Studies of the Mechanism of Nucleosome Dynamics: A Review on Multifactorial Regulation from Computational and Experimental Cases
Abstract
1. Introduction
2. Multiple Regulatory Factors and Associated Mechanism
2.1. Histone Modification
2.1.1. Dynamic Features of Unmodified Histone
2.1.2. Post-Translational Modifications of Histones
2.1.3. The Interplay of Different Histone Modifications
2.2. DNA Methylation
2.2.1. DNA Methylation Effect on DNA Properties
2.2.2. DNA Methylation Effect on the DNA–Histone Complex
2.3. Interactions of Nucleosome-Interacting Factors
2.3.1. Transcription Factors
2.3.2. Nucleosome Remodeling Proteins
2.3.3. Cations
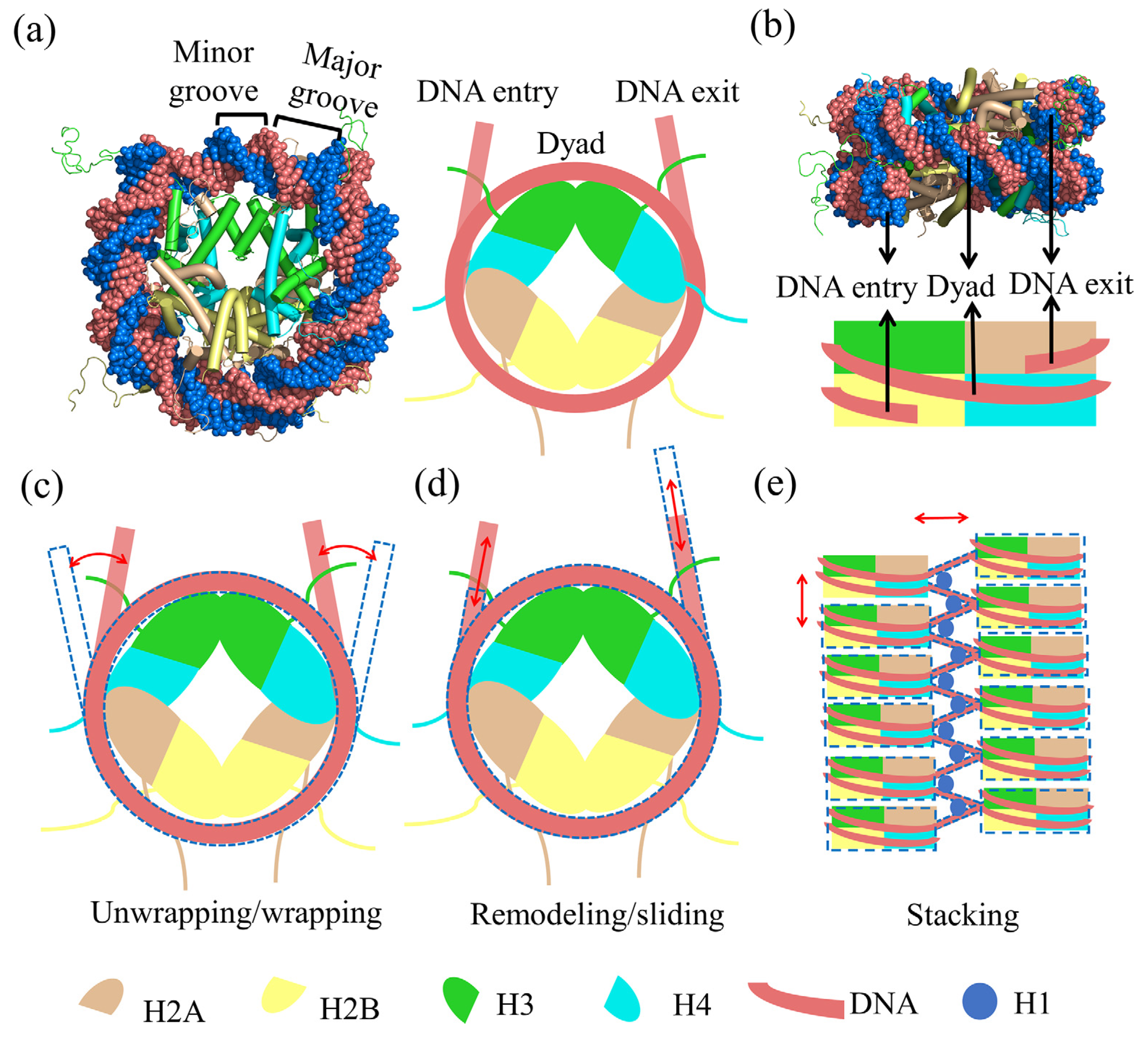
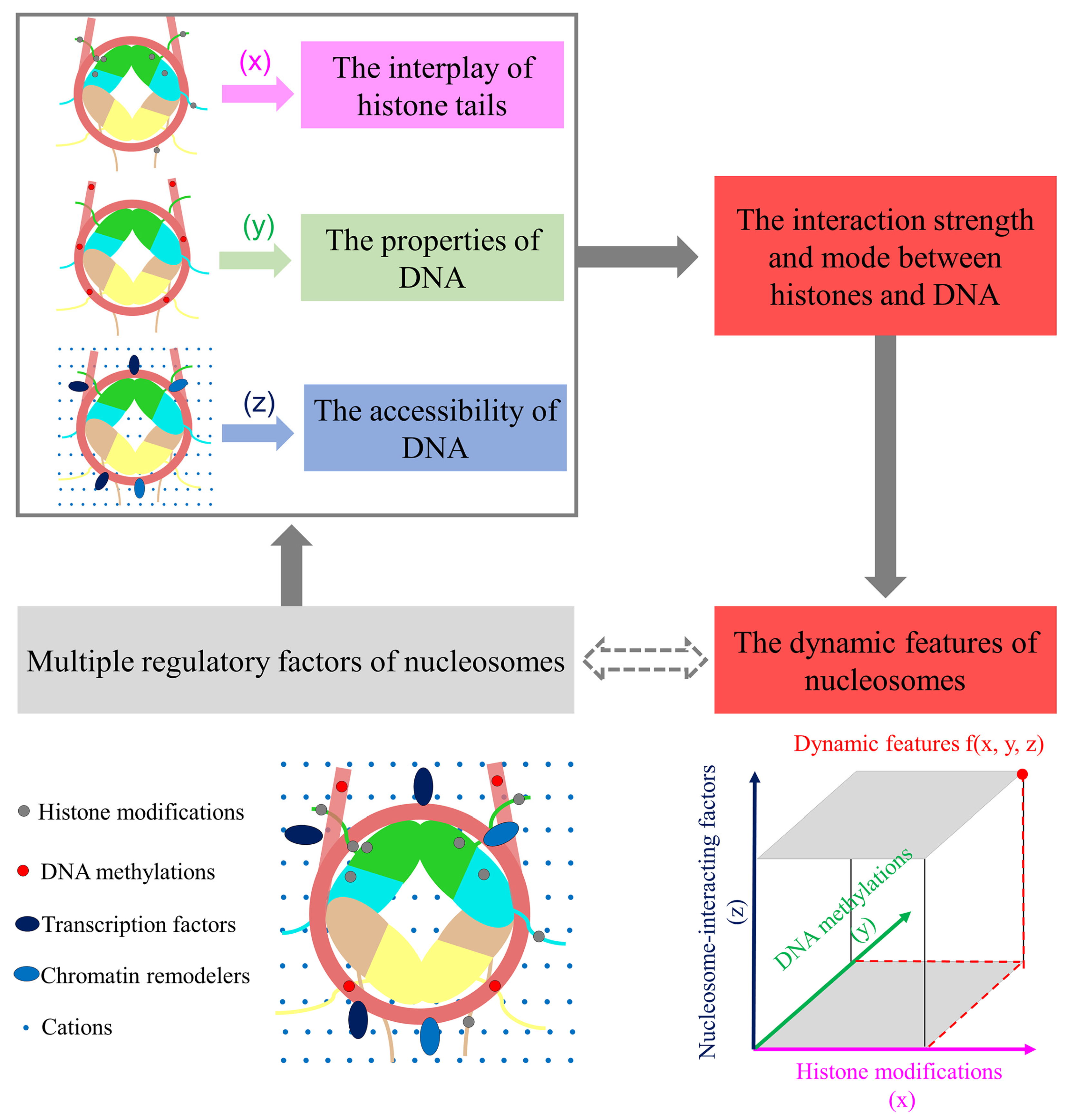
3. The Crosstalk of Multiple Factors and Effects on the High-Order Structure of Nucleosomes
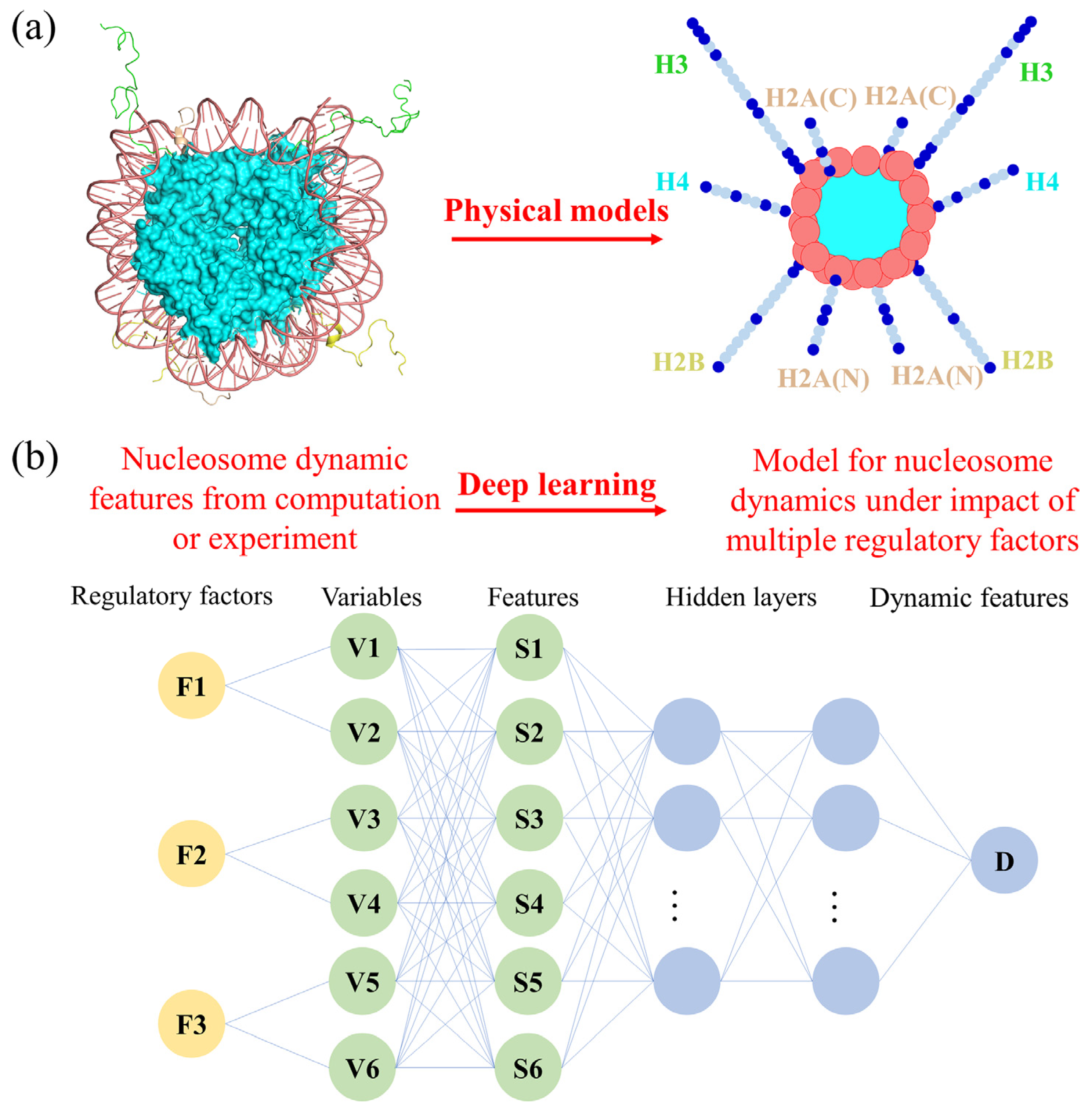
4. Further Mechanism Research through the Integration of Different Techniques
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Olins, D.E.; Olins, A.L. Chromatin history: Our view from the bridge. Nat. Rev. Mol. Cell Biol. 2003, 4, 809–814. [Google Scholar] [CrossRef]
- Arents, G.; Burlingame, R.W.; Wang, B.C.; Love, W.E.; Moudrianakis, E.N. The nucleosomal core histone octamer at 3.1 A resolution: A tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 1991, 88, 10148–10152. [Google Scholar] [CrossRef]
- Arents, G.; Moudrianakis, E.N. The histone fold: A ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 1995, 92, 11170–11174. [Google Scholar] [CrossRef]
- Flaus, A.; Luger, K.; Tan, S.; Richmond, T.J. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. USA 1996, 93, 1370–1375. [Google Scholar] [CrossRef] [PubMed]
- Mcginty, R.K.; Tan, S. Nucleosome structure and function. Chem. Rev. 2015, 115, 2255–2273. [Google Scholar] [CrossRef] [PubMed]
- Hergeth, S.P.; Schneider, R. The H1 linker histones: Multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015, 16, 1439–1453. [Google Scholar] [CrossRef]
- Nodelman, I.M.; Bowman, G.D. Biophysics of Chromatin Remodeling. Annu. Rev. Biophys. 2021, 50, 73–93. [Google Scholar] [CrossRef]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Du Preez, L.L.; Patterton, H.G. Secondary structures of the core histone N-terminal tails: Their role in regulating chromatin structure. Subcell. Biochem. 2013, 61, 37–55. [Google Scholar] [PubMed]
- Woodcock, C.L. A milestone in the odyssey of higher-order chromatin structure. Nat. Struct. Mol. Biol. 2005, 12, 639–640. [Google Scholar] [CrossRef]
- Finch, J.T.; Klug, A. Solenoidal model for superstructure in chromatin. Proc. Natl. Acad. Sci. USA 1976, 73, 1897–1901. [Google Scholar] [CrossRef]
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New insights into nucleosome and chromatin structure: An ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012, 13, 436–447. [Google Scholar] [CrossRef]
- Fierz, B.; Poirier, M.G. Biophysics of Chromatin Dynamics. Annu. Rev. Biophys. 2019, 48, 321–345. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D.; Lorch, Y. Primary Role of the Nucleosome. Mol. Cell 2020, 79, 371–375. [Google Scholar] [CrossRef]
- Ohno, M.; Priest, D.G.; Taniguchi, Y. Nucleosome-level 3D organization of the genome. Biochem. Soc. Trans. 2018, 46, 491–501. [Google Scholar] [CrossRef]
- Polach, K.J.; Widom, J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995, 254, 130–149. [Google Scholar] [CrossRef]
- Markert, J.; Luger, K. Nucleosomes Meet Their Remodeler Match. Trends Biochem. Sci. 2021, 46, 41–50. [Google Scholar] [CrossRef]
- Ludwigsen, J.; Hepp, N.; Klinker, H.; Pfennig, S.; Mueller-Planitz, F. Remodeling and Repositioning of Nucleosomes in Nucleosomal Arrays. Methods Mol. Biol. 2018, 1805, 349–370. [Google Scholar] [PubMed]
- Schalch, T.; Duda, S.; Sargent, D.F.; Richmond, T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 2005, 436, 138–141. [Google Scholar] [CrossRef] [PubMed]
- Dorigo, B.; Schalch, T.; Kulangara, A.; Duda, S.; Schroeder, R.R.; Richmond, T.J. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 2004, 306, 1571–1573. [Google Scholar] [CrossRef]
- Bilokapic, S.; Strauss, M.; Halic, M. Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol. 2018, 25, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Broströmer, E.; Xing, D.; Jin, J.; Chong, S.; Ge, H.; Wang, S.; Gu, C.; Yang, L.; Gao, Y.Q.; et al. Probing allostery through DNA. Science 2013, 339, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Hager, G.L.; Mcnally, J.G.; Misteli, T. Transcription dynamics. Mol. Cell 2009, 35, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, K.; Felekyan, S.; Kuhnemuth, R.; Dimura, M.; Toth, K.; Seidel, C.A.M.; Langowski, J. Dynamics of the nucleosomal histone H3 N-terminal tail revealed by high precision single-molecule FRET. Nucleic Acids Res. 2020, 48, 1551–1571. [Google Scholar] [CrossRef] [PubMed]
- Stutzer, A.; Liokatis, S.; Kiesel, A.; Schwarzer, D.; Sprangers, R.; Soding, J.; Selenko, P.; Fischle, W. Modulations of DNA Contacts by Linker Histones and Post-translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Mol. Cell 2016, 61, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Funke, J.J.; Ketterer, P.; Lieleg, C.; Schunter, S.; Korber, P.; Dietz, H. Uncovering the forces between nucleosomes using DNA origami. Sci. Adv. 2016, 2, e1600974. [Google Scholar] [CrossRef]
- Huertas, J.; Cojocaru, V. Breaths, Twists, and Turns of Atomistic Nucleosomes. J. Mol. Biol. 2021, 433, 166744. [Google Scholar] [CrossRef]
- Perišić, O.; Schlick, T. Computational strategies to address chromatin structure problems. Phys. Biol. 2016, 13, 035006. [Google Scholar] [CrossRef]
- Moller, J.; De Pablo, J.J. Bottom-Up Meets Top-Down: The Crossroads of Multiscale Chromatin Modeling. Biophys. J. 2020, 118, 2057–2065. [Google Scholar] [CrossRef]
- Ozer, G.; Luque, A.; Schlick, T. The chromatin fiber: Multiscale problems and approaches. Curr. Opin. Struct. Biol. 2015, 31, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Desvoyes, B.; Sanchez, M.P.; Ramirez-Parra, E.; Gutierrez, C. Impact of nucleosome dynamics and histone modifications on cell proliferation during Arabidopsis development. Heredity 2010, 105, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Hauer, M.H.; Gasser, S.M. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017, 31, 2204–2221. [Google Scholar] [CrossRef]
- Stack, E.C.; Del Signore, S.J.; Luthi-Carter, R.; Soh, B.Y.; Goldstein, D.R.; Matson, S.; Goodrich, S.; Markey, A.L.; Cormier, K.; Hagerty, S.W.; et al. Modulation of nucleosome dynamics in Huntington’s disease. Hum. Mol. Genet. 2007, 16, 1164–1175. [Google Scholar] [CrossRef]
- Wang, G.; Vasquez, K.M. Dynamic alternative DNA structures in biology and disease. Nat. Rev. Genet. 2023, 24, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Blossey, R.; Schiessel, H. The dynamics of the nucleosome: Thermal effects, external forces and ATP. FEBS J. 2011, 278, 3619–3632. [Google Scholar] [CrossRef] [PubMed]
- Bergman, Y.; Cedar, H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013, 20, 274–281. [Google Scholar] [CrossRef]
- Chen, K.; Xi, Y.; Pan, X.; Li, Z.; Kaestner, K.; Tyler, J.; Dent, S.; He, X.; Li, W. DANPOS: Dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013, 23, 341–351. [Google Scholar] [CrossRef]
- Mines, R.C.; Lipniacki, T.; Shen, X. Slow nucleosome dynamics set the transcriptional speed limit and induce RNA polymerase II traffic jams and bursts. PLoS Comput. Biol. 2022, 18, e1009811. [Google Scholar] [CrossRef]
- Rao, S.; Han, A.L.; Zukowski, A.; Kopin, E.; Sartorius, C.A.; Kabos, P.; Ramachandran, S. Transcription factor-nucleosome dynamics from plasma cfDNA identifies ER-driven states in breast cancer. Sci. Adv. 2022, 8, eabm4358. [Google Scholar] [CrossRef]
- Armeev, G.A.; Gribkova, A.K.; Pospelova, I.; Komarova, G.A.; Shaytan, A.K. Linking chromatin composition and structural dynamics at the nucleosome level. Curr. Opin. Struct. Biol. 2019, 56, 46–55. [Google Scholar] [CrossRef]
- Zhou, K.; Gaullier, G.; Luger, K. Nucleosome structure and dynamics are coming of age. Nat. Struct. Mol. Biol. 2019, 26, 3–13. [Google Scholar] [CrossRef]
- Iwasaki, W.; Miya, Y.; Horikoshi, N.; Osakabe, A.; Taguchi, H.; Tachiwana, H.; Shibata, T.; Kagawa, W.; Kurumizaka, H. Contribution of histone N-terminal tails to the structure and stability of nucleosomes. FEBS Open Bio 2013, 3, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Morales, V.; Richard-Foy, H. Role of histone N-terminal tails and their acetylation in nucleosome dynamics. Mol. Cell Biol. 2000, 20, 7230–7237. [Google Scholar] [CrossRef]
- Ohtomo, H.; Kurita, J.I.; Sakuraba, S.; Li, Z.; Arimura, Y.; Wakamori, M.; Tsunaka, Y.; Umehara, T.; Kurumizaka, H.; Kono, H.; et al. The N-terminal Tails of Histones H2A and H2B Adopt Two Distinct Conformations in the Nucleosome with Contact and Reduced Contact to DNA. J. Mol. Biol. 2021, 433, 167110. [Google Scholar] [CrossRef]
- Rabdano, S.O.; Shannon, M.D.; Izmailov, S.A.; Gonzalez Salguero, N.; Zandian, M.; Purusottam, R.N.; Poirier, M.G.; Skrynnikov, N.R.; Jaroniec, C.P. Histone H4 Tails in Nucleosomes: A Fuzzy Interaction with DNA. Angew. Chem. Int. Ed. Engl. 2021, 60, 6480–6487. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikova, A.A.; Porter-Goff, M.E.; Muthurajan, U.M.; Luger, K.; Hansen, J.C. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface 2013, 10, 20121022. [Google Scholar] [CrossRef] [PubMed]
- Dueva, R.; Akopyan, K.; Pederiva, C.; Trevisan, D.; Dhanjal, S.; Lindqvist, A.; Farnebo, M. Neutralization of the Positive Charges on Histone Tails by RNA Promotes an Open Chromatin Structure. Cell Chem. Biol. 2019, 26, 1436–1449.e5. [Google Scholar] [CrossRef]
- Bendandi, A.; Patelli, A.S.; Diaspro, A.; Rocchia, W. The role of histone tails in nucleosome stability: An electrostatic perspective. Comput. Struct. Biotechnol. J. 2020, 18, 2799–2809. [Google Scholar] [CrossRef]
- Huertas, J.; Scholer, H.R.; Cojocaru, V. Histone tails cooperate to control the breathing of genomic nucleosomes. PLoS Comput. Biol. 2021, 17, e1009013. [Google Scholar] [CrossRef]
- Lequieu, J.; Schwartz, D.C.; De Pablo, J.J. In silico evidence for sequence-dependent nucleosome sliding. Proc. Natl. Acad. Sci. USA 2017, 114, E9197–E9205. [Google Scholar] [CrossRef]
- Hamiche, A.; Kang, J.G.; Dennis, C.; Xiao, H.; Wu, C. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. USA 2001, 98, 14316–14321. [Google Scholar] [CrossRef] [PubMed]
- Lorch, Y.; Kornberg, R.D.; Maier-Davis, B. Role of the histone tails in histone octamer transfer. Nucleic Acids Res. 2023, gkad079. [Google Scholar] [CrossRef]
- Bowman, G.D.; Poirier, M.G. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015, 115, 2274–2295. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, A.T.; Hansen, J.C. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 2000, 9, 37–61. [Google Scholar] [CrossRef] [PubMed]
- Ikebe, J.; Sakuraba, S.; Kono, H. H3 Histone Tail Conformation within the Nucleosome and the Impact of K14 Acetylation Studied Using Enhanced Sampling Simulation. PLoS Comput. Biol. 2016, 12, e1004788. [Google Scholar] [CrossRef] [PubMed]
- Meas, R.; Mao, P. Histone ubiquitylation and its roles in transcription and DNA damage response. DNA Repair 2015, 36, 36–42. [Google Scholar] [CrossRef]
- Krajewski, W.A. Histone Modifications, Internucleosome Dynamics, and DNA Stresses: How They Cooperate to “Functionalize” Nucleosomes. Front. Genet. 2022, 13, 873398. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Simon, M.; North, J.A.; Shimko, J.C.; Forties, R.A.; Ferdinand, M.B.; Manohar, M.; Zhang, M.; Fishel, R.; Ottesen, J.J.; Poirier, M.G. Histone fold modifications control nucleosome unwrapping and disassembly. Proc. Natl. Acad. Sci. USA 2011, 108, 12711–12716. [Google Scholar] [CrossRef]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef]
- Forties, R.A.; North, J.A.; Javaid, S.; Tabbaa, O.P.; Fishel, R.; Poirier, M.G.; Bundschuh, R. A quantitative model of nucleosome dynamics. Nucleic Acids Res. 2011, 39, 8306–8313. [Google Scholar] [CrossRef] [PubMed]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef]
- Norouzi, D.; Zhurkin, V.B. Dynamics of Chromatin Fibers: Comparison of Monte Carlo Simulations with Force Spectroscopy. Biophys. J. 2018, 115, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Lequieu, J.; Cordoba, A.; Schwartz, D.C.; De Pablo, J.J. Tension-Dependent Free Energies of Nucleosome Unwrapping. ACS Cent. Sci. 2016, 2, 660–666. [Google Scholar] [CrossRef]
- Chen, W.; Liu, Y.; Zhu, S.; Chen, G.; Han, J.J. Inter-nucleosomal communication between histone modifications for nucleosome phasing. PLoS Comput. Biol. 2018, 14, e1006416. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Gutierrez-Arcelus, M.; Lappalainen, T.; Montgomery, S.B.; Buil, A.; Ongen, H.; Yurovsky, A.; Bryois, J.; Giger, T.; Romano, L.; Planchon, A.; et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2013, 2, e00523. [Google Scholar] [CrossRef]
- Li, S.; Peng, Y.; Panchenko, A.R. DNA methylation: Precise modulation of chromatin structure and dynamics. Curr. Opin. Struct. Biol. 2022, 75, 102430. [Google Scholar] [CrossRef]
- Dhar, G.A.; Saha, S.; Mitra, P.; Nag Chaudhuri, R. DNA methylation and regulation of gene expression: Guardian of our health. Nucleus 2021, 64, 259–270. [Google Scholar] [CrossRef]
- Hervouet, E.; Peixoto, P.; Delage-Mourroux, R.; Boyer-Guittaut, M.; Cartron, P.F. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin. Epigenet. 2018, 10, 17. [Google Scholar] [CrossRef]
- Gao, F.; Das, S.K. Epigenetic regulations through DNA methylation and hydroxymethylation: Clues for early pregnancy in decidualization. Biomol. Concepts 2014, 5, 95–107. [Google Scholar] [CrossRef]
- Kinde, B.; Gabel, H.W.; Gilbert, C.S.; Griffith, E.C.; Greenberg, M.E. Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2. Proc. Natl. Acad. Sci. USA 2015, 112, 6800–6806. [Google Scholar] [CrossRef]
- Li, J.; Wu, X.; Zhou, Y.; Lee, M.; Guo, L.; Han, W.; Mo, W.; Cao, W.M.; Sun, D.; Xie, R.; et al. Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res. 2018, 46, 2883–2900. [Google Scholar] [CrossRef] [PubMed]
- Mendonca, A.; Chang, E.H.; Liu, W.; Yuan, C. Hydroxymethylation of DNA influences nucleosomal conformation and stability in vitro. Biochim. Biophys. Acta 2014, 1839, 1323–1329. [Google Scholar] [CrossRef]
- Battistini, F.; Dans, P.D.; Terrazas, M.; Castellazzi, C.L.; Portella, G.; Labrador, M.; Villegas, N.; Brun-Heath, I.; Gonzalez, C.; Orozco, M. The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function. PLoS Comput. Biol. 2021, 17, e1009547. [Google Scholar] [CrossRef]
- Hognon, C.; Besancenot, V.; Gruez, A.; Grandemange, S.; Monari, A. Cooperative Effects of Cytosine Methylation on DNA Structure and Dynamics. J. Phys. Chem. B 2019, 123, 7365–7371. [Google Scholar] [CrossRef]
- Horberg, J.; Reymer, A. A sequence environment modulates the impact of methylation on the torsional rigidity of DNA. Chem. Commun. 2018, 54, 11885–11888. [Google Scholar] [CrossRef] [PubMed]
- Severin, P.M.; Zou, X.; Gaub, H.E.; Schulten, K. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res. 2011, 39, 8740–8751. [Google Scholar] [CrossRef]
- Kameda, T.; Suzuki, M.M.; Awazu, A.; Togashi, Y. Structural dynamics of DNA depending on methylation pattern. Phys. Rev. E 2021, 103, 012404. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.T.; Gouveia, L.; Kanna, C.R.; Warmlander, S.K.; Platts, J.A.; Kamerlin, S.C. Understanding the structural and dynamic consequences of DNA epigenetic modifications: Computational insights into cytosine methylation and hydroxymethylation. Epigenetics 2014, 9, 1604–1612. [Google Scholar] [CrossRef] [PubMed]
- Buitrago, D.; Labrador, M.; Arcon, J.P.; Lema, R.; Flores, O.; Esteve-Codina, A.; Blanc, J.; Villegas, N.; Bellido, D.; Gut, M.; et al. Impact of DNA methylation on 3D genome structure. Nat. Commun. 2021, 12, 3243. [Google Scholar] [CrossRef] [PubMed]
- Chodavarapu, R.K.; Feng, S.; Bernatavichute, Y.V.; Chen, P.Y.; Stroud, H.; Yu, Y.; Hetzel, J.A.; Kuo, F.; Kim, J.; Cokus, S.J.; et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010, 466, 388–392. [Google Scholar] [CrossRef]
- Felle, M.; Hoffmeister, H.; Rothammer, J.; Fuchs, A.; Exler, J.H.; Langst, G. Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res. 2011, 39, 6956–6969. [Google Scholar] [CrossRef]
- Jimenez-Useche, I.; Yuan, C. The effect of DNA CpG methylation on the dynamic conformation of a nucleosome. Biophys. J. 2012, 103, 2502–2512. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Lee, T.H. Effects of DNA methylation on the structure of nucleosomes. J. Am. Chem. Soc. 2012, 134, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Portella, G.; Battistini, F.; Orozco, M. Understanding the connection between epigenetic DNA methylation and nucleosome positioning from computer simulations. PLoS Comput. Biol. 2013, 9, e1003354. [Google Scholar] [CrossRef]
- Li, S.; Peng, Y.; Landsman, D.; Panchenko, A.R. DNA methylation cues in nucleosome geometry, stability and unwrapping. Nucleic Acids Res. 2022, 50, 1864–1874. [Google Scholar] [CrossRef] [PubMed]
- Choy, J.S.; Wei, S.; Lee, J.Y.; Tan, S.; Chu, S.; Lee, T.H. DNA methylation increases nucleosome compaction and rigidity. J. Am. Chem. Soc. 2010, 132, 1782–1783. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Barozzi, I.; Simonatto, M.; Bonifacio, S.; Yang, L.; Rohs, R.; Ghisletti, S.; Natoli, G. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell 2014, 54, 844–857. [Google Scholar] [CrossRef]
- Fernandez Garcia, M.; Moore, C.D.; Schulz, K.N.; Alberto, O.; Donague, G.; Harrison, M.M.; Zhu, H.; Zaret, K.S. Structural Features of Transcription Factors Associating with Nucleosome Binding. Mol. Cell 2019, 75, 921–932.e6. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Takada, S. Nucleosome allostery in pioneer transcription factor binding. Proc. Natl. Acad. Sci. USA 2020, 117, 20586–20596. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.K.; Grand, R.S.; Isbel, L.; Cavadini, S.; Kozicka, Z.; Kempf, G.; Bunker, R.D.; Schenk, A.D.; Graff-Meyer, A.; Pathare, G.R.; et al. Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 2020, 368, 1460–1465. [Google Scholar] [CrossRef]
- Newman, J.A.; Aitkenhead, H.; Gavard, A.E.; Rota, I.A.; Handel, A.E.; Hollander, G.A.; Gileadi, O. The crystal structure of human forkhead box N1 in complex with DNA reveals the structural basis for forkhead box family specificity. J. Biol. Chem. 2020, 295, 2948–2958. [Google Scholar] [CrossRef] [PubMed]
- Kar, A.; Gutierrez-Hartmann, A. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 522–543. [Google Scholar] [CrossRef]
- Fairall, L.; Schwabe, J.W.; Chapman, L.; Finch, J.T.; Rhodes, D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature 1993, 366, 483–487. [Google Scholar] [CrossRef]
- Lu, D.; Searles, M.A.; Klug, A. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature 2003, 426, 96–100. [Google Scholar] [CrossRef]
- Hayes, J.J.; Wolffe, A.P. The interaction of transcription factors with nucleosomal DNA. Bioessays 1992, 14, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Farnung, L.; Kaasinen, E.; Sahu, B.; Yin, Y.; Wei, B.; Dodonova, S.O.; Nitta, K.R.; Morgunova, E.; Taipale, M.; et al. The interaction landscape between transcription factors and the nucleosome. Nature 2018, 562, 76–81. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Teves, S.S.; Weber, C.M.; Henikoff, S. Transcribing through the nucleosome. Trends Biochem. Sci. 2014, 39, 577–586. [Google Scholar] [CrossRef]
- Zaret, K.S. Pioneer Transcription Factors Initiating Gene Network Changes. Annu. Rev. Genet. 2020, 54, 367–385. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Drouin, J. Pioneer factors as master regulators of the epigenome and cell fate. Nat. Rev. Mol. Cell Biol. 2022, 23, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Soufi, A.; Garcia, M.F.; Jaroszewicz, A.; Osman, N.; Pellegrini, M.; Zaret, K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161, 555–568. [Google Scholar] [CrossRef]
- Dodonova, S.O.; Zhu, F.; Dienemann, C.; Taipale, J.; Cramer, P. Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 2020, 580, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Sheu, K.M.; Cheng, Q.J.; Hoffmann, A.; Enciso, G. Stochastic models of nucleosome dynamics reveal regulatory rules of stimulus-induced epigenome remodeling. Cell Rep. 2022, 40, 111076. [Google Scholar] [CrossRef]
- Huertas, J.; Maccarthy, C.M.; Scholer, H.R.; Cojocaru, V. Nucleosomal DNA Dynamics Mediate Oct4 Pioneer Factor Binding. Biophys. J. 2020, 118, 2280–2296. [Google Scholar] [CrossRef]
- Maccarthy, C.M.; Huertas, J.; Ortmeier, C.; Vom Bruch, H.; Tan, D.S.; Reinke, D.; Sander, A.; Bergbrede, T.; Jauch, R.; Scholer, H.R.; et al. OCT4 interprets and enhances nucleosome flexibility. Nucleic Acids Res. 2022, 50, 10311–10327. [Google Scholar] [CrossRef]
- Sundaramoorthy, R. Nucleosome remodelling: Structural insights into ATP-dependent remodelling enzymes. Essays Biochem. 2019, 63, 45–58. [Google Scholar]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Yan, L.; Wu, H.; Li, X.; Gao, N.; Chen, Z. Structures of the ISWI–nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol. 2019, 26, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Marfella, C.G.; Imbalzano, A.N. The Chd family of chromatin remodelers. Mutat. Res. 2007, 618, 30–40. [Google Scholar] [CrossRef]
- Mashtalir, N.; D’avino, A.R.; Michel, B.C.; Luo, J.; Pan, J.; Otto, J.E.; Zullow, H.J.; Mckenzie, Z.M.; Kubiak, R.L.; St Pierre, R.; et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175, 1272–1288. e20. [Google Scholar] [CrossRef] [PubMed]
- Eustermann, S.; Schall, K.; Kostrewa, D.; Lakomek, K.; Strauss, M.; Moldt, M.; Hopfner, K.P. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 2018, 556, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Chen, Z. A Unifying Mechanism of DNA Translocation Underlying Chromatin Remodeling. Trends Biochem. Sci. 2020, 45, 217–227. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, B. Theory of Active Chromatin Remodeling. Phys. Rev. Lett. 2019, 123, 208102. [Google Scholar] [CrossRef]
- Brandani, G.B.; Takada, S. Chromatin remodelers couple inchworm motion with twist-defect formation to slide nucleosomal DNA. PLoS Comput. Biol. 2018, 14, e1006512. [Google Scholar] [CrossRef]
- Brandani, G.B.; Niina, T.; Tan, C.; Takada, S. DNA sliding in nucleosomes via twist defect propagation revealed by molecular simulations. Nucleic Acids Res. 2018, 46, 2788–2801. [Google Scholar] [CrossRef] [PubMed]
- Bilokapic, S.; Strauss, M.; Halic, M. Structural rearrangements of the histone octamer translocate DNA. Nat. Commun. 2018, 9, 1330. [Google Scholar] [CrossRef]
- Bhardwaj, S.K.; Hailu, S.G.; Olufemi, L.; Brahma, S.; Kundu, S.; Hota, S.K.; Persinger, J.; Bartholomew, B. Dinucleosome specificity and allosteric switch of the ISW1a ATP-dependent chromatin remodeler in transcription regulation. Nat. Commun. 2020, 11, 5913. [Google Scholar] [CrossRef] [PubMed]
- Romani, A.; Scarpa, A. Regulation of cell magnesium. Arch. Biochem. Biophys. 1992, 298, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sissi, C.; Palumbo, M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009, 37, 702–711. [Google Scholar] [CrossRef]
- Grubbs, R.D.; Maguire, M.E. Magnesium as a regulatory cation: Criteria and evaluation. Magnesium 1987, 6, 113–127. [Google Scholar]
- Inoue, S.; Sugiyama, S.; Travers, A.A.; Ohyama, T. Self-assembly of double-stranded DNA molecules at nanomolar concentrations. Biochemistry 2007, 46, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, J.; Ohyama, T. Selective association between nucleosomes with identical DNA sequences. Nucleic Acids Res. 2013, 41, 1544–1554. [Google Scholar] [CrossRef]
- De Frutos, M.; Raspaud, E.; Leforestier, A.; Livolant, F. Aggregation of nucleosomes by divalent cations. Biophys. J. 2001, 81, 1127–1132. [Google Scholar] [CrossRef]
- Ohyama, T. New Aspects of Magnesium Function: A Key Regulator in Nucleosome Self-Assembly, Chromatin Folding and Phase Separation. Int. J. Mol. Sci. 2019, 20, 4232. [Google Scholar] [CrossRef]
- Sun, T.; Minhas, V.; Mirzoev, A.; Korolev, N.; Lyubartsev, A.P.; Nordenskiold, L. A Bottom-Up Coarse-Grained Model for Nucleosome-Nucleosome Interactions with Explicit Ions. J. Chem. Theory Comput. 2022, 18, 3948–3960. [Google Scholar] [CrossRef]
- Andreeva, T.V.; Maluchenko, N.V.; Sivkina, A.L.; Chertkov, O.V.; Valieva, M.E.; Kotova, E.Y.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Na(+) and K(+) Ions Differently Affect Nucleosome Structure, Stability, and Interactions with Proteins. Microsc. Microanal. 2022, 28, 243–253. [Google Scholar] [CrossRef]
- Kosarim, N.A.; Armeev, G.A.; Kirpichnikov, M.P.; Shaytan, A.K. Analysis of Ion Atmosphere Around Nucleosomes Using Supercomputer MD Simulations. Supercomput. Front. Innov. 2022, 9, 56–67. [Google Scholar]
- Gebala, M.; Johnson, S.L.; Narlikar, G.J.; Herschlag, D. Ion counting demonstrates a high electrostatic field generated by the nucleosome. eLife 2019, 8, e44993. [Google Scholar] [CrossRef]
- Bartke, T.; Vermeulen, M.; Xhemalce, B.; Robson, S.C.; Mann, M.; Kouzarides, T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010, 143, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Brandani, G.B.; Tan, C.; Takada, S. The kinetic landscape of nucleosome assembly: A coarse-grained molecular dynamics study. PLoS Comput. Biol. 2021, 17, e1009253. [Google Scholar] [CrossRef] [PubMed]
- Brandani, G.B.; Gopi, S.; Yamauchi, M.; Takada, S. Molecular dynamics simulations for the study of chromatin biology. Curr. Opin. Struct. Biol. 2022, 77, 102485. [Google Scholar] [CrossRef]
- Jeltsch, A.; Broche, J.; Bashtrykov, P. Molecular Processes Connecting DNA Methylation Patterns with DNA Methyltransferases and Histone Modifications in Mammalian Genomes. Genes 2018, 9, 566. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.K.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.P.; Allis, C.D.; et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ahn, J.H.; Wang, G.G. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol. Life Sci. 2019, 76, 2899–2916. [Google Scholar]
- Li, J.; Bergmann, L.; Rafael De Almeida, A.; Webb, K.M.; Gogol, M.M.; Voigt, P.; Liu, Y.; Liang, H.; Smolle, M.M. H3K36 methylation and DNA-binding both promote Ioc4 recruitment and Isw1b remodeler function. Nucleic Acids Res. 2022, 50, 2549–2565. [Google Scholar] [CrossRef] [PubMed]
- Tallant, C.; Valentini, E.; Fedorov, O.; Overvoorde, L.; Ferguson, F.M.; Filippakopoulos, P.; Svergun, D.I.; Knapp, S.; Ciulli, A. Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC. Structure 2015, 23, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Winogradoff, D.; Dalal, Y.; Papoian, G.A. The Oligomerization Landscape of Histones. Biophys. J. 2019, 116, 1845–1855. [Google Scholar] [CrossRef]
- Alvarado, W.; Moller, J.; Ferguson, A.L.; De Pablo, J.J. Tetranucleosome Interactions Drive Chromatin Folding. ACS. Cent. Sci. 2021, 7, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Farr, S.E.; Woods, E.J.; Joseph, J.A.; Garaizar, A.; Collepardo-Guevara, R. Nucleosome plasticity is a critical element of chromatin liquid-liquid phase separation and multivalent nucleosome interactions. Nat. Commun. 2021, 12, 2883. [Google Scholar] [CrossRef]
- Agbleke, A.A.; Amitai, A.; Buenrostro, J.D.; Chakrabarti, A.; Chu, L.; Hansen, A.S.; Koenig, K.M.; Labade, A.S.; Liu, S.; Nozaki, T.; et al. Advances in Chromatin and Chromosome Research: Perspectives from Multiple Fields. Mol. Cell 2020, 79, 881–901. [Google Scholar] [CrossRef]
- Öztürk, M.A.; De, M.; Cojocaru, V.; Wade, R.C. Chromatosome Structure and Dynamics from Molecular Simulations. Annu. Rev. Phys. Chem. 2020, 71, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Lequieu, J.; Cordoba, A.; Moller, J.; De Pablo, J.J. 1CPN: A coarse-grained multi-scale model of chromatin. J. Chem. Phys. 2019, 150, 215102. [Google Scholar] [CrossRef]
- Han, G.S.; Li, Q.; Li, Y. Nucleosome positioning based on DNA sequence embedding and deep learning. BMC Genom. 2022, 23, 301. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Lin, X.; Zhang, B. Stability and folding pathways of tetra-nucleosome from six-dimensional free energy surface. Nat. Commun. 2021, 12, 1091. [Google Scholar] [CrossRef]
| Regulatory Factors | Location/Subtype | Modifications 1/Protein Name |
|---|---|---|
| Histone modification | H1.2 (R54) | Me, Cit |
| Histone modification | H2A (Q105) | Me |
| Histone modification | H3 (R42) | Me |
| Histone modification | H3 (K56) | Me, Ac, Formyl, Succ |
| Histone modification | H3 (K64) | Me, Ac |
| Histone modification | H3 (K79) | Me, Ac, Formyl, Succ |
| Histone modification | H3 (T118) | Ph |
| Histone modification | H3 (K122) | Me, Ac, Formyl |
| Histone modification | H4 (K91) | Ac, Ubi, Succ, Bu, Cit, Prop |
| DNA Methylation | CpG | 5mC |
| DNA Hydroxymethylation | CpG | 5hmC |
| Transcription factor | Group I | Foxa3/Oct4/Sox2/Pu1/Ascl1/Klf4/Gata3 |
| Transcription factor | Group IIA | Myog/cMyc/Max/Crem/Cebpα/Usf1 |
| Transcription factor | Group IIB | Tbx1/Brachyury/NFkB p50/Gal4/TALE-PBC Pbx1/Ubx |
| Chromatin remodelers | ISWI | Isw1/Isw2 |
| Chromatin remodelers | CHD | Chd1/NuRD |
| Chromatin remodelers | SWI/SNF | Sth1/Snf2/Swi2/Swi3/Swp73/Snf5/ARPs |
| Chromatin remodelers | INO80 | Ino80/Swr1 |
| Cations | monovalent | Na+/K+ |
| Cations | multivalent | Mg2+/Ca2+/Co3+ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, D.; Huang, Y.; Bai, C. Studies of the Mechanism of Nucleosome Dynamics: A Review on Multifactorial Regulation from Computational and Experimental Cases. Polymers 2023, 15, 1763. https://doi.org/10.3390/polym15071763
Shi D, Huang Y, Bai C. Studies of the Mechanism of Nucleosome Dynamics: A Review on Multifactorial Regulation from Computational and Experimental Cases. Polymers. 2023; 15(7):1763. https://doi.org/10.3390/polym15071763
Chicago/Turabian StyleShi, Danfeng, Yuxin Huang, and Chen Bai. 2023. "Studies of the Mechanism of Nucleosome Dynamics: A Review on Multifactorial Regulation from Computational and Experimental Cases" Polymers 15, no. 7: 1763. https://doi.org/10.3390/polym15071763
APA StyleShi, D., Huang, Y., & Bai, C. (2023). Studies of the Mechanism of Nucleosome Dynamics: A Review on Multifactorial Regulation from Computational and Experimental Cases. Polymers, 15(7), 1763. https://doi.org/10.3390/polym15071763






