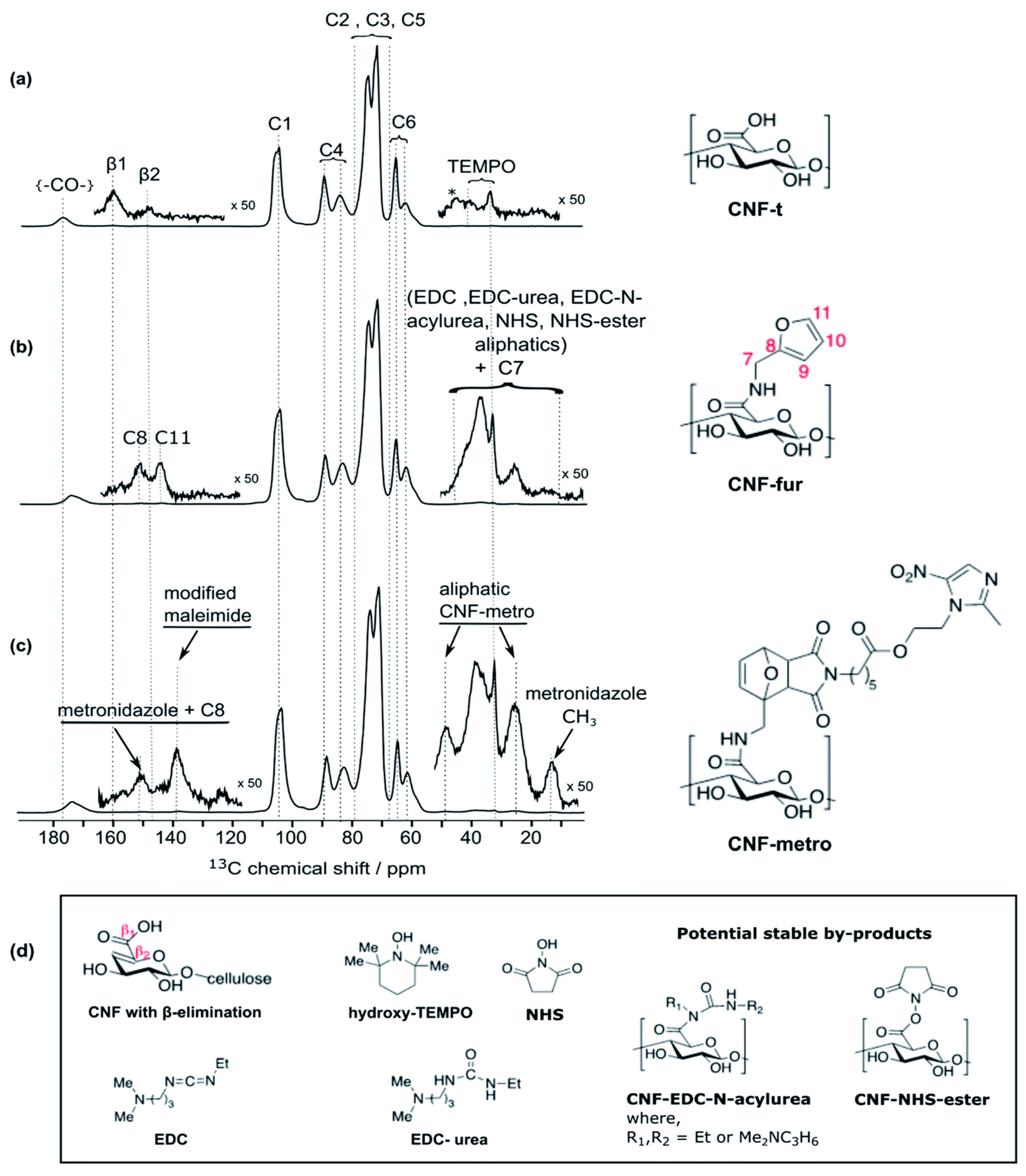
6. Future Opportunities for Nanocellulose as a Drug Delivery Carrier
7. Summary, Concluding Remarks, and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baghaei, B.; Skrifvars, M. All-Cellulose Composites: A Review of Recent Studies on Structure, Properties and Applications. Molecules 2020, 25, 2836. [Google Scholar] [CrossRef]
- Esen, E.; Meier, M.A.R. Sustainable Functionalization of 2,3-Dialdehyde Cellulose via the Passerini Three-Component Reaction. ACS Sustain. Chem. Eng. 2020, 8, 15755–15760. [Google Scholar] [CrossRef]
- Khandelwal, M.; Alan, H.W. Hierarchical Organisation in the Most Abundant Biopolymer–Cellulose. MRS Online Proc. Libr. 2013, 1504, mrsf12-1504-v02-03. [Google Scholar] [CrossRef]
- Klemm, D.; Heublein, B.; Fink, H.-P.; Bohn, A. Cellulose: Fascinating Biopolymer and Sustainable Raw Material. Angew. Chem. Int. Ed. 2005, 44, 3358–3393. [Google Scholar] [CrossRef] [PubMed]
- Kumar Gupta, P.; Sai Raghunath, S.; Venkatesh Prasanna, D.; Venkat, P.; Shree, V.; Chithananthan, C.; Choudhary, S.; Surender, K.; Geetha, K. An Update on Overview of Cellulose, Its Structure and Applications. In Cellulose; Rodríguez Pascual, A.E., Eugenio Martín, M., Eds.; IntechOpen: London, UK, 2019; ISBN 978-1-83968-056-4. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Wang, B.; Ma, M.-G.; Wang, B. Review of Recent Development on Preparation, Properties, and Applications of Cellulose-Based Functional Materials. Int. J. Polym. Sci. 2018, 2018, 8973643. [Google Scholar] [CrossRef]
- Liu, W.; Du, H.; Liu, H.; Xie, H.; Xu, T.; Zhao, X.; Liu, Y.; Zhang, X.; Si, C. Highly Efficient and Sustainable Preparation of Carboxylic and Thermostable Cellulose Nanocrystals via FeCl3-Catalyzed Innocuous Citric Acid Hydrolysis. ACS Sustain. Chem. Eng. 2020, 8, 16691–16700. [Google Scholar] [CrossRef]
- Onwukamike, K.N.; Grelier, S.; Grau, E.; Cramail, H.; Meier, M.A.R. Critical Review on Sustainable Homogeneous Cellulose Modification: Why Renewability Is Not Enough. ACS Sustain. Chem. Eng. 2019, 7, 1826–1840. [Google Scholar] [CrossRef]
- Rahmatika, A.M.; Toyoda, Y.; Nguyen, T.T.; Goi, Y.; Kitamura, T.; Morita, Y.; Kume, K.; Ogi, T. Cellulose Nanofiber and Magnetic Nanoparticles as Building Blocks Constructing Biomass-Based Porous Structured Particles and Their Protein Adsorption Performance. ACS Sustain. Chem. Eng. 2020, 8, 18686–18695. [Google Scholar] [CrossRef]
- Ummartyotin, S.; Manuspiya, H. A Critical Review on Cellulose: From Fundamental to an Approach on Sensor Technology. Renew. Sustain. Energy Rev. 2015, 41, 402–412. [Google Scholar] [CrossRef]
- Xia, Z.; Li, J.; Zhang, J.; Zhang, X.; Zheng, X.; Zhang, J. Processing and Valorization of Cellulose, Lignin and Lignocellulose Using Ionic Liquids. J. Bioresour. Bioprod. 2020, 5, 79–95. [Google Scholar] [CrossRef]
- Updegraff, D.M. Semimicro Determination of Cellulose Inbiological Materials. Anal. Biochem. 1969, 32, 420–424. [Google Scholar] [CrossRef]
- Holtzapple, M.T. CELLULOSE. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Caballero, B., Ed.; Academic Press: Oxford, UK, 2003; pp. 998–1007. ISBN 978-0-12-227055-0. [Google Scholar]
- Aravamudhan, A.; Ramos, D.M.; Nada, A.A.; Kumbar, S.G. Chapter 4—Natural Polymers: Polysaccharides and Their Derivatives for Biomedical Applications. In Natural and Synthetic Biomedical Polymers; Kumbar, S.G., Laurencin, C.T., Deng, M., Eds.; Elsevier: Oxford, UK, 2014; pp. 67–89. ISBN 978-0-12-396983-5. [Google Scholar]
- Zhang, Z.; Ortiz, O.; Goyal, R.; Kohn, J. Chapter 23—Biodegradable Polymers. In Principles of Tissue Engineering, 4th ed.; Lanza, R., Langer, R., Vacanti, J., Eds.; Academic Press: Boston, MA, USA, 2014; pp. 441–473. ISBN 978-0-12-398358-9. [Google Scholar]
- Costa, A.F.S.; Almeida, F.C.G.; Vinhas, G.M.; Sarubbo, L.A. Production of Bacterial Cellulose by Gluconacetobacter Hansenii Using Corn Steep Liquor As Nutrient Sources. Front. Microbiol. 2017, 8, 2027. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Thümmler, K.; Volkert, B.; Hettrich, K.; Schmidt, I.; Fischer, K. Properties and Applications of Cellulose Acetate. Macromol. Symp. 2008, 262, 89–96. [Google Scholar] [CrossRef]
- Ahmadi, P.; Jahanban-Esfahlan, A.; Ahmadi, A.; Tabibiazar, M.; Mohammadifar, M. Development of Ethyl Cellulose-Based Formulations: A Perspective on the Novel Technical Methods. Food Rev. Int. 2020, 18, 1–48. [Google Scholar] [CrossRef]
- Wasilewska, K.; Winnicka, K. Ethylcellulose–A Pharmaceutical Excipient with Multidirectional Application in Drug Dosage Forms Development. Materials 2019, 12, 3386. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, L.; Zhang, X.; Zhu, Y.; Wu, Y.; Li, Y.; Li, B.; Liu, S.; Zhao, J.; Ma, Z. Ethyl Cellulose Nanodispersions as Stabilizers for Oil in Water Pickering Emulsions. Sci. Rep. 2017, 7, 12079. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.A.; Gray, D.G. The Surface Tension of Aqueous Hydroxypropyl Cellulose Solutions. J. Colloid Interface Sci. 1978, 67, 255–265. [Google Scholar] [CrossRef]
- Gosecki, M.; Setälä, H.; Virtanen, T.; Ryan, A.J. A Facile Method to Control the Phase Behavior of Hydroxypropyl Cellulose. Carbohydr. Polym. 2021, 251, 117015. [Google Scholar] [CrossRef]
- Weißenborn, E.; Braunschweig, B. Hydroxypropyl Cellulose as a Green Polymer for Thermo-Responsive Aqueous Foams. Soft Matter 2019, 15, 2876–2883. [Google Scholar] [CrossRef]
- Mbituyimana, B.; Mao, L.; Hu, S.; Ullah, M.W.; Chen, K.; Fu, L.; Zhao, W.; Shi, Z.; Yang, G. Bacterial cellulose/glycolic acid/glycerol composite membrane as a system to deliver glycolic acid for anti-aging treatment. J. Bioresour. Bioprod. 2021, 6, 129–141. [Google Scholar] [CrossRef]
- Karrasch, A.; Jäger, C.; Saake, B.; Potthast, A.; Rosenau, T. Solid-State NMR Studies of Methyl Celluloses. Part 2: Determination of Degree of Substitution and O−6 vs. O−2/O−3 Substituent Distribution in Commercial Methyl Cellulose Samples. Cellulose 2009, 16, 1159–1166. [Google Scholar] [CrossRef]
- Karrasch, A.; Jäger, C.; Karakawa, M.; Nakatsubo, F.; Potthast, A.; Rosenau, T. Solid-State NMR Studies of Methyl Celluloses. Part 1: Regioselectively Substituted Celluloses as Standards for Establishing an NMR Data Basis. Cellulose 2008, 16, 129. [Google Scholar] [CrossRef]
- Luchs, J.I.; Nelinson, D.S.; Macy, J.I.; for the LAC-07-01 Study Group. Efficacy of Hydroxypropyl Cellulose Ophthalmic Inserts (LACRISERT) in Subsets of Patients with Dry Eye Syndrome: Findings From a Patient Registry. Cornea 2010, 29, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.; D’Aversa, G.; Perry, H.D.; Wittpenn, J.R.; Nelinson, D.S. Correlating Patient-Reported Response to Hydroxypropyl Cellulose Ophthalmic Insert (LACRISERT®) Therapy with Clinical Outcomes: Tools for Predicting Response. Curr. Eye Res. 2010, 35, 880–887. [Google Scholar] [CrossRef]
- Makarov, I.S.; Golova, L.K.; Vinogradov, M.I.; Egorov, Y.E.; Kulichikhin, V.G.; Mikhailov, Y.M. New Hydrated Cellulose Fiber Based on Flax Cellulose. Russ. J. Gen. Chem. 2021, 91, 1807–1815. [Google Scholar] [CrossRef]
- Mettler, M.S.; Paulsen, A.D.; Vlachos, D.G.; Dauenhauer, P.J. Pyrolytic Conversion of Cellulose to Fuels: Levoglucosan Deoxygenation via Elimination and Cyclization within Molten Biomass. Energy Environ. Sci. 2012, 5, 7864–7868. [Google Scholar] [CrossRef]
- Jiang, X.; Bai, Y.; Chen, X.; Liu, W. A Review on Raw Materials, Commercial Production and Properties of Lyocell Fiber. J. Bioresour. Bioprod. 2020, 5, 16–25. [Google Scholar] [CrossRef]
- Dhingra, D.; Michael, M.; Rajput, H.; Patil, R.T. Dietary Fibre in Foods: A Review. J. Food Sci. Technol. 2012, 49, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Courtenay, J.C.; Johns, M.A.; Galembeck, F.; Deneke, C.; Lanzoni, E.M.; Costa, C.A.; Scott, J.L.; Sharma, R.I. Surface Modified Cellulose Scaffolds for Tissue Engineering. Cellulose 2017, 24, 253–267. [Google Scholar] [CrossRef]
- de Oliveira Barud, H.G.; da Silva, R.R.; da Silva Barud, H.; Tercjak, A.; Gutierrez, J.; Lustri, W.R.; de Oliveira, O.B.; Ribeiro, S.J.L. A Multipurpose Natural and Renewable Polymer in Medical Applications: Bacterial Cellulose. Carbohydr. Polym. 2016, 153, 406–420. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Sabapathi, S.N. Cellulose Nanocrystals: Synthesis, Functional Properties, and Applications. Nanotechnol. Sci. Appl. 2015, 8, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.A.; Abd Mutalib, M.; Mohd Hir, Z.A.; Zain, M.F.M.; Mohamad, A.B.; Minggu, L.J.; Awang, N.A.; Salleh, W.N. An Overview on Cellulose-Based Material in Tailoring Bio-Hybrid Nanostructured Photocatalysts for Water Treatment and Renewable Energy Applications. Int. J. Biol. Macromol. 2017, 103, 1232–1256. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Sagarika, V.K.; Sabu, C.; Kalarikkal, N.; Thomas, S. Cellulose Nanocomposites: Fabrication and Biomedical Applications. J. Bioresour. Bioprod. 2020, 5, 223–237. [Google Scholar] [CrossRef]
- Duchemin, B.; Thuault, A.; Vicente, A.; Rigaud, B.; Fernandez, C.; Eve, S. Ultrastructure of Cellulose Crystallites in Flax Textile Fibres. Cellulose 2012, 19, 1837–1854. [Google Scholar] [CrossRef]
- Newman, R.H.; Hill, S.J.; Harris, P.J. Wide-Angle X-Ray Scattering and Solid-State Nuclear Magnetic Resonance Data Combined to Test Models for Cellulose Microfibrils in Mung Bean Cell Walls. Plant Physiol. 2013, 163, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, C.J.; Cameron, G.J.; Šturcová, A.; Apperley, D.C.; Altaner, C.; Wess, T.J.; Jarvis, M.C. Microfibril Diameter in Celery Collenchyma Cellulose: X-Ray Scattering and NMR Evidence. Cellulose 2007, 14, 235–246. [Google Scholar] [CrossRef]
- Zuluaga, R.; Putaux, J.L.; Cruz, J.; Vélez, J.; Mondragon, I.; Gañán, P. Cellulose Microfibrils from Banana Rachis: Effect of Alkaline Treatments on Structural and Morphological Features. Carbohydr. Polym. 2009, 76, 51–59. [Google Scholar] [CrossRef]
- Reif, B.; Ashbrook, S.E.; Emsley, L.; Hong, M. Solid-State NMR Spectroscopy. Nat. Rev. Methods Primer 2021, 1, 23. [Google Scholar] [CrossRef] [PubMed]
- Borisov, A.S.; Hazendonk, P.; Hayes, P.G. Solid-State Nuclear Magnetic Resonance Spectroscopy: A Review of Modern Techniques and Applications for Inorganic Polymers. J. Inorg. Organomet. Polym. Mater. 2010, 20, 183–212. [Google Scholar] [CrossRef]
- Brown, S.P. Advanced Solid-State NMR Methods for Characterising Structure and Self-Assembly in Supramolecular Chemistry, Polymers and Hydrogels. Curr. Opin. Colloid Interface Sci. 2018, 33, 86–98. [Google Scholar] [CrossRef]
- El Hariri El Nokab, M.; van der Wel, P.C.A. Use of Solid-State NMR Spectroscopy for Investigating Polysaccharide-Based Hydrogels: A Review. Carbohydr. Polym. 2020, 240, 116276. [Google Scholar] [CrossRef]
- Weingarth, M.; Baldus, M. Solid-State NMR-Based Approaches for Supramolecular Structure Elucidation. Acc. Chem. Res. 2013, 46, 2037–2046. [Google Scholar] [CrossRef] [PubMed]
- El Hariri El Nokab, M.; Sebakhy, K. Solid State NMR Spectroscopy a Valuable Technique for Structural Insights of Advanced Thin Film Materials: A Review. Nanomaterials 2021, 11, 1494. [Google Scholar] [CrossRef] [PubMed]
- Polenova, T.; Gupta, R.; Goldbourt, A. Magic Angle Spinning NMR Spectroscopy: A Versatile Technique for Structural and Dynamic Analysis of Solid-Phase Systems. Anal. Chem. 2015, 87, 5458–5469. [Google Scholar] [CrossRef]
- Foston, M. Advances in Solid-State NMR of Cellulose. Curr. Opin. Biotechnol. 2014, 27, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Yunoki, S.; Shikano, T.; Fujiwara, M.; Erata, T.; Takai, M. CP/MAS 13 C NMR Study of Cellulose and Cellulose Derivatives. 1. Complete Assignment of the CP/MAS 13C NMR Spectrum of the Native Cellulose. J. Am. Chem. Soc. 2002, 124, 7506–7511. [Google Scholar] [CrossRef] [PubMed]
- Okushita, K.; Komatsu, T.; Chikayama, E.; Kikuchi, J. Statistical Approach for Solid-State NMR Spectra of Cellulose Derived from a Series of Variable Parameters. Polym. J. 2012, 44, 895–900. [Google Scholar] [CrossRef]
- Pandey, M.K.; Qadri, Z.; Ramachandran, R. Understanding Cross-Polarization (CP) NMR Experiments through Dipolar Truncation. J. Chem. Phys. 2013, 138, 114108. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, R.; Iuliucci, R.J.; Behnke, G.; Brown, R.; Shoup, D.; Riedel, T.M.; Plavchak, C.; Lininger, B.E.; Spehar, J.M. Moving towards Fast Characterization of Polymorphic Drugs by Solid-State NMR Spectroscopy. J. Pharm. Biomed. Anal. 2018, 148, 163–169. [Google Scholar] [CrossRef]
- Southern, S.A.; Bryce, D.L. Chapter One—Recent Advances in NMR Crystallography and Polymorphism. In Annual Reports on NMR Spectroscopy; Webb, G.A., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 102, pp. 1–80. ISBN 0066-4103. [Google Scholar]
- Kolodziejski, W.; Klinowski, J. Kinetics of Cross-Polarization in Solid-State NMR: A Guide for Chemists. Chem. Rev. 2002, 102, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Grunin, Y.B.; Grunin, L.Y.; Nikol’skaya, E.A.; Talantsev, V.I. Microstructure of Cellulose: NMR Relaxation Study. Polym. Sci. Ser. A 2012, 54, 201–208. [Google Scholar] [CrossRef]
- Tian, D.; Li, T.; Zhang, R.; Wu, Q.; Chen, T.; Sun, P.; Ramamoorthy, A. Conformations and Intermolecular Interactions in Cellulose/Silk Fibroin Blend Films: A Solid-State NMR Perspective. J. Phys. Chem. B 2017, 121, 6108–6116. [Google Scholar] [CrossRef] [PubMed]
- Wanrosli, W.D.; Rohaizu, R.; Ghazali, A. Synthesis and Characterization of Cellulose Phosphate from Oil Palm Empty Fruit Bunches Microcrystalline Cellulose. Carbohydr. Polym. 2011, 84, 262–267. [Google Scholar] [CrossRef]
- Casaburi, A.; Montoya Rojo, Ú.; Cerrutti, P.; Vázquez, A.; Foresti, M.L. Carboxymethyl Cellulose with Tailored Degree of Substitution Obtained from Bacterial Cellulose. Food Hydrocoll. 2018, 75, 147–156. [Google Scholar] [CrossRef]
- Haslinger, S.; Hietala, S.; Hummel, M.; Maunu, S.L.; Sixta, H. Solid-State NMR Method for the Quantification of Cellulose and Polyester in Textile Blends. Carbohydr. Polym. 2019, 207, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Dai, Y.; Wang, C.; Tan, L. Quantitative and Structure Analysis of Cellulose in Tobacco by 13C CP / MAS NMR Spectroscopy. Beitr. Table Int. Tob. Res. 2016, 27, 126–135. [Google Scholar] [CrossRef][Green Version]
- Masuda, K.; Adachi, M.; Hirai, A.; Yamamoto, H.; Kaji, H.; Horii, F. Solid-State 13C and 1H Spin Diffusion NMR Analyses of the Microfibril Structure for Bacterial Cellulose. Solid State Nucl. Magn. Reson. 2003, 23, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Terrett, O.M.; Lyczakowski, J.J.; Yu, L.; Iuga, D.; Franks, W.T.; Brown, S.P.; Dupree, R.; Dupree, P. Molecular Architecture of Softwood Revealed by Solid-State NMR. Nat. Commun. 2019, 10, 4978. [Google Scholar] [CrossRef] [PubMed]
- Bernardinelli, O.D.; Lima, M.A.; Rezende, C.A.; Polikarpov, I.; deAzevedo, E.R. Quantitative 13C MultiCP Solid-State NMR as a Tool for Evaluation of Cellulose Crystallinity Index Measured Directly inside Sugarcane Biomass. Biotechnol. Biofuels 2015, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Zuckerstätter, G.; Terinte, N.; Sixta, H.; Schuster, K.C. Novel Insight into Cellulose Supramolecular Structure through 13C CP-MAS NMR Spectroscopy and Paramagnetic Relaxation Enhancement. Carbohydr. Polym. 2013, 93, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Durand, H.; Zeno, E.; Balsollier, C.; Watbled, B.; Sillard, C.; Fort, S.; Baussanne, I.; Belgacem, N.; Lee, D.; et al. The Surface Chemistry of a Nanocellulose Drug Carrier Unravelled by MAS-DNP. Chem. Sci. 2020, 11, 3868–3877. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, W.; Plog, A.; Xu, Y.; Buntkowsky, G.; Gutmann, T.; Zhang, K. Multi-Responsive Cellulose Nanocrystal–Rhodamine Conjugates: An Advanced Structure Study by Solid-State Dynamic Nuclear Polarization (DNP) NMR. Phys. Chem. Chem. Phys. 2014, 16, 26322–26329. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, S.; Alves, L.; Lindman, B.; Topgaard, D. Polarization Transfer Solid-State NMR: A New Method for Studying Cellulose Dissolution. RSC Adv. 2014, 4, 31836–31839. [Google Scholar] [CrossRef]
- Wang, T.; Yang, H.; Kubicki, J.D.; Hong, M. Cellulose Structural Polymorphism in Plant Primary Cell Walls Investigated by High-Field 2D Solid-State NMR Spectroscopy and Density Functional Theory Calculations. Biomacromolecules 2016, 17, 2210–2222. [Google Scholar] [CrossRef]
- White, P.B.; Wang, T.; Park, Y.B.; Cosgrove, D.J.; Hong, M. Water–Polysaccharide Interactions in the Primary Cell Wall of Arabidopsis Thaliana from Polarization Transfer Solid-State NMR. J. Am. Chem. Soc. 2014, 136, 10399–10409. [Google Scholar] [CrossRef] [PubMed]
- Sparrman, T.; Svenningsson, L.; Sahlin-Sjövold, K.; Nordstierna, L.; Westman, G.; Bernin, D. A Revised Solid-State NMR Method to Assess the Crystallinity of Cellulose. Cellulose 2019, 26, 8993–9003. [Google Scholar] [CrossRef]
- Ghosh, M.; Prajapati, B.P.; Suryawanshi, R.K.; Kishor Dey, K.; Kango, N. Study of the Effect of Enzymatic Deconstruction on Natural Cellulose by NMR Measurements. Chem. Phys. Lett. 2019, 727, 105–115. [Google Scholar] [CrossRef]
- Svenningsson, L.; Sparrman, T.; Bialik, E.; Bernin, D.; Nordstierna, L. Molecular Orientation Distribution of Regenerated Cellulose Fibers Investigated with Rotor Synchronized Solid State NMR Spectroscopy. Cellulose 2019, 26, 4681–4692. [Google Scholar] [CrossRef]
- Wang, S.; Sun, P.; Zhang, R.; Lu, A.; Liu, M.; Zhang, L. Cation/Macromolecule Interaction in Alkaline Cellulose Solution Characterized with Pulsed Field-Gradient Spin-Echo NMR Spectroscopy. Phys. Chem. Chem. Phys. 2017, 19, 7486–7490. [Google Scholar] [CrossRef] [PubMed]
- Kirui, A.; Ling, Z.; Kang, X.; Dickwella Widanage, M.C.; Mentink-Vigier, F.; French, A.D.; Wang, T. Atomic Resolution of Cotton Cellulose Structure Enabled by Dynamic Nuclear Polarization Solid-State NMR. Cellulose 2019, 26, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Lee, D.; Dubois, L.; Bardet, M.; Hediger, S.; De Paëpe, G. Rapid Natural-Abundance 2D 13C–13C Correlation Spectroscopy Using Dynamic Nuclear Polarization Enhanced Solid-State NMR and Matrix-Free Sample Preparation. Angew. Chem. Int. Ed. 2012, 51, 11766–11769. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Kirui, A.; Deligey, F.; Mentink-Vigier, F.; Zhou, Y.; Zhang, B.; Wang, T. Solid-State NMR of Unlabeled Plant Cell Walls: High-Resolution Structural Analysis without Isotopic Enrichment. Biotechnol. Biofuels 2021, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Idström, A.; Schantz, S.; Sundberg, J.; Chmelka, B.F.; Gatenholm, P.; Nordstierna, L. 13C NMR Assignments of Regenerated Cellulose from Solid-State 2D NMR Spectroscopy. Carbohydr. Polym. 2016, 151, 480–487. [Google Scholar] [CrossRef]
- Kita, Y.; Kusumi, R.; Kimura, T.; Kitaoka, M.; Nishiyama, Y.; Wada, M. Surface Structural Analysis of Selectively 13C-Labeled Cellulose II by Solid-State NMR Spectroscopy. Cellulose 2020, 27, 1899–1907. [Google Scholar] [CrossRef]
- Huang, H.; Ge, H.; Song, J.; Yao, Y.; Chen, Q.; Xu, M. NMR Study on the Roles of Li+ in the Cellulose Dissolution Process. ACS Sustain. Chem. Eng. 2019, 7, 618–624. [Google Scholar] [CrossRef]
- Phyo, P.; Wang, T.; Yang, Y.; O’Neill, H.; Hong, M. Direct Determination of Hydroxymethyl Conformations of Plant Cell Wall Cellulose Using 1 H Polarization Transfer Solid-State NMR. Biomacromolecules 2018, 19, 1485–1497. [Google Scholar] [CrossRef] [PubMed]
- Manríquez, R.; López-Dellamary, F.A.; Frydel, J.; Emmler, T.; Breitzke, H.; Buntkowsky, G.; Limbach, H.-H.; Shenderovich, I.G. Solid-State NMR Studies of Aminocarboxylic Salt Bridges in L -Lysine Modified Cellulose. J. Phys. Chem. B 2009, 113, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Lemke, C.H.; Dong, R.Y.; Michal, C.A.; Hamad, W.Y. New Insights into Nano-Crystalline Cellulose Structure and Morphology Based on Solid-State NMR. Cellulose 2012, 19, 1619–1629. [Google Scholar] [CrossRef]
- Felby, C.; Thygesen, L.G.; Kristensen, J.B.; Jørgensen, H.; Elder, T. Cellulose–Water Interactions during Enzymatic Hydrolysis as Studied by Time Domain NMR. Cellulose 2008, 15, 703–710. [Google Scholar] [CrossRef]
- Östlund, Å.; Idström, A.; Olsson, C.; Larsson, P.T.; Nordstierna, L. Modification of Crystallinity and Pore Size Distribution in Coagulated Cellulose Films. Cellulose 2013, 20, 1657–1667. [Google Scholar] [CrossRef]
- Liitiä, T.; Maunu, S.L.; Hortling, B.; Tamminen, T.; Pekkala, O.; Varhimo, A. Cellulose Crystallinity and Ordering of Hemicelluloses in Pine and Birch Pulps as Revealed by Solid-State NMR Spectroscopic Methods. Cellulose 2003, 10, 307–316. [Google Scholar] [CrossRef]
- Liitiä, T.; Maunu, S.L.; Hortling, B. Solid State NMR Studies on Inhomogeneous Structure of Fibre Wall in Kraft Pulp. Holzforschung 2001, 55, 503–510. [Google Scholar] [CrossRef]
- Siller, M.; Amer, H.; Bacher, M.; Roggenstein, W.; Rosenau, T.; Potthast, A. Effects of Periodate Oxidation on Cellulose Polymorphs. Cellulose 2015, 22, 2245–2261. [Google Scholar] [CrossRef]
- Modica, A.; Rosselli, S.; Catinella, G.; Sottile, F.; Catania, C.A.; Cavallaro, G.; Lazzara, G.; Botta, L.; Spinella, A.; Bruno, M. Solid State 13C-NMR Methodology for the Cellulose Composition Studies of the Shells of Prunus Dulcis and Their Derived Cellulosic Materials. Carbohydr. Polym. 2020, 240, 116290. [Google Scholar] [CrossRef] [PubMed]
- Meza-Contreras, J.C.; Manriquez-Gonzalez, R.; Gutiérrez-Ortega, J.A.; Gonzalez-Garcia, Y. XRD and Solid State 13C-NMR Evaluation of the Crystallinity Enhancement of 13C-Labeled Bacterial Cellulose Biosynthesized by Komagataeibacter Xylinus under Different Stimuli: A Comparative Strategy of Analyses. Carbohydr. Res. 2018, 461, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Foston, M.B.; Hubbell, C.A.; Ragauskas, A.J. Cellulose Isolation Methodology for NMR Analysis of Cellulose Ultrastructure. Materials 2011, 4, 1985–2002. [Google Scholar] [CrossRef] [PubMed]
- Palme, A.; Idström, A.; Nordstierna, L.; Brelid, H. Chemical and Ultrastructural Changes in Cotton Cellulose Induced by Laundering and Textile Use. Cellulose 2014, 21, 4681–4691. [Google Scholar] [CrossRef]
- Fu, L.; McCallum, S.A.; Miao, J.; Hart, C.; Tudryn, G.J.; Zhang, F.; Linhardt, R.J. Rapid and Accurate Determination of the Lignin Content of Lignocellulosic Biomass by Solid-State NMR. Fuel 2015, 141, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Kirui, A.; Dickwella Widanage, M.C.; Mentink-Vigier, F.; Cosgrove, D.J.; Wang, T. Lignin-Polysaccharide Interactions in Plant Secondary Cell Walls Revealed by Solid-State NMR. Nat. Commun. 2019, 10, 347. [Google Scholar] [CrossRef]
- Simmons, T.J.; Mortimer, J.C.; Bernardinelli, O.D.; Pöppler, A.-C.; Brown, S.P.; deAzevedo, E.R.; Dupree, R.; Dupree, P. Folding of Xylan onto Cellulose Fibrils in Plant Cell Walls Revealed by Solid-State NMR. Nat. Commun. 2016, 7, 13902. [Google Scholar] [CrossRef] [PubMed]
- Coseri, S.; Biliuta, G.; Simionescu, B.C.; Stana-Kleinschek, K.; Ribitsch, V.; Harabagiu, V. Oxidized Cellulose—Survey of the Most Recent Achievements. Carbohydr. Polym. 2013, 93, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Kim, U.-J.; Kuga, S.; Wada, M.; Okano, T.; Kondo, T. Periodate Oxidation of Crystalline Cellulose. Biomacromolecules 2000, 1, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Eyholzer, C.; Bordeanu, N.; Lopez-Suevos, F.; Rentsch, D.; Zimmermann, T.; Oksman, K. Preparation and Characterization of Water-Redispersible Nanofibrillated Cellulose in Powder Form. Cellulose 2010, 17, 19–30. [Google Scholar] [CrossRef]
- Sannigrahi, P.; Miller, S.J.; Ragauskas, A.J. Effects of Organosolv Pretreatment and Enzymatic Hydrolysis on Cellulose Structure and Crystallinity in Loblolly Pine. Carbohydr. Res. 2010, 345, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, B.; Rosenau, T.; Potthast, A. Effect of Sonochemical Treatments on the Integrity and Oxidation State of Cellulose. Carbohydr. Polym. 2013, 92, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Kwak, J.H.; Wang, Y.; Franz, J.A.; White, J.M.; Holladay, J.E. Effects of Crystallinity on Dilute Acid Hydrolysis of Cellulose by Cellulose Ball-Milling Study. Energy Fuels 2006, 20, 807–811. [Google Scholar] [CrossRef]
- Sirvio, J.; Hyvakko, U.; Liimatainen, H.; Niinimaki, J.; Hormi, O. Periodate Oxidation of Cellulose at Elevated Temperatures Using Metal Salts as Cellulose Activators. Carbohydr. Polym. 2011, 83, 1293–1297. [Google Scholar] [CrossRef]
- Park, S.; Baker, J.O.; Himmel, M.E.; Parilla, P.A.; Johnson, D.K. Cellulose Crystallinity Index: Measurement Techniques and Their Impact on Interpreting Cellulase Performance. Biotechnol. Biofuels 2010, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Johnson, D.K.; Ishizawa, C.I.; Parilla, P.A.; Davis, M.F. Measuring the Crystallinity Index of Cellulose by Solid State 13C Nuclear Magnetic Resonance. Cellulose 2009, 16, 641–647. [Google Scholar] [CrossRef]
- Isogai, A.; Saito, T.; Fukuzumi, H. TEMPO-Oxidized Cellulose Nanofibers. Nanoscale 2011, 3, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Abitbol, T.; Rivkin, A.; Cao, Y.; Nevo, Y.; Abraham, E.; Ben-Shalom, T.; Lapidot, S.; Shoseyov, O. Nanocellulose, a Tiny Fiber with Huge Applications. Curr. Opin. Biotechnol. 2016, 39, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, A. Nanocellulose Processing Properties and Potential Applications. Curr. For. Rep. 2019, 5, 76–89. [Google Scholar] [CrossRef]
- Capadona, J.R.; Van Den Berg, O.; Capadona, L.A.; Schroeter, M.; Rowan, S.J.; Tyler, D.J.; Weder, C. A Versatile Approach for the Processing of Polymer Nanocomposites with Self-Assembled Nanofibre Templates. Nat. Nanotechnol. 2007, 2, 765–769. [Google Scholar] [CrossRef]
- Rezayat, M.; Blundell, R.K.; Camp, J.E.; Walsh, D.A.; Thielemans, W. Green One-Step Synthesis of Catalytically Active Palladium Nanoparticles Supported on Cellulose Nanocrystals. ACS Sustain. Chem. Eng. 2014, 2, 1241–1250. [Google Scholar] [CrossRef]
- Idström, A.; Brelid, H.; Nydén, M.; Nordstierna, L. CP/MAS 13C NMR Study of Pulp Hornification Using Nanocrystalline Cellulose as a Model System. Carbohydr. Polym. 2013, 92, 881–884. [Google Scholar] [CrossRef]
- Liu, J.; Plog, A.; Groszewicz, P.; Zhao, L.; Xu, Y.; Breitzke, H.; Stark, A.; Hoffmann, R.; Gutmann, T.; Zhang, K.; et al. Design of a Heterogeneous Catalyst Based on Cellulose Nanocrystals for Cyclopropanation: Synthesis and Solid-State NMR Characterization. Chem. Eur. J. 2015, 21, 12414–12420. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, T.; Liu, J.; Rothermel, N.; Xu, Y.; Jaumann, E.; Werner, M.; Breitzke, H.; Sigurdsson, S.T.; Buntkowsky, G. Natural Abundance 15N NMR by Dynamic Nuclear Polarization: Fast Analysis of Binding Sites of a Novel Amine-Carboxyl-Linked Immobilized Dirhodium Catalyst. Chem. Eur. J. 2015, 21, 3798–3805. [Google Scholar] [CrossRef]
- Huang, J.-L.; Li, C.-J.; Gray, D.G. Cellulose Nanocrystals Incorporating Fluorescent Methylcoumarin Groups. ACS Sustain. Chem. Eng. 2013, 1, 1160–1164. [Google Scholar] [CrossRef]
- Celebi, D.; Guy, R.H.; Edler, K.J.; Scott, J.L. Ibuprofen Delivery into and through the Skin from Novel Oxidized Cellulose-Based Gels and Conventional Topical Formulations. Int. J. Pharm. 2016, 514, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Paukkonen, H.; Kunnari, M.; Laurén, P.; Hakkarainen, T.; Auvinen, V.-V.; Oksanen, T.; Koivuniemi, R.; Yliperttula, M.; Laaksonen, T. Nanofibrillar Cellulose Hydrogels and Reconstructed Hydrogels as Matrices for Controlled Drug Release. Int. J. Pharm. 2017, 532, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Plackett, D.; Letchford, K.; Jackson, J.; Burt, H. A Review of Nanocellulose as a Novel Vehicle for Drug Delivery. Nord. Pulp Pap. Res. J. 2014, 29, 105–118. [Google Scholar] [CrossRef]
- El Hariri El Nokab, M.; Lasorsa, A.; Sebakhy, K.O.; Picchioni, F.; van der Wel, P.C.A. Solid-State NMR Spectroscopy Insights for Resolving Different Water Pools in Alginate Hydrogels. Food Hydrocoll. 2022, 127, 107500. [Google Scholar] [CrossRef]
- Wang, T.; Phyo, P.; Hong, M. Multidimensional Solid-State NMR Spectroscopy of Plant Cell Walls. Solid State Nucl. Magn. Reson. 2016, 78, 56–63. [Google Scholar] [CrossRef]
- Chakraborty, A.; Fernando, L.D.; Fang, W.; Dickwella Widanage, M.C.; Wei, P.; Jin, C.; Fontaine, T.; Latgé, J.-P.; Wang, T. A Molecular Vision of Fungal Cell Wall Organization by Functional Genomics and Solid-State NMR. Nat. Commun. 2021, 12, 6346. [Google Scholar] [CrossRef]
- Zhao, W.; Fernando, L.D.; Kirui, A.; Deligey, F.; Wang, T. Solid-State NMR of Plant and Fungal Cell Walls: A Critical Review. Solid State Nucl. Magn. Reson. 2020, 107, 101660. [Google Scholar] [CrossRef] [PubMed]
- Santoni, I.; Callone, E.; Sandak, A.; Sandak, J.; Dirè, S. Solid State NMR and IR Characterization of Wood Polymer Structure in Relation to Tree Provenance. Carbohydr. Polym. 2015, 117, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Bergenstråhle-Wohlert, M.; Berglund, L.A.; Brady, J.W.; Larsson, P.T.; Westlund, P.-O.; Wohlert, J. Concentration Enrichment of Urea at Cellulose Surfaces: Results from Molecular Dynamics Simulations and NMR Spectroscopy. Cellulose 2012, 19, 1–12. [Google Scholar] [CrossRef]
- Mori, T.; Chikayama, E.; Tsuboi, Y.; Ishida, N.; Shisa, N.; Noritake, Y.; Moriya, S.; Kikuchi, J. Exploring the Conformational Space of Amorphous Cellulose Using NMR Chemical Shifts. Carbohydr. Polym. 2012, 90, 1197–1203. [Google Scholar] [CrossRef] [PubMed]




| Pulse Sequence | Applications | References |
|---|---|---|
| 1D 1H Static NMR | Detection of cellulose I different phases | [56] |
| 1D 1H a CRAMPS | High-resolution spectra for rigid solids and inter-molecular interactions | [57] |
| 1D 1H Depth | Indicating rigid structures (chain dynamics) | [57] |
| 1D 31P MAS | Structural confirmation of cellulose phosphorylation | [58] |
| 1D 13C CP MAS | Structure analysis of cellulose, quantification of cellulose in blends, determination of degree of substitution | [59,60,61] |
| 1D 13C CP MAS, b DANTE selective excitation pulse trains | Selective excitation of the C4 as a function of the diffusion time | [62] |
| 1D 13C CP-c PSRE MAS | Detecting cellulose crystallite thickness | [38] |
| 1D 13C CP-d PDSD MAS | Cellulose structure analysis and cross-peak determination with lignin | [63] |
| 1D 13C MultiCP MAS | Quantitative cellulose crystallinity | [64] |
| 1D 13C MultiCP MAS with dipolar filter | Filtering spectra for hemicellulose and lignin (spectral editing) | [64] |
| Paramagnetic relaxation enhancement 1D 13C T1 CP MAS | Solvent accessibility of different fractions of wetted cellulose | [65] |
| g DNP enhanced 1D 13C CP MAS | Surface chemistry of cellulose and nanocellulose | [66,67] |
| Polarization transfer solid state NMR (e DP, CP and f INEPT) | Studying of cellulose dissolution | [68] |
| Polarization transfer solid state NMR (T1, T2 filter) | Water proximity to cellulose, hemicellulose and pectin. Water mobility | [69,70] |
| Inversion recovery CP excitation and Saturation recovery | Estimation of 13C T1 relaxation times and crystallinity of cellulose | [71] |
| Relaxation measurements by Torchia-CP | Modified method for measuring 13C T1 relaxation times | [72] |
| Rotor synchronized MAS | Investigating the molecular orientation distribution | [73] |
| 7Li and 23Na h PFG-SE NMR | Studying of cellulose dissolution, provide proximity information between cations/macromolecule | [74] |
| 2D 13C-13C i INADEQUATE by g DNP | Atomic resolution structural analysis without isotopic labeling | [75,76,77] |
| 2D 13C-13C i INADEQUATE by 13C isotopic labeling | Structural connectivity determination | [78,79] |
| 2D 13C-13C d PDSD/j DARR | Structural connectivity determination | [51,78] |
| 2D 1H-6Li,13C k LGHETCOR | Studying of cellulose dissolution, provide proximity information between covalent bonded 1H-13C spins, and probing remote 1H-13C correlations | [80] |
| 2D 13C d PDSD experiments | Multi-bond and long range inter-molecular cross peaks | [69,70] |
| 2D CHHC correlation experiments | Determination of the hydroxylmethyl conformations | [81] |
| 2D l RFDR correlation experiments | Assigning the intra-residue cross peaks | [69] |
| 2D 1H-13C m WISE | Indicating molecular mobility and water localization in blends | [57] |
| 13C-15N n REDOR experiments | Indicate distance between carboxyl carbon and nitrogen of the modification | [82] |
| 2H-13C n REDOR experiments | Detection of the 1H-2H exchangeable regions in cellulose | [83] |
| o CPMG Time domain NMR | Identifying different water phases in hydrolyzed cellulose | [84] |
| o CPMG Cryoporometry NMR | Determination of pore volume, radius and size distribution | [85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Hariri El Nokab, M.; Habib, M.H.; Alassmy, Y.A.; Abduljawad, M.M.; Alshamrani, K.M.; Sebakhy, K.O. Solid State NMR a Powerful Technique for Investigating Sustainable/Renewable Cellulose-Based Materials. Polymers 2022, 14, 1049. https://doi.org/10.3390/polym14051049
El Hariri El Nokab M, Habib MH, Alassmy YA, Abduljawad MM, Alshamrani KM, Sebakhy KO. Solid State NMR a Powerful Technique for Investigating Sustainable/Renewable Cellulose-Based Materials. Polymers. 2022; 14(5):1049. https://doi.org/10.3390/polym14051049
Chicago/Turabian StyleEl Hariri El Nokab, Mustapha, Mohamed H. Habib, Yasser A. Alassmy, Marwan M. Abduljawad, Khalid M. Alshamrani, and Khaled O. Sebakhy. 2022. "Solid State NMR a Powerful Technique for Investigating Sustainable/Renewable Cellulose-Based Materials" Polymers 14, no. 5: 1049. https://doi.org/10.3390/polym14051049
APA StyleEl Hariri El Nokab, M., Habib, M. H., Alassmy, Y. A., Abduljawad, M. M., Alshamrani, K. M., & Sebakhy, K. O. (2022). Solid State NMR a Powerful Technique for Investigating Sustainable/Renewable Cellulose-Based Materials. Polymers, 14(5), 1049. https://doi.org/10.3390/polym14051049