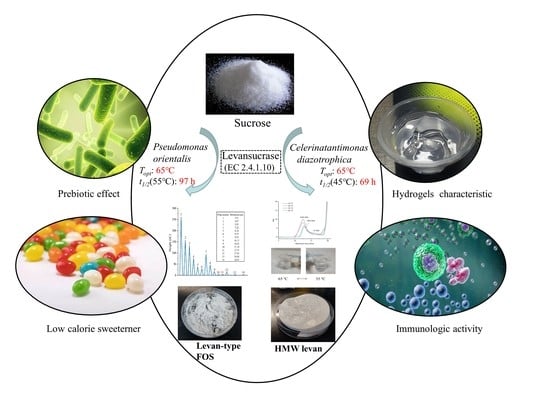
Identification of a Thermostable Levansucrase from Pseudomonas orientalis That Allows Unique Product Specificity at Different Temperatures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Strains
2.2. Expression and Purification of Psor-LS
2.3. Enzyme Activity and the Ratio of Transfructosylation Activity to Hydrolytic Activity (T/H) Assay
2.4. Biochemical Characterization
2.5. Optimization of Levan Production
2.6. Purification of Polysaccharide and FOSs
2.6.1. Purification of Polysaccharides
2.6.2. Purification of FOS
2.7. Nuclear Magnetic Resonance (NMR) Analysis
2.8. MW and Distribution Analysis
2.9. Molecular Dynamics Simulation
3. Results and Discussion
3.1. Computer-Aided Enzyme Screening
3.2. Expression and Purification of Psor-LS
3.3. Effect of pH on the Activity and T/H Ratio of Psor-LS
3.4. Effect of Temperature on the Activity and T/H of Psor-LS
3.5. Thermostability Determination of Psor-LS
3.6. Kinetic Parameters Determination
3.7. The Effect of Sucrose Concentration on the Activity and T/H of Psor-LS
3.8. Effect of Enzyme Concentration on the Levan Production and T/H of Psor-LS
3.9. Biological Production of Psor-LS
3.10. Effect of Temperature on the Product Distribution of Cedi-LS and Psor-LS
3.11. Product Purification and Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lammens, W.; Le Roy, K.; Schroeven, L.; Van Laere, A.; Rabijns, A.; Van den Ende, W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: Functional implications. J. Exp. Bot. 2009, 60, 727–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belghith, K.; Dahech, I.; Belghith, H.; Mejdoub, H. Microbial production of levansucrase for synthesis of fructooligosaccharides and levan. Int. J. Biol. Macromol. 2012, 50, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Ozimek, L.; Kralj, S.; van der Maarel, M.; Dijkhuizen, L. The levansucrase and inulosucrase enzymes of Lactobacillus reuteri 121 catalyse processive and non-processive transglycosylation reactions. Microbiology 2006, 152, 1187–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwar, M.; Kralj, S.; Pique, A.; Leemhuis, H.; van der Maarel, M.; Dijkhuizen, L. Inulin and levan synthesis by probiotic Lactobacillus gasseri strains: Characterization of three novel fructansucrase enzymes and their fructan products. Microbiology 2010, 156, 1264–1274. [Google Scholar] [CrossRef] [Green Version]
- Caputi, L.; Nepogodiev, S.; Malnoy, M.; Rejzek, M.; Field, R.; Benini, S. Biomolecular Characterization of the Levansucrase of Erwinia amylovora, a Promising Biocatalyst for the Synthesis of Fructooligosaccharides. J. Agric. Food Chem. 2013, 61, 12265–12273. [Google Scholar] [CrossRef]
- Vigants, A.; Kruce, R.; Bekers, M.; Zikmanis, P. Response of Zymomonas mobilis levansucrase activity to sodium chloride. Biotechnol. Lett. 1998, 20, 1017–1019. [Google Scholar] [CrossRef]
- Zhang, T.; Li, R.; Qian, H.; Mu, W.; Miao, M.; Jiang, B. Biosynthesis of levan by levansucrase from Bacillus methylotrophicus SK 21.002. Carbohydr. Polym. 2014, 101, 975–981. [Google Scholar] [CrossRef]
- Paul, A.; Samaddar, N.; Dutta, D.; Bagchi, A.; Chakravorty, S.; Chakraborty, W.; Gachhui, R. Mercuric Ion Stabilizes Levansucrase Secreted by Acetobacter nitrogenifigens Strain RG1(T). Protein J. 2011, 30, 262–272. [Google Scholar] [CrossRef]
- Ni, D.; Xu, W.; Bai, Y.; Zhang, W.; Zhang, T.; Mu, W. Biosynthesis of levan from sucrose using a thermostable levansucrase from Lactobacillus reuteri LTH5448. Int. J. Biol. Macromol. 2018, 113, 29–37. [Google Scholar] [CrossRef]
- Gao, S.; Qi, X.; Hart, D.; Gao, H.; An, Y. Expression and Characterization of Levansucrase from Clostridium acetobutylicum. J. Agric. Food Chem. 2017, 65, 867–871. [Google Scholar] [CrossRef]
- Ragab, T.; Shalaby, A.; Awdan, S.; El-Bassyouni, G.; Salama, B.; Helmy, W.; Esawy, M. Role of levan extracted from bacterial honey isolates in curing peptic ulcer: In vivo. Int. J. Biol. Macromol. 2020, 142, 564–573. [Google Scholar] [CrossRef]
- Esawy, M.; Amer, H.; Gamal-Eldeen, A.; El Enshasy, H.; Helmy, W.; Abo-Zeid, M.; Malek, R.; Ahmed, E.; Awad, G. Scaling up, characterization of levan and its inhibitory role in carcinogenesis initiation stage’. Carbohydr. Polym. 2013, 95, 578–587. [Google Scholar] [CrossRef]
- Esawy, M.; Ahmed, E.; Helmy, W.; Mansour, N.; El-Senousy, W.; El-Safty, M. Production of levansucrase from novel honey Bacillus subtilis isolates capable of producing antiviral levans. Carbohydr. Polym. 2011, 86, 823–830. [Google Scholar] [CrossRef]
- Gamal, A.; Abbas, H.; Abdelwahed, N.; Kashef, M.; Mahmoud, K.; Esawy, M.; Ramadan, M. Optimization strategy of Bacillus subtilis MT453867 levansucrase and evaluation of levan role in pancreatic cancer treatment. Int. J. Biol. Macromol. 2021, 182, 1590–1601. [Google Scholar] [CrossRef] [PubMed]
- Raga-Carbajal, E.; Carrillo-Nava, E.; Costas, M.; Porras-Dominguez, J.; Lopez-Munguia, A.; Olvera, C. Size product modulation by enzyme concentration reveals two distinct levan elongation mechanisms in Bacillus subtilis levansucrase. Glycobiology 2016, 26, 377–385. [Google Scholar] [CrossRef] [Green Version]
- Kirtel, O.; Menendez, C.; Versluys, M.; Van den Ende, W.; Hernandez, L.; Oner, E. Levansucrase from Halomonas smyrnensis AAD6(T): First halophilic GH-J clan enzyme recombinantly expressed, purified, and characterized. Appl. Microbiol. Biotechnol. 2018, 102, 9207–9220. [Google Scholar] [CrossRef]
- Santos-Moriano, P.; Fernandez-Arrojo, L.; Poveda, A.; Jimenez-Barbero, J.; Ballesteros, A.; Plou, F. Levan versus fructooligosaccharide synthesis using the levansucrase from Zymomonas mobilis: Effect of reaction conditions. J. Mol. Catal. B Enzym. 2015, 119, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kuhn, R.C.; Filho, F.M. Purification of fructooligosaccharides in an activated charcoal fixed bed column. New Biotechnol. 2010, 27, 862–869. [Google Scholar] [CrossRef]
- Abraham, M.; Murtola, T.; Schulz, R.; P’alla, S.; Smith, J.; Hessa, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Maier, J.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hon, J.; Borko, S.; Stourac, J.; Prokop, Z.; Zendulka, J.; Bednar, D.; Martinek, T.; Damborsky, J. EnzymeMiner: Automated mining of soluble enzymes with diverse structures, catalytic properties and stabilities. Nucleic Acids Res. 2020, 48, W104–W109. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.; de Beer, T.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Yu, S.; Zhang, T.; Jiang, B.; Mu, W. Efficient biosynthesis of levan from sucrose by a novel levansucrase from Brenneria goodwinii. Carbohydr. Polym. 2017, 157, 1732–1740. [Google Scholar] [CrossRef]
- Jadaun, J.; Narnoliya, L.; Agarwal, N.; Singh, S. Catalytic biosynthesis of levan and short-chain fructooligosaccharides from sucrose-containing feedstocks by employing the levansucrase from Leuconostoc mesenteroides MTCC10508. Int. J. Biol. Macromol. 2019, 127, 486–495. [Google Scholar] [CrossRef]
- Sangiliyandi, G.; Chandra Raj, K.; Gunasekaran, P. Elevated temperature and chemical modification selectively abolishes levan forming activity of levansucrase of Zymomonas mobilis. Biotechnol. Lett. 2004, 21, 179–182. [Google Scholar] [CrossRef]
- Ni, D.; Kirtel, O.; Yin, D.; Xu, W.; Chen, Q.; Oner, E.; Mu, W. Improving the catalytic behaviors of Lactobacillus-derived fructansucrases by truncation strategies. Enzyme Microb. Technol. 2021, 149, 109857. [Google Scholar] [CrossRef]
- Ni, D.; Zhang, S.; Kirtel, O.; Xu, W.; Chen, Q.; Oner, E.; Mu, W. Improving the thermostability and catalytic activity of an inulosucrase by rational engineering for the biosynthesis of microbial inulin. J. Agric. Food Chem. 2021, 69, 13125–13134. [Google Scholar] [CrossRef]
- Klaewkla, M.; Pichyangkura, R.; Charoenwongpaiboon, T.; Wangpaiboon, K.; Chunsrivirot, S. Computational design of oligosaccharide producing levansucrase from Bacillus licheniformis RN-01 to improve its thermostability for production of levan-type fructooligosaccharides from sucrose. Int. J. Biol. Macromol. 2020, 160, 252–263. [Google Scholar] [CrossRef]
- Ben Ammar, Y.; Matsubara, T.; Ito, K.; Izuka, M.; Limpaseni, T.; Pongsawasdi, P.; Minamiura, N. Characterization of a thermostable levansucrase from Bacillus sp TH4-2 capable of producing high molecular weight levan at high temperature. J. Biotechnol. 2002, 99, 111–119. [Google Scholar] [CrossRef]
- Salama, B.; Helmy, W.; Ragab, T.; Ali, M.; Taie, H.; Esawy, M. Characterization of a new efficient low molecular weight Bacillus subtilis NRC 16 levansucrase and its levan. J. Basic Microbiol. 2019, 59, 1004–1015. [Google Scholar] [CrossRef]
- Inthanavong, L.; Tian, F.; Khodadadi, M.; Karboune, S. Properties of Geobacillus Stearothermophilus levansucrase as potential biocatalyst for the synthesis of levan and fructooligosaccharides. Biotechnol. Prog. 2013, 29, 1405–1415. [Google Scholar] [CrossRef]
- Li, S.; Yan, Y.; Zhou, Z.; Yu, H.; Zhan, Y.; Zhang, W.; Chen, M.; Lu, W.; Ping, S.; Lin, M. Single amino acid residue changes in subsite −1 of levansucrase from Zymomonas mobilis 10232 strongly influence the enzyme activities and products. Mol. Biol. Rep. 2011, 38, 2437–2443. [Google Scholar] [CrossRef]
- Xu, W.; Ni, D.; Yu, S.; Zhang, T.; Mu, W. Insights into hydrolysis versus transfructosylation: Mutagenesis studies of a novel levansucrase from Brenneria sp. EniD312. Int. J. Biol. Macromol. 2018, 116, 335–345. [Google Scholar] [CrossRef]
- El-Refai, H.; Abdel-Fattah, A.; Mostafa, F. Enzymic synthesis of levan and fructo-oligosaccharides by Bacillus circulans and improvement of levansucrase stability by carbohydrate coupling. World J. Microbiol. Biotechnol. 2009, 25, 821–827. [Google Scholar] [CrossRef]
- Ortiz-Soto, M.; Porras-Dominguez, J.; Rodriguez-Alegria, M.; Morales-Moreno, L.; Diaz-Vilchis, A.; Rudino-Pinera, E.; Beltran-Hernandez, N.; Rivera, H.; Seibel, J.; Lopez Munguia, A. Implications of the mutation S164A on Bacillus subtilis levansucrase product specificity and insights into protein interactions acting upon levan synthesis. Int. J. Biol. Macromol. 2020, 15, 898–908. [Google Scholar] [CrossRef]
- Okuyama, M.; Serizawa, R.; Tanuma, M.; Kikuchi, A.; Sadahiro, J.; Tagami, T.; Lang, W.; Kimura, A. Molecular insight into regioselectivity of transfructosylation catalyzed by GH68 levansucrase and beta-fructofuranosidase. J. Biol. Chem. 2021, 296, 100398. [Google Scholar] [CrossRef]
- Ni, D.; Chen, Z.; Xu, W.; Zhang, W.; Mu, W. Efficient production of inulin and oligosaccharides using thermostable inulosucrase from Lactobacillus jensenii. Int. J. Biol. Macromol. 2020, 165, 1250–1257. [Google Scholar] [CrossRef]
- Kekez, B.; Gojgic-Cvijovic, G.; Jakovljevic, D.; Stefanovic Kojic, J.; Markovic, M.; Beskoski, V.; Vrvic, M. High levan production by Bacillus licheniformis NS032 using ammonium chloride as the sole nitrogen source. Appl. Biochem. Biotechnol. 2015, 175, 3068–3083. [Google Scholar] [CrossRef]
- Oner, E.; Hernandez, L.; Combie, J. Review of Levan polysaccharide: From a century of past experiences to future prospects. Biotechnol. Adv. 2016, 34, 827–844. [Google Scholar] [CrossRef]
- Kang, S.; Lee, J.; Park, Y.; Lee, C.; Kim, S.; Chang, B.; Kim, C.; Seo, J.; Rhee, S.; Jung, S.; et al. Secretory production of Rahnella aquatilis ATCC 33071 levansucrase expressed in Escherichia coli. Int. J. Biol. Macromol. 2004, 14, 1232–1238. [Google Scholar]
- Aramsangtienchai, P.; Kongmon, T.; Pechroj, S.; Srisook, K. Enhanced production and immunomodulatory activity of levan from the acetic acid bacterium, Tanticharoenia sakaeratensis. Int. J. Biol. Macromol. 2020, 163, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, L.; Arrieta, J.; Betancourt, L.; Falcon, V.; Madrazo, J.; Coego, A.; Menendez, C. Levansucrase from Acetobacter diazotrophicus SRT4 is secreted via periplasm by a signal-peptide-dependent pathway. Curr. Microbiol. 1999, 39, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Sattar, F.; Ashfaq, I.; Lindemann, S.; Chen, M.-H.; Van den Ende, W.; Öner, E.; Kirtel, O.; Khaliq, S.; Ghauri, M.; et al. Production and characterization of a high molecular weight levan and fructooligosaccharides from a rhizospheric isolate of Bacillus aryabhattai. LWT 2020, 123, 109093. [Google Scholar] [CrossRef]
- Xavier, J.R.; Ramana, K.V. Optimization of Levan Production by Cold-Active Bacillus licheniformis ANT 179 and Fructooligosaccharide Synthesis by Its Levansucrase. Appl. Biochem. Biotechnol. 2017, 181, 986–1006. [Google Scholar] [CrossRef]










| Microorganisms | Optimal Temp. (°C) | Thermostability | Reference |
|---|---|---|---|
| P. orientalis | 65 | The half-life was 69 h at 45 °C and 7.5 h at 55 °C. | This study |
| B. licheniformis RN-01 | 50 | Half of the initial activity was lost after 1 h at 50 °C. | [29] |
| Bacillus sp. TH4-2 | 60 | Half of the initial activity was lost after 30 min at 60 °C. | [30] |
| G. stearothermophilus | 57 | More than 95% of the initial activity was retained at 4-47 °C for 6 h. | [32] |
| Z. mobilis | 30 | The activity lost at 50 °C for 15 min. | [33] |
| Brenneria sp. EniD312 | 45 | The half-life was 2 h at 45 °C and 1.2 h at 55 °C | [34] |
| B. goodwinii | 40 | The activity lost after 0.5 h of incubation at 50 °C | [24] |
| L. reuteri LTH5448 | 35 | The activity remained 63.8% at 55 °C for 12 h | [9] |
| Bacillus subtilis NRC | 45 | The activity remained 60% at 50 °C for 12 h | [31] |
| B. circulans | 35 | The half-life was 130 min at 50 ℃ | [35] |
| LS | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | |||
|---|---|---|---|---|---|---|
| Hydrolysis | Transfer | Hydrolysis | Transfer | Hydrolysis | Transfer | |
| Cedi-LS | 57 ± 2 | 202 ± 7 | 332 ± 22 | 449 ± 13 | 5.80 | 2.23 |
| Psor-LS | 117 ± 8 | - | 620 ± 12 | - | 5.27 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guang, C.; Zhang, X.; Ni, D.; Zhang, W.; Xu, W.; Mu, W. Identification of a Thermostable Levansucrase from Pseudomonas orientalis That Allows Unique Product Specificity at Different Temperatures. Polymers 2023, 15, 1435. https://doi.org/10.3390/polym15061435
Guang C, Zhang X, Ni D, Zhang W, Xu W, Mu W. Identification of a Thermostable Levansucrase from Pseudomonas orientalis That Allows Unique Product Specificity at Different Temperatures. Polymers. 2023; 15(6):1435. https://doi.org/10.3390/polym15061435
Chicago/Turabian StyleGuang, Cuie, Xiaoqi Zhang, Dawei Ni, Wenli Zhang, Wei Xu, and Wanmeng Mu. 2023. "Identification of a Thermostable Levansucrase from Pseudomonas orientalis That Allows Unique Product Specificity at Different Temperatures" Polymers 15, no. 6: 1435. https://doi.org/10.3390/polym15061435
APA StyleGuang, C., Zhang, X., Ni, D., Zhang, W., Xu, W., & Mu, W. (2023). Identification of a Thermostable Levansucrase from Pseudomonas orientalis That Allows Unique Product Specificity at Different Temperatures. Polymers, 15(6), 1435. https://doi.org/10.3390/polym15061435