Applications of Polymers for Organ-on-Chip Technology in Urology
Abstract
1. Introduction
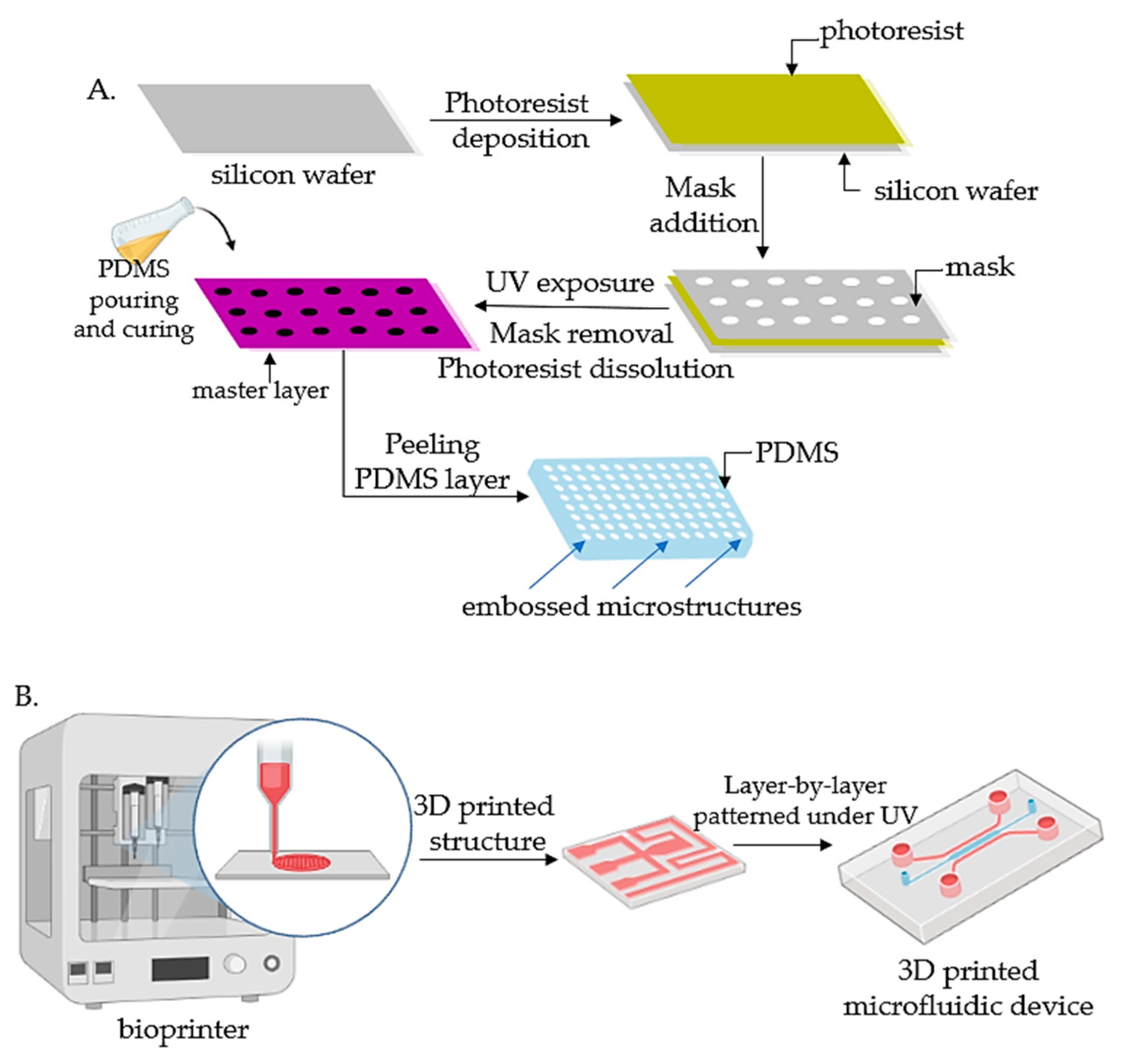
2. Fabrication of Organ-on-Chip
2.1. Principle and Manufacture of Organ-on-Chips
2.1.1. Lithography
2.1.2. Bioprinting
2.2. Material Requirements for the Fabrication of Organ-on-Chips
2.3. Polymers for Organ-on-Chips
3. Kidney-on-Chip
3.1. In Vitro 2D/3D Models for Kidney Disease Modeling
3.2. Kidney-on-Chip Models
3.2.1. Glomerulus-on-Chip
3.2.2. Tubule-on-Chip
3.3. Multiorgan-on-Chip Models That Include Kidney-on-Chip
4. Bladder-on-Chip
4.1. Modeling Bladder Cancer
4.2. Modeling Urinary Tract Infections
5. Prostate-on-Chip
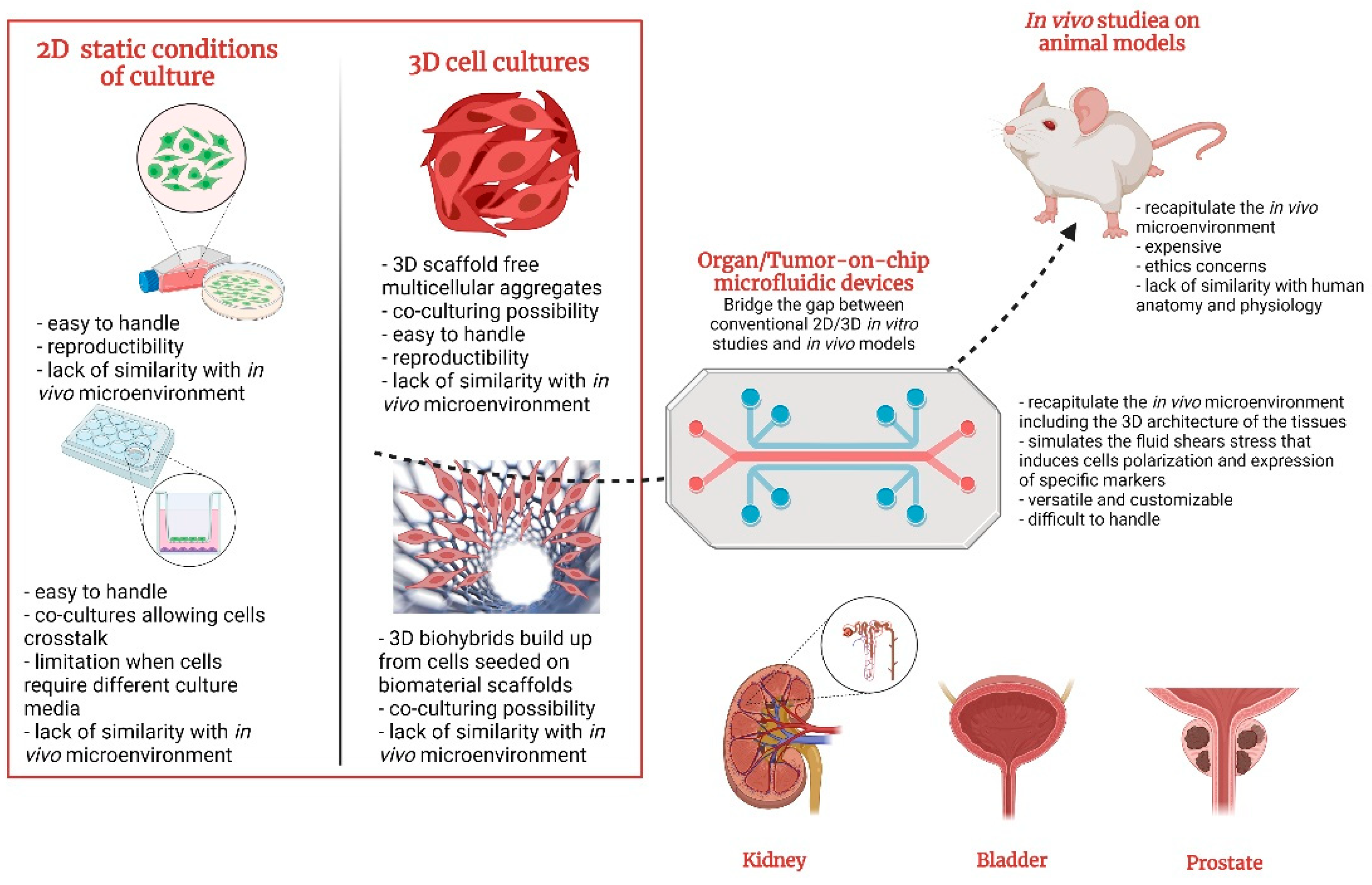
6. Benefits and Challenges of OOCs/TOCs in Urology
7. Conclusions and Future Considerations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Faria, J.; Ahmed, S.; Gerritsen, K.G.; Mihaila, S.M.; Masereeuw, R. Kidney-Based in Vitro Models for Drug-Induced Toxicity Testing. Arch. Toxicol. 2019, 93, 3397–3418. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, F. Kidney-on-a-Chip. In Organ-on-a-Chip; Elsevier: Amsterdam, The Netherlands, 2020; pp. 233–253. [Google Scholar]
- Tiong, H.Y.; Huang, P.; Xiong, S.; Li, Y.; Vathsala, A.; Zink, D. Drug-Induced Nephrotoxicity: Clinical Impact and Preclinical in Vitro Models. Mol. Pharm. 2014, 11, 1933–1948. [Google Scholar] [CrossRef] [PubMed]
- Day, C.-P.; Merlino, G.; Van Dyke, T. Preclinical Mouse Cancer Models: A Maze of Opportunities and Challenges. Cell 2015, 163, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Berlin, J.A.; Glasser, S.C.; Ellenberg, S.S. Adverse Event Detection in Drug Development: Recommendations and Obligations beyond Phase 3. Am. J. Public Health 2008, 98, 1366–1371. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Luo, Z.; Lee, J.; Kim, H.; Lee, K.; Tebon, P.; Feng, Y.; Dokmeci, M.R.; Sengupta, S.; Khademhosseini, A. Organ-on-a-chip for Cancer and Immune Organs Modeling. Adv. Healthc. Mater. 2019, 8, 1801363. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D. The Pharmaceutical Industry and the Future of Drug Development. In Pharmaceuticals in the Environment; Royal Society of Chemistry: London, UK, 2015; pp. 1–33. [Google Scholar] [CrossRef]
- Soo, J.Y.-C.; Jansen, J.; Masereeuw, R.; Little, M.H. Advances in Predictive in Vitro Models of Drug-Induced Nephrotoxicity. Nat. Rev. Nephrol. 2018, 14, 378–393. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, A. The Flaws and Human Harms of Animal Experimentation. Camb. Q. Healthc. Ethics 2015, 24, 407–419. [Google Scholar] [CrossRef]
- Bracken, M.B. Why Animal Studies Are Often Poor Predictors of Human Reactions to Exposure. J. R. Soc. Med. 2009, 102, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Baumans, V. Use of Animals in Experimental Research: An Ethical Dilemma? Gene Ther. 2004, 11, S64–S66. [Google Scholar] [CrossRef]
- Festing, S.; Wilkinson, R. The Ethics of Animal Research: Talking Point on the Use of Animals in Scientific Research. EMBO Rep. 2007, 8, 526–530. [Google Scholar] [CrossRef] [PubMed]
- European Parliament. Available online: Https://Www.Europarl.Europa.Eu/Plenary/En/Vod.Html?Mode=chapter&vodLanguage=EN&vodId=6ea360e5-0dd3-Decd-2a72-642c028c0a34&date=20210708# (accessed on 25 February 2022).
- American Institute for Cancer Research. Available online: https://www.wcrf.org/dietandcancer/worldwide-cancer-data/2020 (accessed on 25 February 2022).
- World Health Organization. Available online: https://gco.iarc.fr/tomorrow/en/dataviz/bars?mode=cancer&group_populations=1&multiple_cancers=1&cancers=39_27_30_29&key=total&show_bar_mode_prop=1&populations=903_904_905_908_909_935&years=2040&sexes=1&types=02020 (accessed on 25 February 2022).
- Trujillo-de Santiago, G.; Flores-Garza, B.G.; Tavares-Negrete, J.A.; Lara-Mayorga, I.M.; González-Gamboa, I.; Zhang, Y.S.; Rojas-Martínez, A.; Ortiz-López, R.; Álvarez, M.M. The Tumor-on-Chip: Recent Advances in the Development of Microfluidic Systems to Recapitulate the Physiology of Solid Tumors. Materials 2019, 12, 2945. [Google Scholar] [CrossRef] [PubMed]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-Dimensional Cell Culture Systems as an in Vitro Platform for Cancer and Stem Cell Modeling. World J. Stem Cells 2019, 11, 1065. [Google Scholar] [CrossRef]
- Przekwas, A.; Somayaji, M.R. Computational Pharmacokinetic Modeling of Organ-on-Chip Devices and Microphysiological Systems. Organ-A-Chip 2020, 311–361. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling Cancer in Microfluidic Human Organs-on-Chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T. The Tumor Microenvironment and Its Role in Promoting Tumor Growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, J.; Huang, S.; Wu, X.; Xie, X.; Wang, J.; Liu, F.; Zhang, M.; Peng, Z.; Hu, N. Tumor-on-a-Chip: From Bioinspired Design to Biomedical Application. Microsyst. Nanoeng. 2021, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-Chips: Into the next Decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef]
- Ingber, D.E. Developmentally Inspired Human ‘Organs on Chips. Development 2018, 145, dev156125. [Google Scholar] [CrossRef]
- Brill-Karniely, Y.; Dror, D.; Duanis-Assaf, T.; Goldstein, Y.; Schwob, O.; Millo, T.; Orehov, N.; Stern, T.; Jaber, M.; Loyfer, N. Triangular Correlation (TrC) between Cancer Aggressiveness, Cell Uptake Capability, and Cell Deformability. Sci. Adv. 2020, 6, eaax2861. [Google Scholar] [CrossRef]
- Rahmanuddin, S.; Korn, R.; Cridebring, D.; Borazanci, E.; Brase, J.; Boswell, W.; Jamil, A.; Cai, W.; Sabir, A.; Motarjem, P. Role of 3D Volumetric and Perfusion Imaging for Detecting Early Changes in Pancreatic Adenocarcinoma. Front. Oncol. 2021, 11, 678617. [Google Scholar] [CrossRef] [PubMed]
- Gilkes, D.M.; Semenza, G.L.; Wirtz, D. Hypoxia and the Extracellular Matrix: Drivers of Tumour Metastasis. Nat. Rev. Cancer 2014, 14, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Ehsan, S.M.; Welch-Reardon, K.M.; Waterman, M.L.; Hughesbce, C.C.; George, S.C. A three-dimensional in vitro model of tumor cell intravasation. Integr. Biol (Camb). 2014, 6, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-A.; King, A.D.; Shih, H.-C.; Peng, C.-C.; Wu, C.-Y.; Liao, W.-H.; Tung, Y.-C. Generation of Oxygen Gradients in Microfluidic Devices for Cell Culture Using Spatially Confined Chemical Reactions. Lab Chip 2011, 11, 3626–3633. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.-C.; Lee, T.-A.; Wu, H.-M.; Ko, P.-L.; Liao, W.-H.; Tung, Y.-C. Microfluidic Collective Cell Migration Assay for Study of Endothelial Cell Proliferation and Migration under Combinations of Oxygen Gradients, Tensions, and Drug Treatments. Sci. Rep. 2019, 9, 8234. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic Organs-on-Chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Barrile, R.; van der Meer, A.D.; Park, H.; Fraser, J.P.; Simic, D.; Teng, F.; Conegliano, D.; Nguyen, J.; Jain, A.; Zhou, M. Organ-on-chip Recapitulates Thrombosis Induced by an Anti-CD154 Monoclonal Antibody: Translational Potential of Advanced Microengineered Systems. Clin. Pharmacol. Ther. 2018, 104, 1240–1248. [Google Scholar] [CrossRef]
- Ferrari, E.; Nebuloni, F.; Rasponi, M.; Occhetta, P. Photo and Soft Lithography for Organ-on-Chip Applications. In Organ-on-a-Chip; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1–19. [Google Scholar]
- Puryear, J.R., III; Yoon, J.-K.; Kim, Y. Advanced Fabrication Techniques of Microengineered Physiological Systems. Micromachines 2020, 11, 730. [Google Scholar] [CrossRef]
- Sticker, D.; Rothbauer, M.; Lechner, S.; Hehenberger, M.-T.; Ertl, P. Multi-Layered, Membrane-Integrated Microfluidics Based on Replica Molding of a Thiol–Ene Epoxy Thermoset for Organ-on-a-Chip Applications. Lab Chip 2015, 15, 4542–4554. [Google Scholar] [CrossRef]
- Tajeddin, A.; Mustafaoglu, N. Design and Fabrication of Organ-on-Chips: Promises and Challenges. Micromachines 2021, 12, 1443. [Google Scholar] [CrossRef]
- Shrestha, J.; Ghadiri, M.; Shanmugavel, M.; Razavi Bazaz, S.; Vasilescu, S.; Ding, L. A Rapidly Prototyped Lung-on-a-Chip Model Using 3D-Printed Molds. Organs-A-Chip 2019, 1, 100001. [Google Scholar] [CrossRef]
- Yang, Q.; Lian, Q.; Xu, F. Perspective: Fabrication of Integrated Organ-on-a-Chip via Bioprinting. Biomicrofluidics 2017, 11, 031301. [Google Scholar] [CrossRef] [PubMed]
- Kasi, D.G.; de Graaf, M.N.; Motreuil-Ragot, P.A.; Frimat, J.-P.; Ferrari, M.D.; Sarro, P.M.; Mastrangeli, M.; van den Maagdenberg, A.M.; Mummery, C.L.; Orlova, V.V. Rapid Prototyping of Organ-on-a-Chip Devices Using Maskless Photolithography. Micromachines 2022, 13, 49. [Google Scholar] [CrossRef]
- Christoffersson, J.; Mandenius, C.-F. Fabrication of a Microfluidic Cell Culture Device Using Photolithographic and Soft Lithographic Techniques. In Cell-Based Assays Using iPSCs for Drug Development and Testing; Springer: Berlin/Heidelberg, Germany, 2019; pp. 227–233. [Google Scholar]
- Ferreira, D.A.; Rothbauer, M.; Conde, J.P.; Ertl, P.; Oliveira, C.; Granja, P.L. A Fast Alternative to Soft Lithography for the Fabrication of Organ-on-a-Chip Elastomeric-Based Devices and Microactuators. Adv. Sci. 2021, 8, 2003273. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.; Ingber, D. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef]
- Chiadò, A.; Palmara, G.; Chiappone, A.; Tanzanu, C.; Pirri, C.F.; Roppolo, I.; Frascella, F. A Modular 3D Printed Lab-on-a-Chip for Early Cancer Detection. Lab Chip 2020, 20, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Samadian, H.; Jafari, S.; Sepand, M.; Alaei, L.; Malvajerd, S.S.; Jaymand, M.; Ghobadinezhad, F.; Jahanshahi, F.; Hamblin, M.; Derakhshankhah, H. 3D Bioprinting Technology to Mimic the Tumor Microenvironment: Tumor-on-a-Chip Concept. Mater. Today Adv. 2021, 12, 100160. [Google Scholar] [CrossRef]
- Lantada, A.D.; Pfleging, W.; Besser, H.; Guttmann, M.; Wissmann, M.; Plewa, K.; Smyrek, P.; Piotter, V.; García-Ruíz, J.P. Research on the Methods for the Mass Production of Multi-Scale Organs-on-Chips. Polymers 2018, 10, 1238. [Google Scholar] [CrossRef] [PubMed]
- Gröger, M.; Dinger, J.; Kiehntopf, M.; Peters, F.T.; Rauen, U.; Mosig, A.S. Preservation of Cell Structure, Metabolism, and Biotransformation Activity of Liver-on-chip Organ Models by Hypothermic Storage. Adv. Healthc. Mater. 2018, 7, 1700616. [Google Scholar] [CrossRef] [PubMed]
- Shaegh, S.A.M.; Pourmand, A.; Nabavinia, M.; Avci, H.; Tamayol, A.; Mostafalu, P.; Ghavifekr, H.B.; Aghdam, E.N.; Dokmeci, M.R.; Khademhosseini, A. Rapid Prototyping of Whole-Thermoplastic Microfluidics with Built-in Microvalves Using Laser Ablation and Thermal Fusion Bonding. Sens. Actuators B Chem. 2018, 255, 100–109. [Google Scholar] [CrossRef]
- Thakare, K.; Jerpseth, L.; Pei, Z.; Elwany, A.; Quek, F.; Qin, H. Bioprinting of Organ-on-Chip Systems: A Literature Review from a Manufacturing Perspective. J. Manuf. Mater. Process. 2021, 5, 91. [Google Scholar] [CrossRef]
- Cao, X.; Ashfaq, R.; Cheng, F.; Maharjan, S.; Li, J.; Ying, G.; Hassan, S.; Xiao, H.; Yue, K.; Zhang, Y.S. A Tumor-on-a-chip System with Bioprinted Blood and Lymphatic Vessel Pair. Adv. Funct. Mater. 2019, 29, 1807173. [Google Scholar] [CrossRef] [PubMed]
- O’Cearbhaill, E. 3D Bioprinting Chips Away at Glioblastomal Resistance. Sci. Transl. Med. 2019, 11, eaax1724. [Google Scholar] [CrossRef]
- Bishop, E.S.; Mostafa, S.; Pakvasa, M.; Luu, H.H.; Lee, M.J.; Wolf, J.M.; Ameer, G.A.; He, T.-C.; Reid, R.R. 3-D Bioprinting Technologies in Tissue Engineering and Regenerative Medicine: Current and Future Trends. Genes Dis. 2017, 4, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Gan, S.; Wang, X.; Liu, W.; Li, X. Applications of 3D Bioprinting in Tissue Engineering: Advantages, Deficiencies, Improvements, and Future Perspectives. J. Mater. Chem. B 2021, 9, 5385–5413. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, T.G.; Manolesou, D.; Dimakakos, E.; Tsoucalas, G.; Vavuranakis, M.; Tousoulis, D. 3D Bioprinting Methods and Techniques: Applications on Artificial Blood Vessel Fabrication. Acta Cardiol. Sin. 2019, 35, 284. [Google Scholar]
- Colosi, C.; Shin, S.R.; Manoharan, V.; Massa, S.; Costantini, M.; Barbetta, A.; Dokmeci, M.R.; Dentini, M.; Khademhosseini, A. Microfluidic Bioprinting of Heterogeneous 3D Tissue Constructs Using Low-viscosity Bioink. Adv. Mater. 2016, 28, 677–684. [Google Scholar] [CrossRef]
- Keenan, T.M.; Folch, A. Biomolecular Gradients in Cell Culture Systems. Lab Chip 2008, 8, 34–57. [Google Scholar] [CrossRef]
- Nagy, J.; Chang, S.H.; Dvorak, A.M.; Dvorak, H.F. Why Are Tumour Blood Vessels Abnormal and Why Is It Important to Know? Br. J. Cancer 2009, 100, 865–869. [Google Scholar] [CrossRef]
- Han, S.; Kim, S.; Chen, Z.; Shin, H.K.; Lee, S.-Y.; Moon, H.E.; Paek, S.H.; Park, S. 3D Bioprinted Vascularized Tumour for Drug Testing. Int. J. Mol. Sci. 2020, 21, 2993. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.G.; Choi, Y.; Jang, J. 3D Bioprinting-Based Vascularized Tissue Models Mimicking Tissue-Specific Architecture and Pathophysiology for in Vitro Studies. Front. Bioeng. Biotechnol. 2021, 9, 685507. [Google Scholar] [CrossRef]
- Datta, P.; Dey, M.; Ataie, Z.; Unutmaz, D.; Ozbolat, I.T. 3D Bioprinting for Reconstituting the Cancer Microenvironment. NPJ Precis. Oncol. 2020, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Ryu, H.; Lee, B.; Ha, D.-H.; Ahn, M.; Kim, S.; Kim, J.Y.; Jeon, N.L.; Cho, D.-W. Development of a Functional Airway-on-a-Chip by 3D Cell Printing. Biofabrication 2018, 11, 015002. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Kim, G. Intestinal Villi Model with Blood Capillaries Fabricated Using Collagen-Based Bioink and Dual-Cell-Printing Process. ACS Appl. Mater. Interfaces 2018, 10, 41185–41196. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Pr, A.K.; Yoo, J.J.; Zahran, F.; Atala, A.; Lee, S.J. A Photo-crosslinkable Kidney ECM-derived Bioink Accelerates Renal Tissue Formation. Adv. Healthc. Mater. 2019, 8, 1800992. [Google Scholar] [CrossRef]
- Fransen, M.F.; Addario, G.; Bouten, C.V.; Halary, F.; Moroni, L.; Mota, C. Bioprinting of Kidney in Vitro Models: Cells, Biomaterials, and Manufacturing Techniques. Essays Biochem. 2021, 65, 587–602. [Google Scholar]
- Sun, M.; Sun, X.; Wang, Z.; Guo, S.; Yu, G.; Yang, H. Synthesis and Properties of Gelatin Methacryloyl (GelMA) Hydrogels and Their Recent Applications in Load-Bearing Tissue. Polymers 2018, 10, 1290. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.; Kamble, S.C.; Kashikar, N.C. Polymeric Bioinks for 3D Hepatic Printing. Chemistry 2021, 3, 164–181. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.-R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V. Bioprinting 3D Microfibrous Scaffolds for Engineering Endothelialized Myocardium and Heart-on-a-Chip. Biomaterials 2016, 110, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Lind, J.U.; Busbee, T.A.; Valentine, A.D.; Pasqualini, F.S.; Yuan, H.; Yadid, M.; Park, S.-J.; Kotikian, A.; Nesmith, A.P.; Campbell, P.H. Instrumented Cardiac Microphysiological Devices via Multimaterial Three-Dimensional Printing. Nat. Mater. 2017, 16, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Burkholder-Wenger, A.C.; Golzar, H.; Wu, Y.; Tang, X.S. Development of a Hybrid Nanoink for 3D Bioprinting of Heterogeneous Tumor Models. ACS Biomater. Sci. Eng. 2022, 8, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wenger, A.; Golzar, H.; Tang, X.S. 3D Bioprinting of Bicellular Liver Lobule-Mimetic Structures via Microextrusion of Cellulose Nanocrystal-Incorporated Shear-Thinning Bioink. Sci. Rep. 2020, 10, 20648. [Google Scholar] [CrossRef] [PubMed]
- Lampin, M.; Warocquier-Clérout, R.; Legris, C.; Degrange, M.; Sigot-Luizard, M. Correlation between Substratum Roughness and Wettability, Cell Adhesion, and Cell Migration. J. Biomed. Mater. Res. Off. J. Soc. Biomater. Jpn. Soc. Biomater. 1997, 36, 99–108. [Google Scholar] [CrossRef]
- Majhy, B.; Priyadarshini, P.; Sen, A. Effect of Surface Energy and Roughness on Cell Adhesion and Growth–Facile Surface Modification for Enhanced Cell Culture. RSC Adv. 2021, 11, 15467–15476. [Google Scholar] [CrossRef]
- Chen, L.; Yan, C.; Zheng, Z. Functional Polymer Surfaces for Controlling Cell Behaviors. Mater. Today 2018, 21, 38–59. [Google Scholar] [CrossRef]
- Patel, S.; Thakar, R.G.; Wong, J.; McLeod, S.D.; Li, S. Control of Cell Adhesion on Poly (Methyl Methacrylate). Biomaterials 2006, 27, 2890–2897. [Google Scholar] [CrossRef] [PubMed]
- Riau, A.K.; Mondal, D.; Yam, G.H.; Setiawan, M.; Liedberg, B.; Venkatraman, S.S.; Mehta, J.S. Surface Modification of PMMA to Improve Adhesion to Corneal Substitutes in a Synthetic Core–Skirt Keratoprosthesis. ACS Appl. Mater. Interfaces 2015, 7, 21690–21702. [Google Scholar] [CrossRef]
- Jastrzebska, E.; Zuchowska, A.; Flis, S.; Sokolowska, P.; Bulka, M.; Dybko, A.; Brzozka, Z. Biological Characterization of the Modified Poly (Dimethylsiloxane) Surfaces Based on Cell Attachment and Toxicity Assays. Biomicrofluidics 2018, 12, 044105. [Google Scholar] [CrossRef] [PubMed]
- Premnath, P.; Tavangar, A.; Tan, B.; Venkatakrishnan, K. Tuning Cell Adhesion by Direct Nanostructuring Silicon into Cell Repulsive/Adhesive Patterns. Exp. Cell Res. 2015, 337, 44–52. [Google Scholar] [CrossRef]
- Azadbakht, M.; Madaeni, S.S.; Sahebjamee, F. Biocompatibility of Polyethersulfone Membranes for Cell Culture Systems. Eng. Life Sci. 2011, 11, 629–635. [Google Scholar] [CrossRef]
- Ruben, B.; Elisa, M.; Leandro, L.; Victor, M.; Gloria, G.; Marina, S.; Pandiyan, R.; Nadhira, L. Oxygen Plasma Treatments of Polydimethylsiloxane Surfaces: Effect of the Atomic Oxygen on Capillary Flow in the Microchannels. Micro Nano Lett. 2017, 12, 754–757. [Google Scholar] [CrossRef]
- Ribas, J.; Sadeghi, H.; Manbachi, A.; Leijten, J.; Brinegar, K.; Zhang, Y.S.; Ferreira, L.; Khademhosseini, A. Cardiovascular Organ-on-a-Chip Platforms for Drug Discovery and Development. Appl. Vitro Toxicol. 2016, 2, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Takayama, S. Organ-on-a-Chip and the Kidney. Kidney Res. Clin. Pract. 2015, 34, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered Liver-on-a-Chip Platform to Mimic Liver Functions and Its Biomedical Applications: A Review. Micromachines 2019, 10, 676. [Google Scholar] [CrossRef]
- Zamprogno, P.; Wüthrich, S.; Achenbach, S.; Thoma, G.; Stucki, J.D.; Hobi, N.; Schneider-Daum, N.; Lehr, C.-M.; Huwer, H.; Geiser, T. Second-Generation Lung-on-a-Chip with an Array of Stretchable Alveoli Made with a Biological Membrane. Commun. Biol. 2021, 4, 168. [Google Scholar] [CrossRef] [PubMed]
- Ogończyk, D.; Jankowski, P.; Garstecki, P. A Method for Simultaneous Polishing and Hydrophobization of Polycarbonate for Microfluidic Applications. Polymers 2020, 12, 2490. [Google Scholar] [CrossRef]
- He, Y.; Guo, Y.; He, R.; Jin, T.; Chen, F.; Fu, Q.; Zhou, N.; Shen, J. Towards High Molecular Weight Poly (Bisphenol a Carbonate) with Excellent Thermal Stability and Mechanical Properties by Solid-State Polymerization. Chin. J. Polym. Sci. 2015, 33, 1176–1185. [Google Scholar] [CrossRef]
- Aazmi, A.; Zhou, H.; Li, Y.; Yu, M.; Xu, X.; Wu, Y.; Ma, L.; Zhang, B.; Yang, H. Engineered Vasculature for Organ-on-a-Chip Systems. Engineering 2021, 9, 131–147. [Google Scholar] [CrossRef]
- Ma, Y.; Pan, J.-Z.; Zhao, S.-P.; Lou, Q.; Zhu, Y.; Fang, Q. Microdroplet Chain Array for Cell Migration Assays. Lab Chip 2016, 16, 4658–4665. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, K.; Yasukawa, A.; Kobayashi, Y. Wettability Characteristics of Poly (Ethylene Terephthalate) Films Treated by Atmospheric Pressure Plasma and Ultraviolet Excimer Light. Polym. J. 2011, 43, 545–551. [Google Scholar] [CrossRef][Green Version]
- Bin, Y.; Oishi, K.; Yoshida, K.; Matsuo, M. Mechanical Properties of Poly (Ethylene Terephthalate) Estimated in Terms of Orientation Distribution of Crystallites and Amorphous Chain Segments under Simultaneous Biaxially Stretching. Polym. J. 2004, 36, 888–898. [Google Scholar] [CrossRef]
- Xiang, Y.; Wen, H.; Yu, Y.; Li, M.; Fu, X.; Huang, S. Gut-on-Chip: Recreating Human Intestine in Vitro. J. Tissue Eng. 2020, 11, 2041731420965318. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.-Y.; Ingber, D.E. Human Kidney Proximal Tubule-on-a-Chip for Drug Transport and Nephrotoxicity Assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Deng, R.; Hao Tong, W.; Huan, L.; Chan Way, N.; IslamBadhan, A.; Iliescu, C.; Yu, H. A Perfusion Incubator Liver Chip for 3D Cell Culture with Application on Chronic Hepatotoxicity Testing. Sci. Rep. 2017, 7, 14528. [Google Scholar] [CrossRef] [PubMed]
- Laput, O.; Vasenina, I.; Salvadori, M.C.; Savkin, K.; Zuza, D.; Kurzina, I. Low-Temperature Plasma Treatment of Polylactic Acid and PLA/HA Composite Material. J. Mater. Sci. 2019, 54, 11726–11738. [Google Scholar] [CrossRef]
- Ko, H.-S.; Lee, S.; Lee, D.; Jho, J.Y. Mechanical Properties and Bioactivity of Poly (Lactic Acid) Composites Containing Poly (Glycolic Acid) Fiber and Hydroxyapatite Particles. Nanomaterials 2021, 11, 249. [Google Scholar] [CrossRef]
- Pensabene, V.; Costa, L.; Terekhov, A.Y.; Gnecco, J.S.; Wikswo, J.P.; Hofmeister, W.H. Ultrathin Polymer Membranes with Patterned, Micrometric Pores for Organs-on-Chips. ACS Appl. Mater. Interfaces 2016, 8, 22629–22636. [Google Scholar] [CrossRef] [PubMed]
- Janvikul, W.; Uppanan, P.; Thavornyutikarn, B.; Kosorn, W.; Kaewkong, P. Effects of Surface Topography, Hydrophilicity and Chemistry of Surface-Treated PCL Scaffolds on Chondrocyte Infiltration and ECM Production. Procedia Eng. 2013, 59, 158–165. [Google Scholar] [CrossRef]
- Dziadek, M.; Menaszek, E.; Zagrajczuk, B.; Pawlik, J.; Cholewa-Kowalska, K. New Generation Poly (ε-Caprolactone)/Gel-Derived Bioactive Glass Composites for Bone Tissue Engineering: Part I. Material Properties. Mater. Sci. Eng. C 2015, 56, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Pensabene, V.; Crowder, S.W.; Balikov, D.A.; Lee, J.B.; Sung, H.-J. Optimization of Electrospun Fibrous Membranes for in Vitro Modeling of Blood-Brain Barrier; IEEE: Piscataway, NJ, USA, 2016; pp. 125–128. [Google Scholar]
- Wloch, J.; Terzyk, A.P.; Wisniewski, M.; Kowalczyk, P. Nanoscale Water Contact Angle on Polytetrafluoroethylene Surfaces Characterized by Molecular Dynamics-Atomic Force Microscopy Imaging. Langmuir 2018, 34, 4526–4534. [Google Scholar] [CrossRef] [PubMed]
- Uehara, H.; Jounai, K.; Endo, R.; Okuyama, H.; Kanamoto, T.; Porter, R.S. High Modulus Films of Polytetrafluoroethylene Prepared by Two-Stage Drawing of Reactor Powder. Polym. J. 1997, 29, 198–200. [Google Scholar] [CrossRef][Green Version]
- Ahadian, S.; Civitarese, R.; Bannerman, D.; Mohammadi, M.H.; Lu, R.; Wang, E.; Davenport-Huyer, L.; Lai, B.; Zhang, B.; Zhao, Y. Organ-on-a-chip Platforms: A Convergence of Advanced Materials, Cells, and Microscale Technologies. Adv. Healthc. Mater. 2018, 7, 1700506. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.; Özkan, A.; Oh, C.; Mahajan, G.; Prantil-Baun, R.; Ingber, D.E. Predicting Drug Concentrations in PDMS Microfluidic Organ Chips. Lab Chip 2021, 21, 3509–3519. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Duan, B. Materials and Their Biomedical Applications. In Encyclopedia of Biomedical Engineering; Elsevier: Amsterdam, The Netherlands, 2019; pp. 135–152. [Google Scholar]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-Chip: Recent Breakthroughs and Future Prospects. Biomed. Eng. Online 2020, 19, 9. [Google Scholar] [CrossRef]
- Osório, L.A.; Silva, E.; Mackay, R.E. A Review of Biomaterials and Scaffold Fabrication for Organ-on-a-Chip (OOAC) Systems. Bioengineering 2021, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Arif, U.; Haider, S.; Haider, A.; Khan, N.; Alghyamah, A.A.; Jamila, N.; Khan, M.I.; Almasry, W.A.; Kang, I.-K. Biocompatible Polymers and Their Potential Biomedical Applications: A Review. Curr. Pharm. Des. 2019, 25, 3608–3619. [Google Scholar] [CrossRef] [PubMed]
- Raj, M.K.; Chakraborty, S. PDMS Microfluidics: A Mini Review. J. Appl. Polym. Sci. 2020, 137, 48958. [Google Scholar] [CrossRef]
- Fujii, T. PDMS-Based Microfluidic Devices for Biomedical Applications. Microelectron. Eng. 2002, 61, 907–914. [Google Scholar] [CrossRef]
- Carter, S.-S.D.; Atif, A.-R.; Kadekar, S.; Lanekoff, I.; Engqvist, H.; Varghese, O.P.; Tenje, M.; Mestres, G. PDMS Leaching and Its Implications for On-Chip Studies Focusing on Bone Regeneration Applications. Organs-Chip 2020, 2, 100004. [Google Scholar] [CrossRef]
- Bunge, F.; den Driesche, S.V.; Vellekoop, M.J. Microfluidic Platform for the Long-Term on-Chip Cultivation of Mammalian Cells for Lab-on-a-Chip Applications. Sensors 2017, 17, 1603. [Google Scholar] [CrossRef] [PubMed]
- Gezer, P.G.; Brodsky, S.; Hsiao, A.; Liu, G.L.; Kokini, J.L. Modification of the Hydrophilic/Hydrophobic Characteristic of Zein Film Surfaces by Contact with Oxygen Plasma Treated PDMS and Oleic Acid Content. Colloids Surf. B Biointerfaces 2015, 135, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Choi, D.S.; Nguyen, Y.H.; Chang, J.; Qin, L. Studying Cancer Stem Cell Dynamics on PDMS Surfaces for Microfluidics Device Design. Sci. Rep. 2013, 3, 2332. [Google Scholar] [CrossRef] [PubMed]
- Chuah, Y.J.; Kuddannaya, S.; Lee, M.H.A.; Zhang, Y.; Kang, Y. The Effects of Poly (Dimethylsiloxane) Surface Silanization on the Mesenchymal Stem Cell Fate. Biomater. Sci. 2015, 3, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Pitingolo, G.; Riaud, A.; Nastruzzi, C.; Taly, V. Gelatin-Coated Microfluidic Channels for 3d Microtissue Formation: On-Chip Production and Characterization. Micromachines 2019, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, B.; Scott, J.; Vilander, G.; Marx, L.; Quirk, M.; Lindberg, J.; Koerner, K. The Role of Lean Process Improvement in Implementation of Evidence-Based Practices in Behavioral Health Care. J. Behav. Health Serv. Res. 2015, 42, 504–518. [Google Scholar] [CrossRef]
- Khetani, S.; Yong, K.W.; Ozhukil Kollath, V.; Eastick, E.; Azarmanesh, M.; Karan, K.; Sen, A.; Sanati-Nezhad, A. Engineering Shelf-Stable Coating for Microfluidic Organ-on-a-Chip Using Bioinspired Catecholamine Polymers. ACS Appl. Mater. Interfaces 2020, 12, 6910–6923. [Google Scholar] [CrossRef]
- Park, S.E.; Georgescu, A.; Oh, J.M.; Kwon, K.W.; Huh, D. Polydopamine-Based Interfacial Engineering of Extracellular Matrix Hydrogels for the Construction and Long-Term Maintenance of Living Three-Dimensional Tissues. ACS Appl. Mater. Interfaces 2019, 11, 23919–23925. [Google Scholar] [CrossRef]
- Mikhail, A.S.; Ranger, J.J.; Liu, L.; Longenecker, R.; Thompson, D.B.; Sheardown, H.D.; Brook, M.A. Rapid and Efficient Assembly of Functional Silicone Surfaces Protected by PEG: Cell Adhesion to Peptide-Modified PDMS. J. Biomater. Sci. Polym. Ed. 2010, 21, 821–842. [Google Scholar] [CrossRef]
- Henry, O.Y.; Villenave, R.; Cronce, M.J.; Leineweber, W.D.; Benz, M.A.; Ingber, D.E. Organs-on-Chips with Integrated Electrodes for Trans-Epithelial Electrical Resistance (TEER) Measurements of Human Epithelial Barrier Function. Lab Chip 2017, 17, 2264–2271. [Google Scholar] [CrossRef]
- Wang, Y.I.; Abaci, H.E.; Shuler, M.L. Microfluidic Blood–Brain Barrier Model Provides in Vivo-like Barrier Properties for Drug Permeability Screening. Biotechnol. Bioeng. 2017, 114, 184–194. [Google Scholar] [CrossRef]
- Domansky, K.; Sliz, J.D.; Wen, N.; Hinojosa, C.; Thompson, G.; Fraser, J.P.; Hamkins-Indik, T.; Hamilton, G.A.; Levner, D.; Ingber, D.E. SEBS Elastomers for Fabrication of Microfluidic Devices with Reduced Drug Absorption by Injection Molding and Extrusion. Microfluid. Nanofluidics 2017, 21, 107. [Google Scholar] [CrossRef]
- Domansky, K.; Leslie, D.C.; McKinney, J.; Fraser, J.P.; Sliz, J.D.; Hamkins-Indik, T.; Hamilton, G.A.; Bahinski, A.; Ingber, D.E. Clear Castable Polyurethane Elastomer for Fabrication of Microfluidic Devices. Lab Chip 2013, 13, 3956–3964. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Chen, X.; Kang, Q.; Yan, X. Biomedical Application of Functional Materials in Organ-on-a-Chip. Front. Bioeng. Biotechnol. 2020, 8, 823. [Google Scholar] [CrossRef] [PubMed]
- Pasman, T.; Grijpma, D.; Stamatialis, D.; Poot, A. Flat and Microstructured Polymeric Membranes in Organs-on-Chips. J. R. Soc. Interface 2018, 15, 20180351. [Google Scholar] [CrossRef] [PubMed]
- Busek, M.; Nøvik, S.; Aizenshtadt, A.; Amirola-Martinez, M.; Combriat, T.; Grünzner, S.; Krauss, S. Thermoplastic Elastomer (TPE)–Poly (Methyl Methacrylate)(PMMA) Hybrid Devices for Active Pumping PDMS-Free Organ-on-a-Chip Systems. Biosensors 2021, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Jung, S.H.; Lee, M.S.; Park, T.-E.; Ahn, S.; Kang, J.H. Robust Chemical Bonding of PMMA Microfluidic Devices to Porous PETE Membranes for Reliable Cytotoxicity Testing of Drugs. Lab Chip 2019, 19, 3706–3713. [Google Scholar] [CrossRef]
- Echave, M.C.; Burgo, L.S.; Pedraz, J.L.; Orive, G. Gelatin as Biomaterial for Tissue Engineering. Curr. Pharm. Des. 2017, 23, 3567–3584. [Google Scholar] [CrossRef]
- Santoro, M.; Tatara, A.M.; Mikos, A.G. Gelatin Carriers for Drug and Cell Delivery in Tissue Engineering. J. Control. Release 2014, 190, 210–218. [Google Scholar] [CrossRef]
- Lee, H.; Cho, D.-W. One-Step Fabrication of an Organ-on-a-Chip with Spatial Heterogeneity Using a 3D Bioprinting Technology. Lab Chip 2016, 16, 2618–2625. [Google Scholar] [CrossRef]
- Lam, T.; Dehne, T.; Krüger, J.P.; Hondke, S.; Endres, M.; Thomas, A.; Lauster, R.; Sittinger, M.; Kloke, L. Photopolymerizable Gelatin and Hyaluronic Acid for Stereolithographic 3D Bioprinting of Tissue-engineered Cartilage. J. Biomed. Mater. Res. B Appl. Biomater. 2019, 107, 2649–2657. [Google Scholar] [CrossRef] [PubMed]
- Nawroth, J.C.; Scudder, L.L.; Halvorson, R.T.; Tresback, J.; Ferrier, J.P.; Sheehy, S.P.; Cho, A.; Kannan, S.; Sunyovszki, I.; Goss, J.A. Automated Fabrication of Photopatterned Gelatin Hydrogels for Organ-on-Chips Applications. Biofabrication 2018, 10, 025004. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Hunziker, W.; Choudhury, D. Engineering Microfluidic Organoid-on-a-Chip Platforms. Micromachines 2019, 10, 165. [Google Scholar] [CrossRef]
- Benton, G.; Arnaoutova, I.; George, J.; Kleinman, H.K.; Koblinski, J. Matrigel: From Discovery and ECM Mimicry to Assays and Models for Cancer Research. Adv. Drug Deliv. Rev. 2014, 79, 3–18. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Huang, R.; Huang, Z.; Hu, B.; Zheng, W.; Yang, G.; Jiang, X. Evaluation of the Effect of the Structure of Bacterial Cellulose on Full Thickness Skin Wound Repair on a Microfluidic Chip. Biomacromolecules 2015, 16, 780–789. [Google Scholar] [CrossRef]
- Anton-Sales, I.; Beekmann, U.; Laromaine, A.; Roig, A.; Kralisch, D. Opportunities of Bacterial Cellulose to Treat Epithelial Tissues. Curr. Drug Targets 2019, 20, 808–822. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Huang, J. Drug-Induced Nephrotoxicity: Pathogenic Mechanisms, Biomarkers and Prevention Strategies. Curr. Drug Metab. 2018, 19, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, J.L.; Li, X.C. Proximal Nephron. Compr. Physiol. 2013, 3, 1079. [Google Scholar]
- McCormick, J.; Ellison, D. Distal Convoluted Tubule. Compr. Physiol. 2015, 5, 45–98. [Google Scholar] [PubMed]
- Nolin, T.D.; Himmelfarb, J. Mechanisms of Drug-Induced Nephrotoxicity. Advers. Drug React. 2010, 196, 111–130. [Google Scholar]
- Mu, X.; Zheng, W.; Xiao, L.; Zhang, W.; Jiang, X. Engineering a 3D Vascular Network in Hydrogel for Mimicking a Nephron. Lab Chip 2013, 13, 1612–1618. [Google Scholar] [CrossRef] [PubMed]
- Wilmer, M.J.; Ng, C.P.; Lanz, H.L.; Vulto, P.; Suter-Dick, L.; Masereeuw, R. Kidney-on-a-Chip Technology for Drug-Induced Nephrotoxicity Screening. Trends Biotechnol. 2016, 34, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, V.; Rbaibi, Y.; Pastor-Soler, N.M.; Carattino, M.D.; Weisz, O.A. Shear Stress-Dependent Regulation of Apical Endocytosis in Renal Proximal Tubule Cells Mediated by Primary Cilia. Proc. Natl. Acad. Sci. USA 2014, 111, 8506–8511. [Google Scholar] [CrossRef] [PubMed]
- Miravète, M.; Dissard, R.; Klein, J.; Gonzalez, J.; Caubet, C.; Pecher, C.; Pipy, B.; Bascands, J.-L.; Mercier-Bonin, M.; Schanstra, J.P. Renal Tubular Fluid Shear Stress Facilitates Monocyte Activation toward Inflammatory Macrophages. Am. J. Physiol.-Ren. Physiol. 2012, 302, F1409–F1417. [Google Scholar] [CrossRef] [PubMed]
- Van de Water, B.; Jaspers, J.; Maasdam, D.H.; Mulder, G.J.; Nagelkerke, J.F. In Vivo and in Vitro Detachment of Proximal Tubular Cells and F-Actin Damage: Consequences for Renal Function. Am. J. Physiol.-Ren. Physiol. 1994, 267, F888–F899. [Google Scholar] [CrossRef] [PubMed]
- Ross, E.J.; Gordon, E.R.; Sothers, H.; Darji, R.; Baron, O.; Haithcock, D.; Prabhakarpandian, B.; Pant, K.; Myers, R.M.; Cooper, S.J. Three Dimensional Modeling of Biologically Relevant Fluid Shear Stress in Human Renal Tubule Cells Mimics in Vivo Transcriptional Profiles. Sci. Rep. 2021, 11, 14053. [Google Scholar] [CrossRef] [PubMed]
- Musah, S.; Mammoto, A.; Ferrante, T.C.; Jeanty, S.S.; Hirano-Kobayashi, M.; Mammoto, T.; Roberts, K.; Chung, S.; Novak, R.; Ingram, M. Mature Induced-Pluripotent-Stem-Cell-Derived Human Podocytes Reconstitute Kidney Glomerular-Capillary-Wall Function on a Chip. Nat. Biomed. Eng. 2017, 1, 0069. [Google Scholar] [CrossRef] [PubMed]
- Roye, Y.; Bhattacharya, R.; Mou, X.; Zhou, Y.; Burt, M.A.; Musah, S. A Personalized Glomerulus Chip Engineered from Stem Cell-Derived Epithelium and Vascular Endothelium. Micromachines 2021, 12, 967. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tao, T.; Su, W.; Yu, H.; Yu, Y.; Qin, J. A Disease Model of Diabetic Nephropathy in a Glomerulus-on-a-Chip Microdevice. Lab Chip 2017, 17, 1749–1760. [Google Scholar] [CrossRef]
- Ng, C.P.; Zhuang, Y.; Lin, A.W.H.; Teo, J.C.M. A Fibrin-Based Tissue-Engineered Renal Proximal Tubule for Bioartificial Kidney Devices: Development, Characterization and in Vitro Transport Study. Int. J. Tissue Eng. 2012, 2013, 319476. [Google Scholar] [CrossRef]
- Jansen, J.; Fedecostante, M.; Wilmer, M.; Peters, J.; Kreuser, U.; Van Den Broek, P.; Mensink, R.; Boltje, T.; Stamatialis, D.; Wetzels, J. Bioengineered Kidney Tubules Efficiently Excrete Uremic Toxins. Sci. Rep. 2016, 6, 26715. [Google Scholar] [CrossRef] [PubMed]
- Sciancalepore, A.G.; Sallustio, F.; Girardo, S.; Gioia Passione, L.; Camposeo, A.; Mele, E.; Di Lorenzo, M.; Costantino, V.; Schena, F.P.; Pisignano, D. A Bioartificial Renal Tubule Device Embedding Human Renal Stem/Progenitor Cells. PLoS ONE 2014, 9, e87496. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.J.; Chapron, A.; Chapron, B.D.; Voellinger, J.L.; Lidberg, K.A.; Yeung, C.K.; Wang, Z.; Yamaura, Y.; Hailey, D.W.; Neumann, T. Development of a Microphysiological Model of Human Kidney Proximal Tubule Function. Kidney Int. 2016, 90, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Sochol, R.D.; Gupta, N.R.; Bonventre, J.V. A Role for 3D Printing in Kidney-on-a-Chip Platforms. Curr. Transplant. Rep. 2016, 3, 82–92. [Google Scholar] [CrossRef]
- Homan, K.A.; Kolesky, D.B.; Skylar-Scott, M.A.; Herrmann, J.; Obuobi, H.; Moisan, A.; Lewis, J.A. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Sci. Rep. 2016, 6, 34845. [Google Scholar] [CrossRef] [PubMed]
- Odijk, M.; van der Meer, A.D.; Levner, D.; Kim, H.J.; van der Helm, M.W.; Segerink, L.I.; Frimat, J.-P.; Hamilton, G.A.; Ingber, D.E.; van den Berg, A. Measuring Direct Current Trans-Epithelial Electrical Resistance in Organ-on-a-Chip Microsystems. Lab Chip 2015, 15, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Snouber, L.C.; Letourneur, F.; Chafey, P.; Broussard, C.; Monge, M.; Legallais, C.; Leclerc, E. Analysis of Transcriptomic and Proteomic Profiles Demonstrates Improved Madin–Darby Canine Kidney Cell Function in a Renal Microfluidic Biochip. Biotechnol. Prog. 2012, 28, 474–484. [Google Scholar] [CrossRef]
- Kim, S.; LesherPerez, S.C.; Yamanishi, C.; Labuz, J.M.; Leung, B.; Takayama, S. Pharmacokinetic Profile That Reduces Nephrotoxicity of Gentamicin in a Perfused Kidney-on-a-Chip. Biofabrication 2016, 8, 015021. [Google Scholar] [CrossRef] [PubMed]
- Vormann, M.K.; Gijzen, L.; Hutter, S.; Boot, L.; Nicolas, A.; van den Heuvel, A.; Vriend, J.; Ng, C.P.; Nieskens, T.T.; van Duinen, V. Nephrotoxicity and Kidney Transport Assessment on 3D Perfused Proximal Tubules. AAPS J. 2018, 20, 90. [Google Scholar] [CrossRef] [PubMed]
- Trietsch, S.J.; Israëls, G.D.; Joore, J.; Hankemeier, T.; Vulto, P. Microfluidic Titer Plate for Stratified 3D Cell Culture. Lab Chip 2013, 13, 3548–3554. [Google Scholar] [CrossRef] [PubMed]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A. A Four-Organ-Chip for Interconnected Long-Term Co-Culture of Human Intestine, Liver, Skin and Kidney Equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef]
- Chang, S.-Y.; Weber, E.J.; Sidorenko, V.S.; Chapron, A.; Yeung, C.K.; Gao, C.; Mao, Q.; Shen, D.; Wang, J.; Rosenquist, T.A. Human Liver-Kidney Model Elucidates the Mechanisms of Aristolochic Acid Nephrotoxicity. JCI Insight 2017, 2, e95978. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.D. Anatomy of the Lower Urinary Tract and Male Genitalia. In Campbell-Walsh Urology; Saunders/Elsevier: Philadelphia, PA, USA, 2007. [Google Scholar]
- de Groat, W.C.; Yoshimura, N. Anatomy and Physiology of the Lower Urinary Tract. Handb. Clin. Neurol. 2015, 130, 61–108. [Google Scholar]
- Bolla, S.R.; Odeluga, N.; Jetti, R. Histology, Bladder. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Cumberbatch, M.G.K.; Jubber, I.; Black, P.C.; Esperto, F.; Figueroa, J.D.; Kamat, A.M.; Kiemeney, L.; Lotan, Y.; Pang, K.; Silverman, D.T. Epidemiology of Bladder Cancer: A Systematic Review and Contemporary Update of Risk Factors in 2018. Eur. Urol. 2018, 74, 784–795. [Google Scholar] [CrossRef]
- Lenis, A.T.; Lec, P.M.; Chamie, K. Bladder Cancer: A Review. Jama 2020, 324, 1980–1991. [Google Scholar] [CrossRef]
- Zhu, S.; Zhu, Z.; Ma, A.-H.; Sonpavde, G.P.; Cheng, F.; Pan, C. Preclinical Models for Bladder Cancer Research. Hematol. Clin. 2021, 35, 613–632. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.S.; Jeong, K.-C.; Lee, S.-J.; Seo, H.K. Establishment and Application of Bladder Cancer Patient-Derived Xenografts as a Novel Preclinical Platform. Transl. Cancer Res. 2017, 6, S733–S743. [Google Scholar] [CrossRef]
- Bernardo, C.; Costa, C.; Palmeira, C.; Pinto-Leite, R.; Oliveira, P.; Freitas, R.; Amado, F.; Santos, L.L. What We Have Learned from Urinary Bladder Cancer Models. J. Cancer Metastasis Treat. 2016, 2, 51–58. [Google Scholar]
- Arantes-Rodrigues, R.; Colaco, A.; Pinto-Leite, R.; Oliveira, P.A. In Vitro and in Vivo Experimental Models as Tools to Investigate the Efficacy of Antineoplastic Drugs on Urinary Bladder Cancer. Anticancer Res. 2013, 33, 1273–1296. [Google Scholar]
- DeGraff, D.J.; Robinson, V.L.; Shah, J.B.; Brandt, W.D.; Sonpavde, G.; Kang, Y.; Liebert, M.; Wu, X.-R.; Taylor, J.A. Current Preclinical Models for the Advancement of Translational Bladder Cancer Research. Mol. Cancer Ther. 2013, 12, 121–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, P.; Cao, Y.; Zhang, S.; Zhao, Y.; Liu, X.; Shi, H.; Hu, K.; Zhu, G.; Ma, B.; Niu, H. A Bladder Cancer Microenvironment Simulation System Based on a Microfluidic Co-Culture Model. Oncotarget 2015, 6, 37695. [Google Scholar] [CrossRef]
- Xu, X.-D.; Shao, S.-X.; Cao, Y.-W.; Yang, X.-C.; Shi, H.-Q.; Wang, Y.-L.; Xue, S.-Y.; Wang, X.-S.; Niu, H.-T. The Study of Energy Metabolism in Bladder Cancer Cells in Co-Culture Conditions Using a Microfluidic Chip. Int. J. Clin. Exp. Med. 2015, 8, 12327. [Google Scholar] [PubMed]
- Lee, E.; Kwon, C.; Han, H.; Park, J.; Kim, Y.-C.; Ok, M.-R.; Seok, H.-K.; Jeon, H. Bladder Cancer-on-a-Chip for Analysis of Tumor Transition Mechanism. In Proceedings of the 10th World Biomaterials Congress, Montréal, QC, Canada, 17–22 May 2016. [Google Scholar] [CrossRef]
- Hooton, T.M. Uncomplicated Urinary Tract Infection. N. Engl. J. Med. 2012, 366, 1028–1037. [Google Scholar] [CrossRef]
- Sen, A. Recurrent Cystitis in Non-Pregnant Women. BMJ Clin. Evid. 2008, 2008, 0801. [Google Scholar] [PubMed]
- Foxman, B. The Epidemiology of Urinary Tract Infection. Nat. Rev. Urol. 2010, 7, 653–660. [Google Scholar] [CrossRef]
- Kudinha, T. The Pathogenesis of Escherichia Coli Urinary Tract Infection. In Escherichia coli-Recent Advances on Physiology, Pathogenesis and Biotechnological Applications; IntechOpen: London, UK, 2017; pp. 45–61. [Google Scholar]
- Sharma, K.; Dhar, N.; Thacker, V.V.; Simonet, T.M.; Signorino-Gelo, F.; Knott, G.W.; McKinney, J.D. Dynamic Persistence of UPEC Intracellular Bacterial Communities in a Human Bladder-Chip Model of Urinary Tract Infection. Elife 2021, 10, e66481. [Google Scholar] [CrossRef] [PubMed]
- Aaron, L.; Franco, O.E.; Hayward, S.W. Review of Prostate Anatomy and Embryology and the Etiology of Benign Prostatic Hyperplasia. Urol. Clin. 2016, 43, 279–288. [Google Scholar] [CrossRef]
- Jiang, L.; Ivich, F.; Tran, M.; Frank, S.B.; Miranti, C.K.; Zohar, Y. Development of a Microfluidic-Based Model of a Human Prostate Gland; Chemical and Biological Microsystems Society: Palm Springs, CA, USA, 2018; pp. 1625–1627. [Google Scholar]
- Ivich, F.; Tran, M.; Tahsin, S.; Frank, S.B.; Kraft, A.; Miranti, C.K.; Zohar, Y.; Jiang, L. Application of a Microfluidic-Based Model of a Human Prostate Gland for Cancer Research; IEEE: Piscataway, NJ, USA, 2018; pp. 109–112. [Google Scholar]
- Frank, S.B.; Miranti, C.K. Disruption of Prostate Epithelial Differentiation Pathways and Prostate Cancer Development. Front. Oncol. 2013, 3, 273. [Google Scholar] [CrossRef]
- Lamb, L.E.; Knudsen, B.S.; Miranti, C.K. E-Cadherin-Mediated Survival of Androgen-Receptor-Expressing Secretory Prostate Epithelial Cells Derived from a Stratified in Vitro Differentiation Model. J. Cell Sci. 2010, 123, 266–276. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Qiu, X.; Zhu, Y.; Takeda, D.Y.; Pan, W.; Baca, S.C.; Gusev, A.; Korthauer, K.D.; Severson, T.M.; Ha, G. Prostate Cancer Reactivates Developmental Epigenomic Programs during Metastatic Progression. Nat. Genet. 2020, 52, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, D.; Spring, D.J.; DePinho, R.A. Genetics and Biology of Prostate Cancer. Genes Dev. 2018, 32, 1105–1140. [Google Scholar] [CrossRef] [PubMed]
- Ivich, F. Development of a Microfluidic Model of a Human Prostate Gland for Cancer Research. Ph.D. Thesis, The University of Arizon, Tucson, AZ, USA, 2019. [Google Scholar]
- Liu, Y.; Gill, E.; Huang, Y.Y.S. Microfluidic on-chip biomimicry for 3D cell culture: A fit-for-purpose investigation from the end user standpoint. Future Sci. OA 2017, 3, pFSO173. [Google Scholar] [CrossRef] [PubMed]
- Picollet-d’Hahan, N.; Gerbaud, S.; Kermarrec, F.; Alcaraz, J.-P.; Obeid, P.; Bhajun, R.; Guyon, L.; Sulpice, E.; Cinquin, P.; Dolega, M.E. The Modulation of Attachment, Growth and Morphology of Cancerous Prostate Cells by Polyelectrolyte Nanofilms. Biomaterials 2013, 34, 10099–10108. [Google Scholar] [CrossRef]
- Jiang, L.; Ivich, F.; Tahsin, S.; Tran, M.; Frank, S.; Miranti, C.; Zohar, Y. Human Stroma and Epithelium Co-Culture in a Microfluidic Model of a Human Prostate Gland. Biomicrofluidics 2019, 13, 064116. [Google Scholar] [CrossRef]
- Imparato, G.; Urciuolo, F.; Netti, P.A. Organ on Chip Technology to Model Cancer Growth and Metastasis. Bioengineering 2022, 9, 28. [Google Scholar] [CrossRef]
- Top 10 Emerging Technologies. Available online: https://www.weforum.org/agenda/2016/06/top-10-emerging-technologies-2016/2016 (accessed on 25 February 2022).
- Shourabi, A.Y.; Kashaninejad, N.; Saidi, M.S. An Integrated Microfluidic Concentration Gradient Generator for Mechanical Stimulation and Drug Delivery. J. Sci. Adv. Mater. Devices 2021, 6, 280–290. [Google Scholar] [CrossRef]
- Adamowicz, J.; Pokrywczyńska, M.; Tworkiewicz, J.; Wolski, Z.; Drewa, T. The Relationship of Cancer Stem Cells in Urological Cancers. Cent. Eur. J. Urol. 2013, 66, 273. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-Y.; Evangelista, E.A.; Yang, J.; Kelly, E.J.; Yeung, C.K. Kidney Organoid and Microphysiological Kidney Chip Models to Accelerate Drug Development and Reduce Animal Testing. Front. Pharmacol. 2021, 12, 695920. [Google Scholar] [CrossRef] [PubMed]
- Barisam, M.; Niavol, F.R.; Kinj, M.A.; Saidi, M.S.; Ghanbarian, H.; Kashaninejad, N. Enrichment of Cancer Stem-like Cells by Controlling Oxygen, Glucose and Fluid Shear Stress in a Microfluidic Spheroid Culture Device. J. Sci. Adv. Mater. Devices 2022, 7, 100439. [Google Scholar] [CrossRef]
- Radisic, M.; Loskill, P. Beyond PDMS and Membranes: New Materials for Organ-on-a-Chip Devices. ACS Biomater. Sci. Eng. 2021, 7, 2861–2863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Radisic, M. Organ-on-a-Chip Devices Advance to Market. Lab Chip 2017, 17, 2395–2420. [Google Scholar] [CrossRef] [PubMed]


| Bioink Composition | Bioprinting Method | OOC Model | References |
|---|---|---|---|
| Collagen | extrusion | Lung, gut | [60,61] |
| Gelatin | extrusion | kidney | [62,63] |
| Methacrylate gelatin (GelMa) | extrusion | Vessel, liver | [64,65] |
| Alginate | extrusion | Heart | [66,67] |
| Cellulose | extrusion | tumor, liver | [68,69] |
| Polyethyleneglycol (PEG) Poly ε caprolactone (PCL) | inkjet, extrusion | Colon tumor | [59,65] |
| Polymer | Chemical Structure | Contact Angle with Water | Young’s Modulus | Application |
|---|---|---|---|---|
| Polydimethylsiloxane (PDMS) | 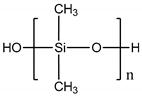 | 107° [78] | Variable from kPa to MPa | Cardiovascular [79] Kidney [80] Liver [81] Lung [82] |
| Poly (bisphenol-A- carbonate) (PC) |  | ~85° [83] | ~1.2 GPa [84] | Tumor vasculature [85] Colon and breast cancer [86] |
| Poly (ethylene terephthalate) (PET) |  | 83° [87] | 4.7 GPa [88] | Gut [89] Kidney [90] Liver [91] |
| Polylactic acid (PLA) |  | ~75° [92] | 3.1 GPa [93] | Endothelial barrier [94] |
| Poly (ε-caprolactone) (PCL) | 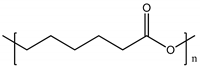 | 120° [95] | ~400 MPa [96] | Blood-brain barrier [97] |
| Polytetrafluoroethylene (PTFE) |  | 108° [98] | 392 MPa [99] | Liver [100] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galateanu, B.; Hudita, A.; Biru, E.I.; Iovu, H.; Zaharia, C.; Simsensohn, E.; Costache, M.; Petca, R.-C.; Jinga, V. Applications of Polymers for Organ-on-Chip Technology in Urology. Polymers 2022, 14, 1668. https://doi.org/10.3390/polym14091668
Galateanu B, Hudita A, Biru EI, Iovu H, Zaharia C, Simsensohn E, Costache M, Petca R-C, Jinga V. Applications of Polymers for Organ-on-Chip Technology in Urology. Polymers. 2022; 14(9):1668. https://doi.org/10.3390/polym14091668
Chicago/Turabian StyleGalateanu, Bianca, Ariana Hudita, Elena Iuliana Biru, Horia Iovu, Catalin Zaharia, Eliza Simsensohn, Marieta Costache, Razvan-Cosmin Petca, and Viorel Jinga. 2022. "Applications of Polymers for Organ-on-Chip Technology in Urology" Polymers 14, no. 9: 1668. https://doi.org/10.3390/polym14091668
APA StyleGalateanu, B., Hudita, A., Biru, E. I., Iovu, H., Zaharia, C., Simsensohn, E., Costache, M., Petca, R.-C., & Jinga, V. (2022). Applications of Polymers for Organ-on-Chip Technology in Urology. Polymers, 14(9), 1668. https://doi.org/10.3390/polym14091668













