Odor Fingerprinting of Chitosan and Source Identification of Commercial Chitosan: HS-GC-IMS, Multivariate Statistical Analysis, and Tracing Path Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
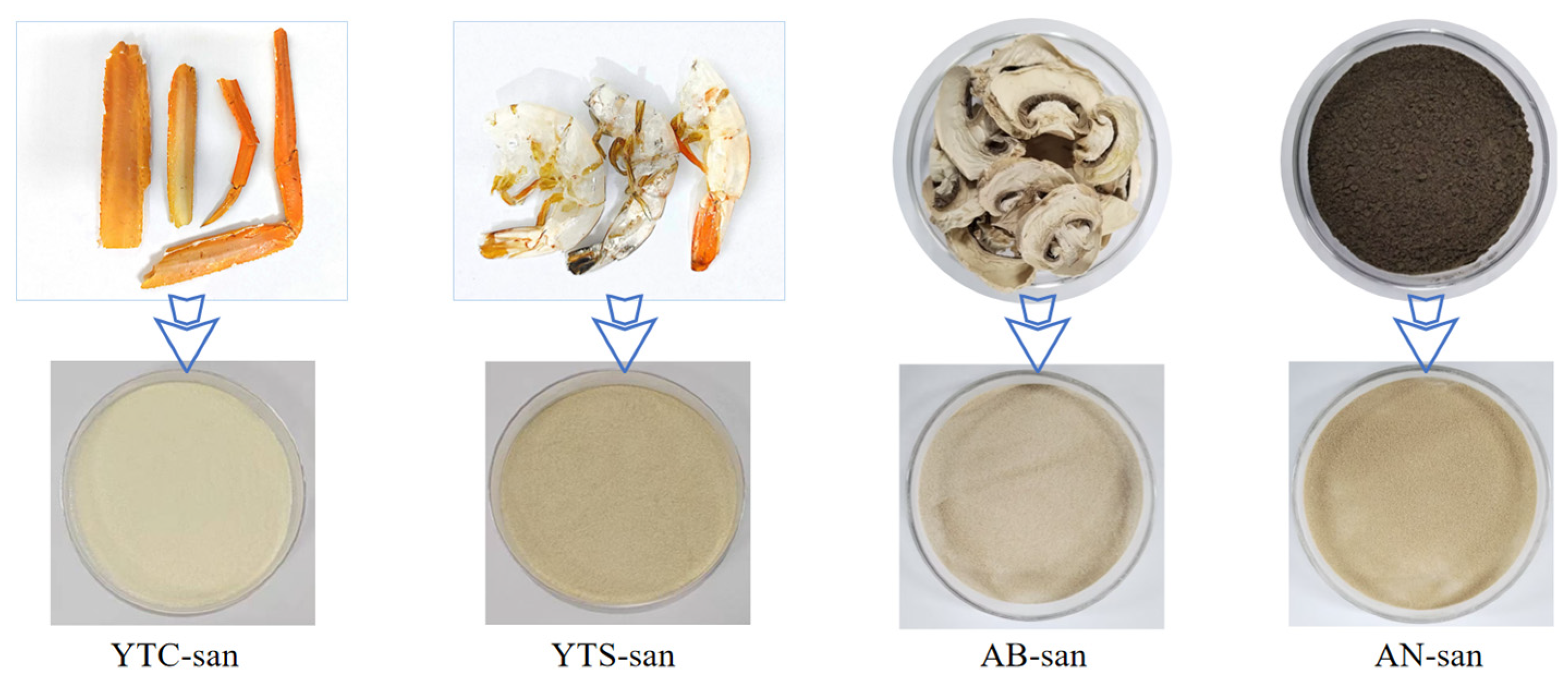
2.2. Chitosan Samples
2.3. HS-GC-IMS Detection and Analysis
2.4. Chemometrics
3. Results and Discussion
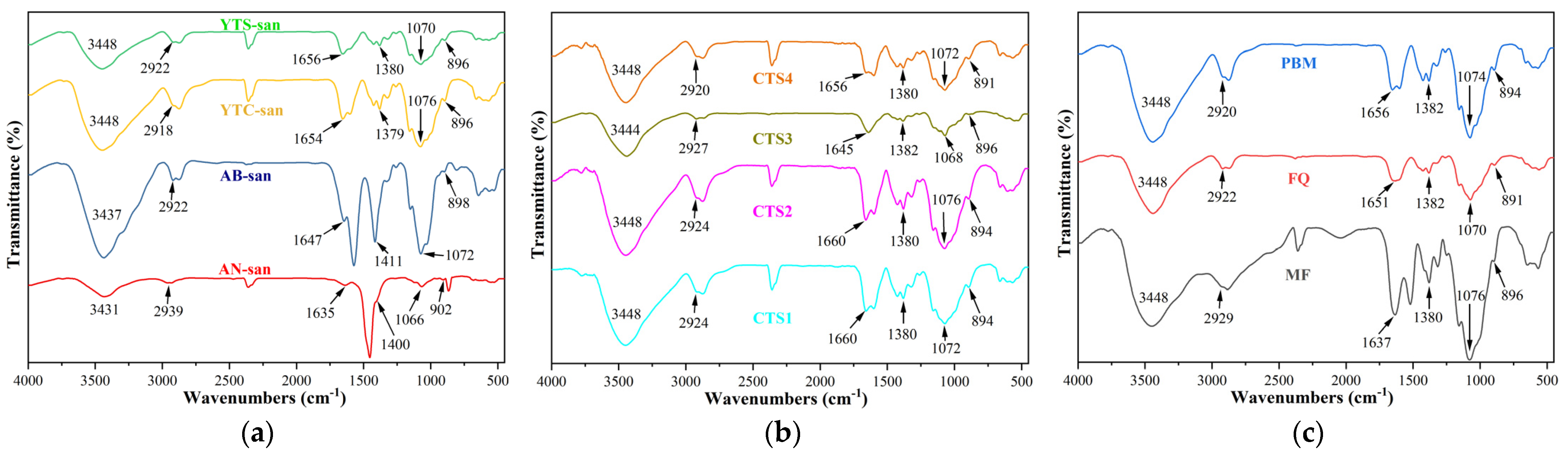
3.1. Structure Analysis
3.2. HS-GC-IMS Topographic Photographs
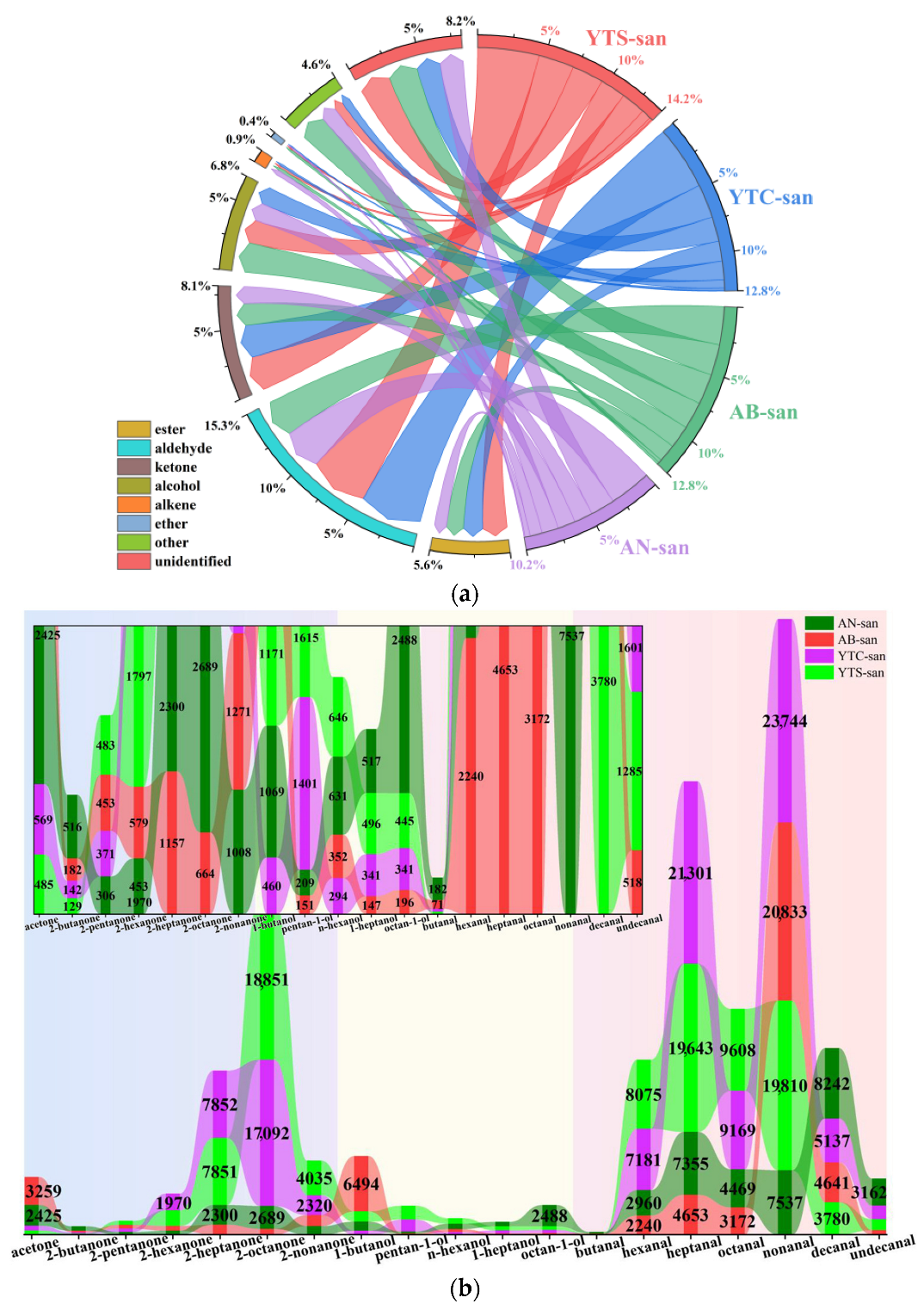
3.3. Analysis of VOCs in Chitosan
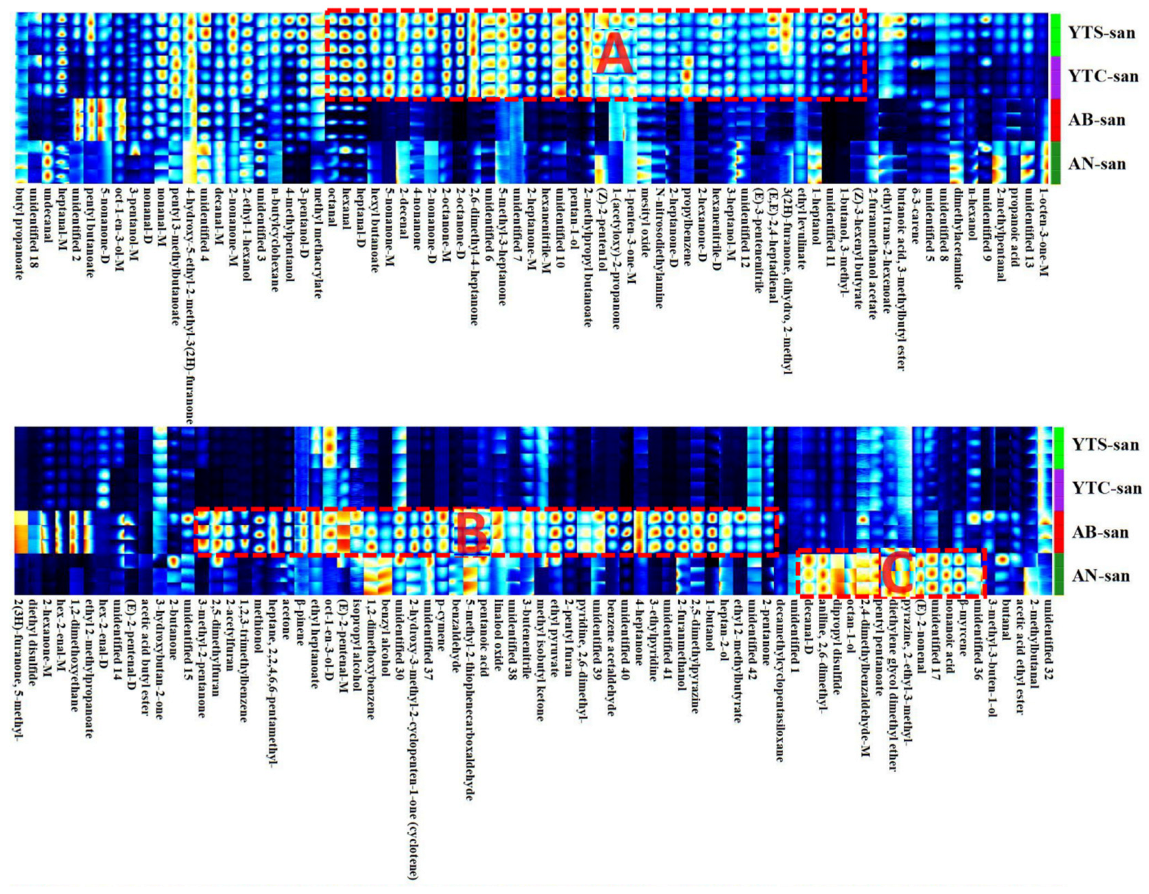
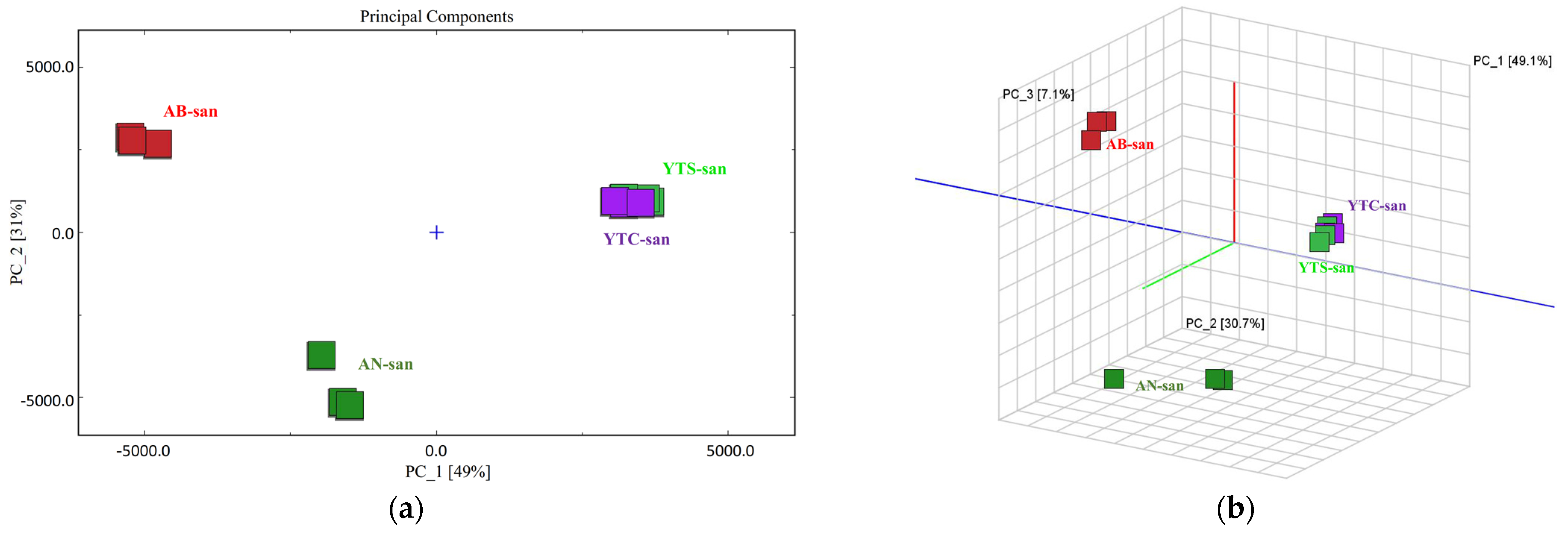
3.4. VOCs Fingerprints and PCA Analysis
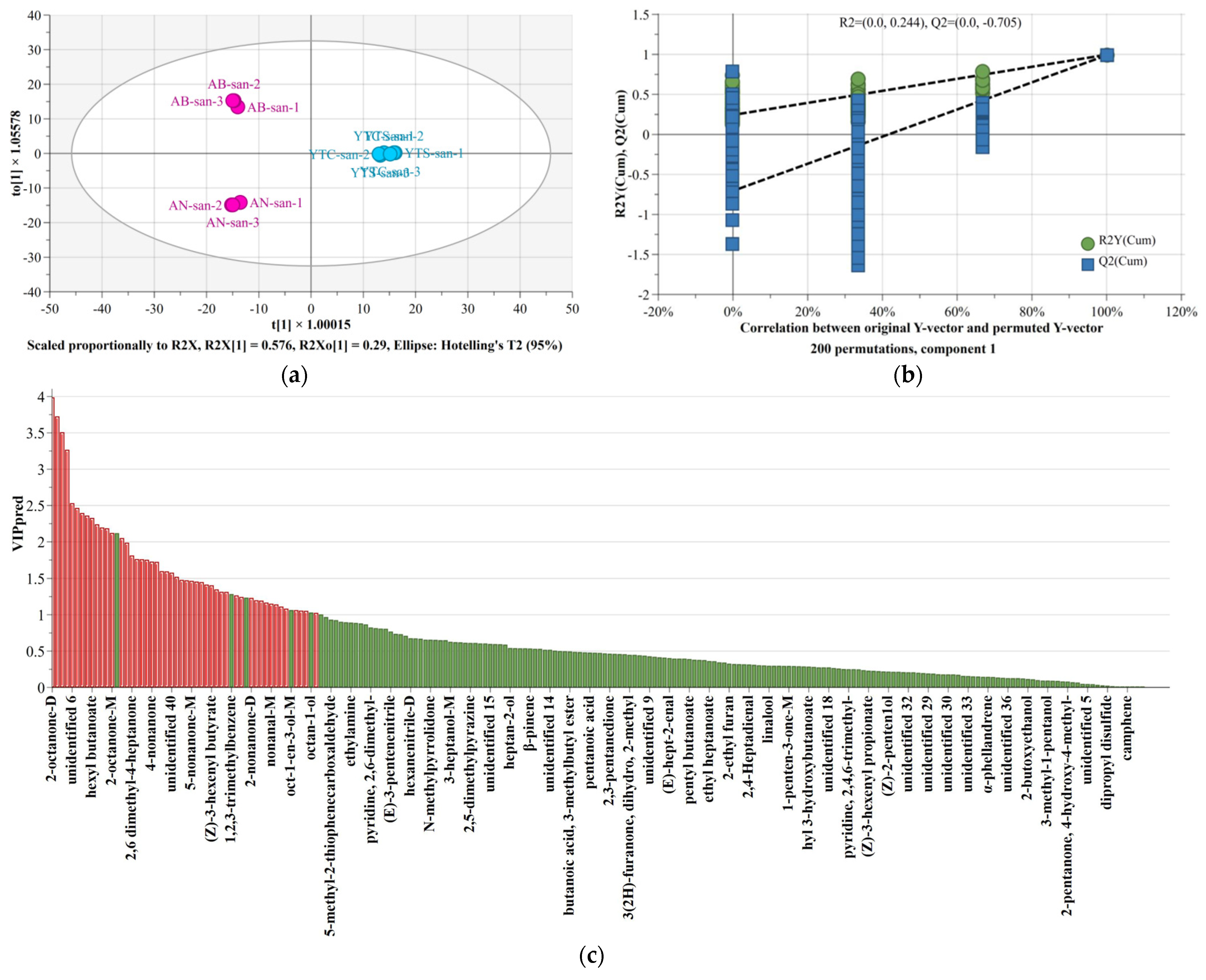
3.5. Establishment and Validation of a Model for Identification of Chitosan
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casadidio, C.; Peregrina, D.V.; Gigliobianco, M.R.; Deng, S.; Censi, R.; Di Martino, P. Chitin and Chitosans: Characteristics, Eco-Friendly Processes, and Applications in Cosmetic Science. Mar. Drugs 2019, 17, 369. [Google Scholar] [CrossRef] [PubMed]
- Rajoka, M.S.R.; Zhao, L.Q.; Mehwish, H.M.; Wu, Y.G.; Mahmood, S. Chitosan and its derivatives: Synthesis, biotechnological applications, and future challenges. Appl. Microbiol. Biotechnol. 2019, 103, 1557–1571. [Google Scholar] [CrossRef] [PubMed]
- Nunes, Y.L.; de Menezes, F.L.; de Sousa, I.G.; Cavalcante, A.L.G.; Cavalcante, F.T.T.; Moreira, K.D.; de Oliveira, A.L.B.; Mota, G.F.; Souza, J.D.E.; Falcão, I.R.D.; et al. Chemical and physical Chitosan modification for designing enzymatic industrial biocatalysts: How to choose the best strategy? Int. J. Biol. Macromol. 2021, 181, 1124–1170. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Rodríguez, A.; Hernández-Viezcas, J.A.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L.; Perales-Pérez, O.; Román-Velázquez, F.R. Synthesis of protonated chitosan flakes for the removal of vanadium(III, IV and V) oxyanions from aqueous solutions. Microchem. J. 2015, 118, 1–11. [Google Scholar] [CrossRef]
- Morin-Crini, N.; Lichtfouse, E.; Torri, G.; Crini, G. Applications of chitosan in food, pharmaceuticals, medicine, cosmetics, agriculture, textiles, pulp and paper, biotechnology, and environmental chemistry. Environ. Chem. Lett. 2019, 17, 1667–1692. [Google Scholar] [CrossRef]
- Brun, P.; Zamuner, A.; Cassari, L.; D’Auria, G.; Falcigno, L.; Franchi, S.; Contini, G.; Marsotto, M.; Battocchio, C.; Iucci, G.; et al. Chitosan Covalently Functionalized with Peptides Mapped on Vitronectin and BMP-2 for Bone Tissue Engineering. Nanomaterials 2021, 11, 2784. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Chen, X. Sustainability: Don’t waste seafood waste. Nature 2015, 524, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.W.; Han, B.Q.; Dong, W.; Yang, Z.; Lv, Y.; Liu, W.S. Effects of carboxymethyl chitosan on the blood system of rats. Biochem. Biophys. Res. Commun. 2011, 408, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Waibel, K.H.; Haney, B.; Moore, M.; Whisman, B.; Gomez, R. Safety of Chitosan Bandages in Shellfish Allergic Patients. Mil Med. 2011, 176, 1153–1156. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, M.J.; Wang, T.; Chen, X.G.; Wang, H. Chitosan-based self-assembled nanomaterials: Their application in drug delivery. VIEW 2021, 2, 20200069. [Google Scholar] [CrossRef]
- Alhulaybi, Z.A. Fabrication and Characterization of Poly(lactic acid)-Based Biopolymer for Surgical Sutures. ChemEngineering 2023, 7, 98. [Google Scholar] [CrossRef]
- Notario-Pérez, F.; Martín-Illana, A.; Cazorla-Luna, R.; Ruiz-Caro, R.; Veiga, M.D. Applications of Ch-itosan in Surgical and Post-Surgical Materials. Mar. Drugs 2022, 20, 396. [Google Scholar] [CrossRef]
- Tan, S.C.; Tan, T.K.; Wong, S.M.; Khor, E. The chitosan yield of zygomycetes at their optimum harvesting time. Carbohydr. Polym. 1996, 30, 239–242. [Google Scholar] [CrossRef]
- Ghormade, V.; Pathan, E.K.; Deshpande, M.V. Can fungi compete with marine sources for chitosan p-roduction? Int. J. Biol. Macromol. 2017, 104, 1415–1421. [Google Scholar] [CrossRef]
- Wang, W.P.; Du, Y.M.; Wang, X.Y. Physical properties of fungal chitosan. World J. Microbiol. Biotechnol. 2008, 24, 2717–2720. [Google Scholar] [CrossRef]
- Nwe, N.; Stevens, W.F. Effect of urea on fungal chitosan production in solid substrate fermentation. Process Biochem. 2004, 39, 1639–1642. [Google Scholar] [CrossRef]
- Circular of the General Administration of China on the Release of the Guiding Principles for the Technical Review of the Registration of Medical Devices of Animal Origin (Revised in 2017). Available online: https://www.nmpa.gov.cn/xxgk/ggtg/ylqxggtg/ylqxqtggtg/20180105112501619.html (accessed on 17 May 2024).
- Q5A(R2) Viral Safety Evaluation of Biotechnology Products Derived from Cell Lines of Human or Animal Origin. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/q5ar2-viral-safety-evaluation-biotechnology-products-derived-cell-lines-human-or-animal-origin (accessed on 17 May 2024).
- Yang, X.L.; Zhang, T.X.; Yang, D.D.; Xie, J.C. Application of gas chromatography-ion mobility spectrometry in the analysis of food volatile components. Acta Chromatogr. 2023, 35, 35–45. [Google Scholar] [CrossRef]
- He, J.; Huang, Q.; Peng, H.X.; Chen, Y.T.; Xu, W.S. Comparison of volatile flavor compounds in plant-based and real pork mince by Headspace-Gas Chromatography-Ion Mobility Spectrometry (HS-GC-IMS). Food Sci. Technol. 2022, 42, e50722. [Google Scholar] [CrossRef]
- Hu, X.; Wang, R.R.; Guo, J.J.; Ge, K.D.; Li, G.Y.; Fu, F.H.; Ding, S.H.; Shan, Y. Changes in the Volatile Components of Candied Kumquats in Different Processing Methodologies with Headspace-Gas Chr-omatography-Ion Mobility Spectrometry. Molecules 2019, 24, 3053. [Google Scholar] [CrossRef]
- Hu, M.Y.; Wang, S.Y.; Liu, Q.; Cao, R.; Xue, Y. Flavor profile of dried shrimp at different processing stages. LWT Food Sci. Technol. 2021, 146, 111403. [Google Scholar] [CrossRef]
- Zhang, Q.; Ding, Y.; Gu, S.; Zhu, S.; Zhou, X.; Ding, Y. Identification of changes in volatile compounds in dry-cured fish during storage using HS-GC-IMS. Food Res. Int. 2020, 137, 109339. [Google Scholar] [CrossRef]
- Weinhold, M.X.; Sauvageau, J.C.M.; Keddig, N.; Matzke, M.; Tartsch, B.; Grunwald, I.; Kübel, C.; Jastorff, B.; Thöming, J. Strategy to improve the characterization of chitosan for sustainable biomedical applications SAR guided multi-dimensional analysis. Green Chem. 2009, 11, 498–509. [Google Scholar] [CrossRef]
- Kumirska, J.; Czerwicka, M.; Kaczyński, Z.; Bychowska, A.; Brzozowski, K.; Thöming, J.; Stepnowski, P. Application of Spectroscopic Methods for Structural Analysis of Chitin and Chitosan. Mar. Drugs 2010, 8, 1567–1636. [Google Scholar] [CrossRef]
- Oliveira, M.A.; Ciarlini, P.C.; Feitosa, J.P.A.; de Paula, R.C.M.; Paula, H.C.B. Chitosan/“angico” gum nanoparticles: Synthesis and characterization. Mater. Sci. Eng. C 2009, 29, 448–451. [Google Scholar] [CrossRef]
- Aldana, A.A.; Martinelli, M.; Strumia, M. Synthesis and Properties of Dendronized Chitosan. Macromol. Symp. 2010, 298, 99–107. [Google Scholar] [CrossRef]
- Gao, Y.Y.; Chen, X.R.; Liao, B.; Ding, X.B.; Zheng, Z.H.; Cheng, X.; Pang, H.; Peng, Y.X. Polyelectrolyte Self-assembly Approach to Smart Nanocontainers. Polym. Bull. 2006, 56, 305–311. [Google Scholar] [CrossRef]
- Santos, V.P.; Marques, N.S.S.; Maia, P.C.S.V.; de Lima, M.A.B.; Franco, L.D.; de Campos-Takaki, G.M. Seafood Waste as Attractive Source of Chitin and Chitosan Production and Their Applications. Int. J. Mol. Sci. 2020, 21, 4290. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zhang, S.S.; Ren, G.D. The Progress of Chitin Extracted from Crustaceans, Insect and Fungi. Food Res. Dev. 2015, 36, 122–126. [Google Scholar] [CrossRef]
- Carrasco-Sandoval, J.; Aranda, M.; Henríquez-Aedo, K.; Fernández, M.; López-Rubio, A.; Fabra, M.J. Impact of molecular weight and deacetylation degree of chitosan on the bioaccessibility of quercetin encapsulated in alginate/chitosan-coated zein nanoparticles. Int. J. Biol. Macromol. 2023, 242, 124876. [Google Scholar] [CrossRef]
- Armaselu, A. New Spectral Applications of the Fourier Transforms in Medicine, Biological and Biomedical Fields. In Fourier Transforms—High-Tech Application and Current Trends; BoD–Books on Demand: Norderstedt, Germany, 2017; pp. 235–252. [Google Scholar] [CrossRef]
- Zam, Z.Z.; Muin, F.; Fataruba, A. Identification of chitosan beads from coconut crab patani variety using Fourier Transform Infrared Spectroscopy (FTIR). J. Phys. Conf. Ser. 2021, 1832, 012014. [Google Scholar] [CrossRef]
- Taaca, K.L.M.; Nakajima, H.; Thumanu, K.; Prieto, E.I.; Vasquez, M.R., Jr. Network formation and differentiation of chitosan-acrylic acid hydrogels using X-ray absorption spectroscopy and multivariate analysis of Fourier transform infrared spectra. J. Electron. Spectrosc. Relat. Phenom. 2023, 267, 147372. [Google Scholar] [CrossRef]
- Espinosa-Andrews, H.; Sandoval-Castilla, O.; Vázquez-Torres, H.; Vernon-Carter, E.J.; Lobato-Calleros, C. Determination of the gum Arabic-chitosan interactions by Fourier Transform Infrared Spectroscopy and characterization of the microstructure and rheological features of their coacervates. Carbohydr. Polym. 2010, 79, 541–546. [Google Scholar] [CrossRef]
- Soncu, E.D.; Özdemir, N.; Arslan, B.; Küçükkaya, S.; Soyer, A. Contribution of surface application of chitosan–thyme and chitosan–rosemary essential oils to the volatile composition, microbial profile, and physicochemical and sensory quality of dry-fermented sausages during storage. Meat Sci. 2020, 166, 108127. [Google Scholar] [CrossRef]
- Xie, C.C.; Wang, F.H.; He, Z.C.; Tang, H.J.; Li, H.Y.; Hou, J.J.; Liu, Y.Z.; Jiang, L.W. Development and characterization of active packaging based on chitosan/chitin nanofibers incorporated with scallion flower extract and its preservation in fresh-cut bananas. Int. J. Biol. Macromol. 2023, 242, 125045. [Google Scholar] [CrossRef]
- Wu, W.D.; Zhan, J.L.; Tang, X.Y.; Li, T.; Duan, S.N. Characterization and identification of pork flavor compounds and their precursors in Chinese indigenous pig breeds by volatile profiling and multivari-ate analysis. Food Chem. 2022, 385, 132543. [Google Scholar] [CrossRef]
- Lu, W.H.; Chen, J.; Li, X.B.; Qi, Y.H.; Jiang, R. Flavor components detection and discrimination of isomers in Huaguo tea using headspace-gas chromatography-ion mobility spectrometry and multivariate statistical analysis. Anal. Chim. Acta 2023, 1243, 340842. [Google Scholar] [CrossRef]
- Natasa, P.K.; Antonio, F.; Natalia, M.; George, Z.; Peter, Q.T.; Luigi, M.; Victoria, F.S.; Erwin, R. A volatilomics analytical protocol employing solid phase microextraction coupled to GC × GC-MS analysis and combined with multivariate chemometrics for the detection of pomegranate juice adulteration. Talanta 2024, 266, 125027. [Google Scholar] [CrossRef]
- Lu, W.H.; Jiang, R.; Li, X.B.; Qi, Y.H.; Ji, Z.Y.; Lv, X.H.; Wang, Y.C.; Feng, C.; Wang, B.Y. Characterization and discrimination of volatile compounds in gel-pen ink via headspace-gas chromatography-ion mobility spectrometry combined with chemometric strategies. Microchem. J. 2022, 182, 107855. [Google Scholar] [CrossRef]
- Li, X.B.; Guo, C.H.; Qi, Y.H.; Lu, W.H.; Xu, G.T.; Wang, B.Y.; Zhang, D.B.; Zhao, S.P.; Ding, M.X. Identification of volatile organic compounds in muscle tissues of different species based on Headspace-Gas-Chromatography Ion-Mobility spectrometry. Leg. Med. 2022, 59, 102132. [Google Scholar] [CrossRef]
- Mario, L.V.; Caterina, D.; Marina, C. Chapter 3—Exploratory Data Analysis. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 28, pp. 55–126. [Google Scholar] [CrossRef]
- Yun, J.; Cui, C.J.; Zhang, S.H.; Zhu, J.J.; Peng, C.Y.; Cai, H.M.; Yang, X.G.; Hou, R.Y. Use of headspace GC/MS combined with chemometric analysis to identify the geographic origins of black tea. Food Chem. 2021, 360, 130033. [Google Scholar] [CrossRef]
- Zhao, S.; Niu, C.; Yang, X.; Xu, X.; Zheng, F.; Liu, C.; Wang, J.; Li, Q. Roles of sunlight exposure on chemosensory characteristic of broad bean paste by untargeted profiling of volatile flavors and multivariate statistical analysis. Food Chem. 2022, 381, 132115. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.-S.; Lu, G.; Song, F.-L.; Meng, M.-Y.; Song, Y.-H.; Ma, H.-N.; Xie, X.-R.; Zhu, Y.-J.; He, S.; Li, X.-B. Odor Fingerprinting of Chitosan and Source Identification of Commercial Chitosan: HS-GC-IMS, Multivariate Statistical Analysis, and Tracing Path Study. Polymers 2024, 16, 1858. https://doi.org/10.3390/polym16131858
Guo J-S, Lu G, Song F-L, Meng M-Y, Song Y-H, Ma H-N, Xie X-R, Zhu Y-J, He S, Li X-B. Odor Fingerprinting of Chitosan and Source Identification of Commercial Chitosan: HS-GC-IMS, Multivariate Statistical Analysis, and Tracing Path Study. Polymers. 2024; 16(13):1858. https://doi.org/10.3390/polym16131858
Chicago/Turabian StyleGuo, Jin-Shuang, Gang Lu, Fu-Lai Song, Ming-Yu Meng, Yu-Hao Song, Hao-Nan Ma, Xin-Rui Xie, Yi-Jia Zhu, Song He, and Xue-Bo Li. 2024. "Odor Fingerprinting of Chitosan and Source Identification of Commercial Chitosan: HS-GC-IMS, Multivariate Statistical Analysis, and Tracing Path Study" Polymers 16, no. 13: 1858. https://doi.org/10.3390/polym16131858
APA StyleGuo, J.-S., Lu, G., Song, F.-L., Meng, M.-Y., Song, Y.-H., Ma, H.-N., Xie, X.-R., Zhu, Y.-J., He, S., & Li, X.-B. (2024). Odor Fingerprinting of Chitosan and Source Identification of Commercial Chitosan: HS-GC-IMS, Multivariate Statistical Analysis, and Tracing Path Study. Polymers, 16(13), 1858. https://doi.org/10.3390/polym16131858




