Producing and Characterizing Polyhydroxyalkanoates from Starch and Chickpea Waste Using Mixed Microbial Cultures in Solid-State Fermentation
Abstract
:1. Introduction
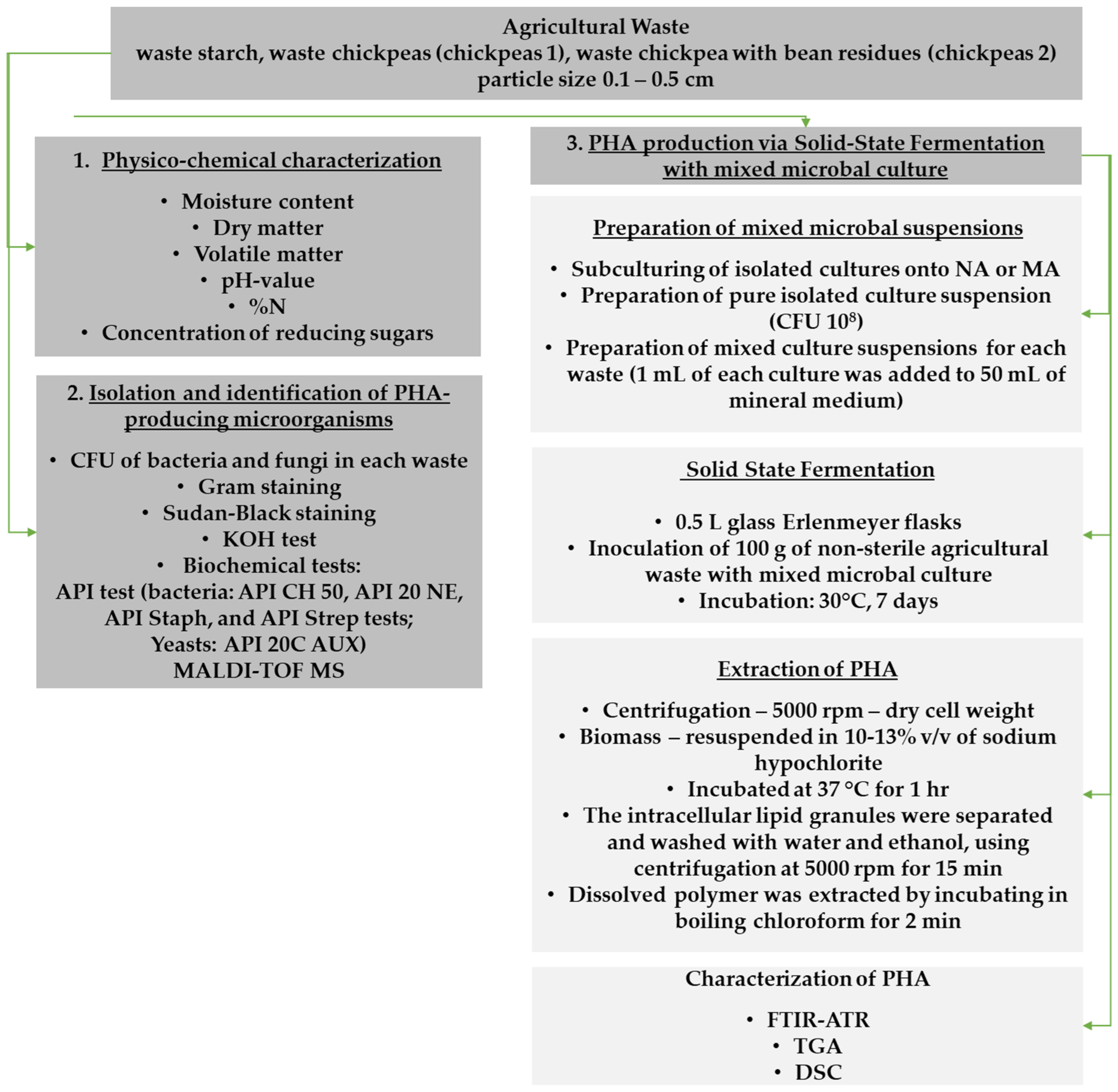
2. Materials and Methods
2.1. Physico-Chemical Characterization of Agricultural Waste
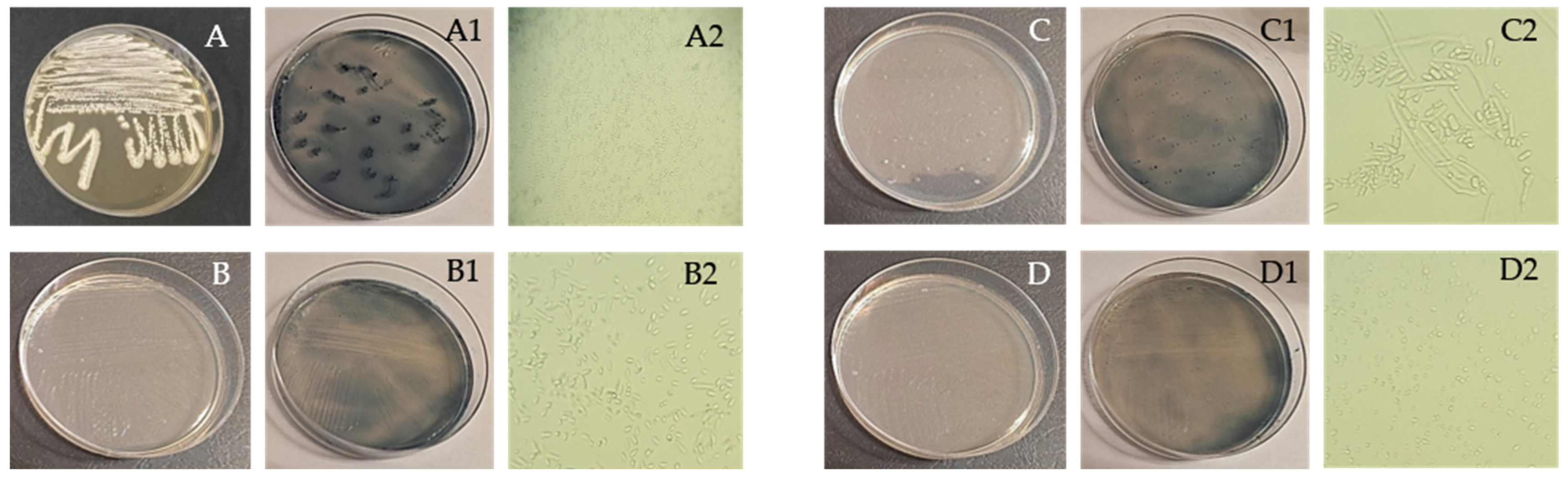
2.2. Isolation and Identification of PHA-Producing Microorganisms
2.3. PHA Production from Agricultural Waste via Solid-State Fermentation with Mixed Microbial Cultures
3. Results and Discussion
3.1. Isolation and Identification of PHA-Producing Microorganisms
3.2. PHA Production from Agricultural Waste via Solid-State Fermentation with Mixed Microbial Cultures
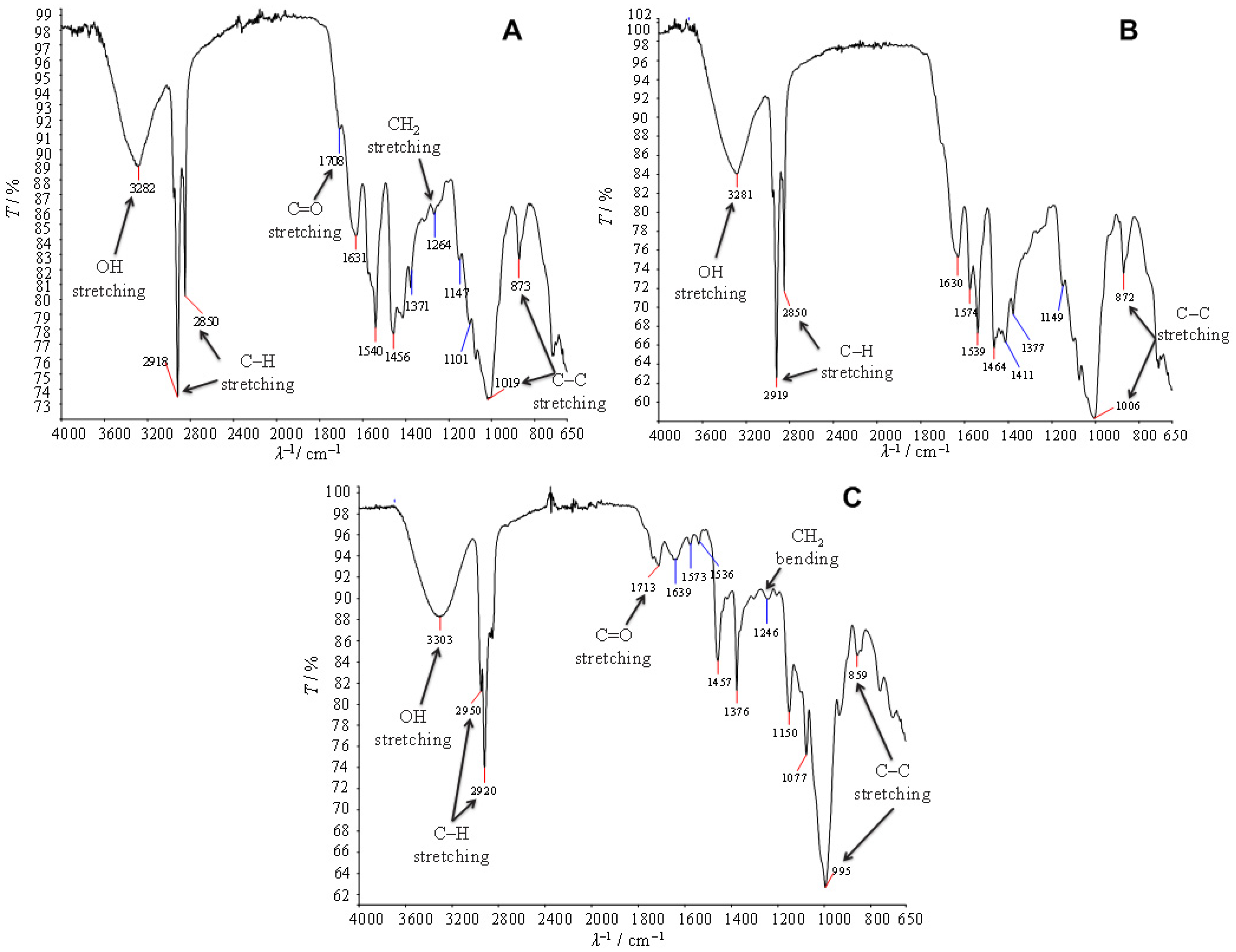
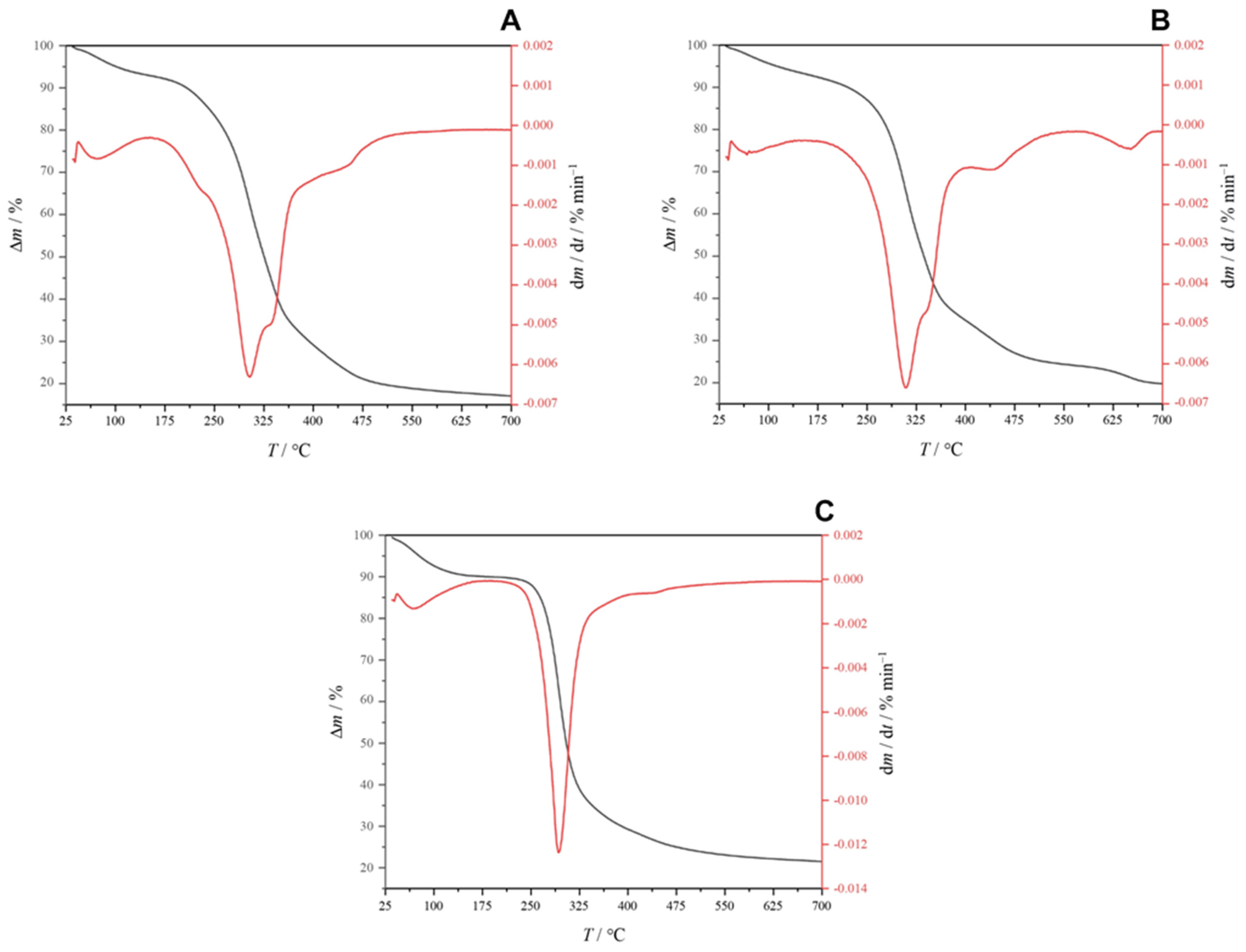
3.3. Characterization of Extracted PHA
3.3.1. FTIR-ATR Spectroscopy Analysis of the Extract
3.3.2. DSC Analysis of the Extract
3.3.3. TGA of the Extract
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Reddy, C.S.; Ghai Rashmi, R.; Kalia, V.C. Polyhydroxyalkanoates: An overview. Bioresour. Technol. 2003, 87, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Sabapathy, P.C.; Devaraj, S.; Meixner, K.; Anburajan, P.; Kathirvel, P.; Ravikumar, Y.; Zabed, H.M.; Qi, X. Recent development sin Polyhydroxyalkanoates (PHAs) production—A review. Bioresour. Technol. 2020, 306, 123132. [Google Scholar] [CrossRef] [PubMed]
- Saharan, R.; Kharb, J. Exploration of Bioplastics: (A Review). Orient. J. Chem. 2022, 38, 840–854. [Google Scholar] [CrossRef]
- European Bioplastics. Available online: https://www.european-bioplastics.org/market/ (accessed on 14 October 2024).
- Panda, P.K.; Dash, P. Preparation, Characterization, and Evaluation of Antibacterial Properties of Poly(3-Hydroxybutyrate-Co-3-Hydroxyvalerate) (PHBV)-Based Films and Coatings. In Biopolymer-Based Films and Coatings: Trends and Challenges, 1st ed.; Bangar, S.P., Siroha, A.K., Eds.; CRC Press: Boca Raton, FL, USA, 2023; Volume 1, pp. 1–18. [Google Scholar]
- Raza, Z.A.; Abid, S.; Banat, I.M. Polyhydroxyalkanoates: Characteristics, production, recent developments and applications. Int. Biodeterior. Biodegrad. 2018, 126, 45–56. [Google Scholar] [CrossRef]
- Shrivastav, A.; Kim, H.-Y.; Kim, Y.-R. Advances in the Applications of Polyhydroxyalkanoate Nanopaticles for Novel Drug Delivery System. BioMed Res. Int. 2013, 2013, 581684. [Google Scholar] [CrossRef]
- Madkour, M.H.; Heinrich, D.; Alghamdi, M.A.; Shabbaj, I.I.; Steinbüchel, A. PHA Recovery from Biomass. Biomacromolecules 2013, 14, 2963–2972. [Google Scholar] [CrossRef]
- Jiang, N.; Wang, M.; Song, L.; Yu, D.; Zhou, S.; Li, Y.; Li, H.; Han, X. Polyhydroxybutyrate production by recombinant Escherichia coli based on genes related to synthesis pathway of PHB from Massilia sp. UMI-21. Microb. Cell Fact. 2023, 22, 129. [Google Scholar] [CrossRef]
- Zhou, W.; Bergsma, S.; Colpa, D.I.; Euverink, G.-J.W.; Krooneman, J. Polyhydroxyalkanoates (PHAs) synthesis and degradation by microbes and applications towards a circular economy. J. Environ. Manag. 2023, 341, 118033. [Google Scholar] [CrossRef]
- Faccin, D.J.L.; Correa, M.P.; Rech, R.; Ayub, M.A.Z.; Secchi, A.R.; Cardozo, N.S.M. Modeling P(3HB) production by Bacillus megaterium. J. Chem. Technol. Biotechnol. 2011, 87, 325–333. [Google Scholar] [CrossRef]
- Haas, C.; El-Najjar, T.; Virgolini, N.; Smerilli, M.; Neureiter, M. High cell-density production of poly(3-hydroxybutyrate) in a membrane bioreactor. New Biotechnol. 2017, 37, 117–122. [Google Scholar] [CrossRef]
- Berezina, N.; Yada, B. Improvement of the poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) production by dual feeding with levulinic acid and sodium propionate in Cupriavidus necator. New Biotechnol. 2016, 31, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Centeno-Leija, S.; Huerta-Beristain, G.; Giles-Gomez, M.; Bolivar, F.; Gosset, G.; Martinez, A. Improving poly-3-hydroxybutyrate production in Escherichia coli by combining the increase in the NADPH pool and acetyl-CoA availability. Antoine Van Leeuwenhoek 2014, 105, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Akaraonye, E.; Moreno, C.; Knowles, J.C.; Keshavarz, T.; Roy, I. Poly(3-hydroxybutyrate) production by Bacillus cereus SPV using sugarcane molasses as the main carbon source. Biotechnol. J. 2011, 7, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Steinbüchel, A.; Debzi, E.-M.; Marchessault, R.H.; Timm, A. Synthesis and production of poly(3-hydroxyvaleric acid)homopolyester by Chromobacterium violaceum. Appl. Microbiol. Biotechnol. 1993, 39, 443–449. [Google Scholar] [CrossRef]
- Steinbüchel, A.; Schmack, G. Large-scale production of poly(3-hydroxyvaleric acid) by fermentation of Chromobacterium violaceum, processing, and characterization of the homopolyester. J. Environ. Polym. Deg. 1995, 3, 243–258. [Google Scholar] [CrossRef]
- Chen, G.-Q.; König, K.-H.; Lafferty, R.M. Production of poly-D(-)-3-hydroxybutyrate and poly-D(-)-3-hydroxyvalerate by strains of Alcaligenes latus. Antonie Van Leeuwenhoek 1991, 60, 61–66. [Google Scholar] [CrossRef]
- Yamane, T.; Chen, X.-F.; Ueda, S. Growth-Associated Production of Poly(3-Hydroxyvalerate) from n-Pentanol by a Methylotrophic Bacterium, Paracoccus denitrificans. Appl. Environ. Microbiol. 1995, 62, 380–384. [Google Scholar] [CrossRef]
- Urtuvia, V.; Ponce, B.; Andler, R.; Peña, C.; Diaz-Barrera, A. Extended batch cultures for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) production by Azotobacter vinelandii OP growing at different aeration rates. Biotech 2022, 12, 304. [Google Scholar] [CrossRef]
- Muigano, M.N.; Anami, S.E.; Onguso, J.M.; Mauti, G.O. Optimized Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) Production by Moderately Haloalkaliphilic Bacterium Halomonas alkalicola Ext. Int. J. Polym. Sci. 2024, 2024, 6667843. [Google Scholar] [CrossRef]
- Akdoğan, M.; Çelik, E. Enhanced production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) biopolymer by recombinant Bacillus megaterium in fed-batch bioreactors. Bioprocess Biosyst. Eng. 2021, 44, 403–416. [Google Scholar] [CrossRef]
- Jian, J.; Li, Z.-J.; Ye, H.-M.; Yuan, M.-Q.; Chen, G.-Q. Metabolic engineering for microbial production of polyhydroxyalkanoates consisting of high 3-hydroxyhexanoate content by recombinant Aeromonas hydrophila. Bioresour. Technol. 2010, 101, 6096–6102. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-H.; Zhou, X.-R.; Liu, Q.; Chen, G.-Q. Biosynthesis of polyhydroxyalkanoate homopolymers by Pseudomonas putida. Appl. Microbiol. Biotechnol. 2011, 89, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, G.; Zhou, X.R.; Chen, G.-Q. Biosynthesis of poly(3-hydroxydecanoate) and 3-hydroxydodecanoate dominating polyhydroxyalkanoates by β-oxidation pathway inhibited Pseudomonas putida. Met. Eng. 2011, 13, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zhang, G.; Wu, Q.; Chen, G.-Q.; Zhang, R. Production of polyhydroxyalkanoates by Pseudomonas nitroreducens. Antonie Van Leeuwenhoek 1991, 75, 345–349. [Google Scholar] [CrossRef]
- Gong, Y.-A.; Wu, H.-S.; Wei, Y.-H.; Sun, Y.-M. Fermentation strategy for the production of poly(3-hydroxyhexanoate) by Aeromonas sp. KC014. Korean J. Chem. Eng. 2008, 25, 1422–1426. [Google Scholar] [CrossRef]
- Paduvari, R.; Bhat, K.S.; Somashekara, D.M. Biosynthesis of 3-hydroxydecanoate (3HD) and 3-hydroxydodecanoate (3HDd) in the polyhydroxyalkanoate copolymeric ester chain by Pseudomonas aeruginosa MCC 5300 using oleic acid as a carbon source. Bioresour. Technol. Rep. 2024, 25, 101690. [Google Scholar] [CrossRef]
- Chung, A.-L.; Jin, H.-L.; Huang, L.-J.; Ye, H.-M.; Chen, J.-C.; Wu, Q.; Chen, G.-Q. Biosynthesis and Characterization of Poly(3-hydroxydodecanoate) by β-Oxidation Inhibited Mutant of Pseudomonas entomophila L48. Biomacromolecules 2011, 12, 3559–3566. [Google Scholar] [CrossRef]
- Barbuzzi, T.; Giuffrida, M.; Impallomeni, G.; Carnazza, S.; Ferreri, A.; Guglielmino, S.P.P.; Ballistreri, A. Microbial Synthesis of Poly(3-hydroxyalkanoates) by Pseudomonas aeruginosa from Fatty Acids: Identification of Higher Monomer Units and Structural Characterization. Biomacromolecules 2004, 5, 2469–2478. [Google Scholar] [CrossRef]
- Impallomeni, G.; Ballistreri, A.; Carnemolla, G.M.; Rizzo, M.G.; Nicolo, M.S.; Guglielmino, S.P.P. Biosynthesis and structural characterization of polyhydroxyalkanoates produced by Pseudomonas aeruginosa ATCC 27853 from long odd-chain fatty acids. Int. J. Biol. Macromol. 2018, 108, 608–614. [Google Scholar] [CrossRef]
- Ray, S.; Kalia, V.C. Co-metabolism of substrates by Bacillus thuringiensis regulates polyhydroxyalkanoate co-polymer composition. Bioresour. Technol. 2017, 224, 743–747. [Google Scholar] [CrossRef]
- Zytner, P.; Kumar, D.; Elsayed, A.; Mohanty, A.; Ramarao, B.V.; Misra, M. A review on polyhydroxyalkanoate (PHA) production through the use of lignocellulosic biomass. RSC Sustain. 2013, 1, 2120–2134. [Google Scholar] [CrossRef]
- Brizga, J.; Hubacek, K.; Feng, K. The Unintended Side Effects of Bioplastics: Carbon, Land, and Water Footprints. One Earth 2020, 3, 45–53. [Google Scholar] [CrossRef]
- Bardhan, S.K.; Gupta, S.; Gorman, M.E.; Haider, M.A. Biorenewable chemicals; Feedstocks, technologies and the conflict with food production. Renew. Sustain. Energy Rev. 2015, 51, 506–520. [Google Scholar] [CrossRef]
- Rosenboom, J.-G.; Langer, R.; Traverso, G. Bioplastics for a circular economy. Nat. Rev. Mat. 2022, 7, 117–137. [Google Scholar] [CrossRef]
- Prado-Acebo, I.; Cubero-Cardoso, J.; Lu-Chao, T.A.; Eibes, G. Integral multi-valorization of agro-industrial wastes: A review. Waste Manag. 2024, 183, 42–52. [Google Scholar] [CrossRef]
- Chauhan, A.; Islam, F.; Imran, A.; Ikram, A.; Zahoor, T.; Khurshid, S.; Shah, M.A. A review on waste valorization, biotechnological utilization and management of potato. Food Sci. Nutr. 2023, 11, 5573–5785. [Google Scholar] [CrossRef]
- Sanabria, D.P.; Farfan, H.I.; Quintana, D.N.; Abril, I.V.; Castro, H.F.; Paredes, R.A.; Roa, K.L. Synthesis of starch powder from differents organic waster: A green approach to a valuable material. IOP Conf. Ser. Mat. Sci. Eng. 2021, 1154, 012041. [Google Scholar] [CrossRef]
- Dorantes-Fuertes, M.-G.; López-Méndez, M.C.; Martínez-Castellanos, G.; Meléndez-Armenta, R.A.; Kimenéz-Martínez, H.-E. Starch Extraction Methods in Tubers and Roots: A Systematic Review. Agronomy 2024, 14, 865. [Google Scholar] [CrossRef]
- Fraser-Reid, B.O.; Tatsuta, K.; Thiem, J. Glycoscience, 2nd ed.; Springer: Berlin, Germany, 2008; p. 1437. [Google Scholar]
- Hirdyani, H. Nutritional composition of Chickpea (Cicerarietinum-L) and value added products. Indian J. Community Health 2014, 26, 102–106. [Google Scholar]
- Aouine, M.; Elalami, D.; Haggoud, A.; Koraichi, S.I.; Roumeas, L.; Barakat, A. Dry chemo-mechanical pretreatment of chickpea straw: Effect and optimization of experimental parameters to improve hydrolysis yields. Bioresour. Technol. Rep. 2022, 18, 101011. [Google Scholar] [CrossRef]
- Jose, S.; Pandit, P.; Pandey, R. Chickpea husk—A potential agro waste for coloration and functional finishing of textiles. Ind. Crops Prod. 2019, 142, 111833. [Google Scholar] [CrossRef]
- Wu, Y.; Yao, S.; Narale, B.A.; Shanmugam, A.; Mettu, S.; Ashokkumar, M. Ultrasonic Processing of Food Waste to Generate Value-Added Products. Foods 2022, 11, 2035. [Google Scholar] [CrossRef] [PubMed]
- Khattab, M.M.; Dahman, Y. Production and recovery of poly-3-hydroxybutyrate bioplastics using agro-industrial residues of hemp hurd biomass. Bioprocess Biosyst. Eng. 2019, 42, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Zhang, D.; Wang, H. Polyhydroxyalkanoates (PHA) production from fermented thermal-hydrolyzed sludge by mixed microbial cultures: The link between phosphorus and PHA yields. Waste Manag. 2019, 96, 149–157. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, H.; Chang, J.; Sun, J.; Tu, W.; Wang, H. Effect of thermal hydrolysis pretreatment on volatile fatty acids production in sludge acidificaion and subsequent polyhydroxyalkanoates production. Biore. Technol. 2019, 279, 92–100. [Google Scholar] [CrossRef]
- Paul, A.; Jia, L.; Majumder, E.L.-W.; Yoo, C.G.; Rajendran, K.; Villarreal, E.; Kumar, D. Poly(3hydroxybutyrate) production from industrial hemp waste pretreated with chemicalfree hydrothermal process. Bioresour. Technol. 2023, 381, 129161. [Google Scholar] [CrossRef]
- Tripathi, A.D.; Mishra, P.K.; Darani, K.K.; Agarwal, A.; Paul, V. Hydrothermal treatment of lignocellulose waste for the production of polyhydroxyalkanoates copolymer with potential application in food packaging. Trends Food Sci. Technol. 2022, 123, 223–250. [Google Scholar] [CrossRef]
- Naitam, M.G.; Tomar, G.S.; Kaushik, R.; Singh, S.; Nain, L. Agro-Industrial Waste as Potential Renewable Feedstock for Biopolymer Poly-Hydroxyalkanoates (PHA) Production. Enz. Eng. 2022, 11, 190. [Google Scholar]
- Titz, M.; Kettl, K.-H.; Shahzad, K.; Koller, M.; Schnitzer, H.; Narodoslawsky, M. Process optimization for efficient biomediated PHA production from animal-based waste streams. Clean Technol. Environ. Policy 2012, 14, 495–503. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, L.; Liao, Q.; Ran, Q.; Ran, Y.; Hu, F.; Gao, M.; She, Z.; Zhao, Y.; Jin, C.; et al. Polyhydroxyalkanoate (PHA) production with acid or alkali pretreated sludge acidogenic liquid as carbon source: Substrate metabolism and monomer composition. Process Saf. Environ. Prot. 2020, 142, 156–164. [Google Scholar] [CrossRef]
- Saratale, G.D.; Oh, M.-K. Characterization of poly-3-hydroxybutyrate (PHB) produced from Ralstonia eutropha using alkali-pretreated biomass feedstock. Int. J. Biol. Macromol. 2015, 80, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Sachan, R.S.K.; Devgon, I.; Karnwal, A.; Mahmoud, A.E.D. Valorization of sugar extracted from wheat straw for eco-friendly polyhydroxyalkanoate (PHA) production by Bacillus megaterium MTCC 453. Bioresour. Technol. Rep. 2024, 25, 101770. [Google Scholar] [CrossRef]
- Radecka, I.; Irorere, V.; Jiang, G.; Hill, D.; Williams, C.; Adamus, G.; Kwiecién, M.; Marek, A.A.; Zawadiak, J.; Johnston, B.; et al. Oxidized Polyethylene Wax as a Potential Carbon Source for PHA Production. Materials 2016, 9, 367. [Google Scholar] [CrossRef] [PubMed]
- Barrameda, H.J.C.; Requiso, P.J.; Alfafara, C.G.; Nayve, F.R.P., Jr.; Ventura, R.L.G.; Ventura, J.-R.S. Hydrolysate production from sugarcane bagasse using steam explosion and sequential steam explosion-dilute acid pretreatment for polyhydroxyalkanoate fermentation. Bioact. Carbohydr. Diet. Fibre 2023, 30, 100376. [Google Scholar] [CrossRef]
- Mabazza, K.A.A.; Requiso, P.J.; Alfafara, C.G.; Nayve, F.R.P., Jr.; Ventura, J.-R.S. Steam Explosion and Sequential Steam Explosion—Dilute Acid Pretreatment Opotimization of Banana Pseudostem for Polyhydroxybutyrate (PHB) Production. Philipp. J. Sci. 2020, 149, 285–297. [Google Scholar] [CrossRef]
- Perez, N.A.D.; Requiso, P.J.; Alfafara, C.G.; Capunitan, J.A.; Nayve, F.R.P., Jr.; Ventura, J.-R.S. Pretreatment Optimization of Corn Stover with Subsequent Enzymatic Hydrolysis for Polyhydroxybutyrate (PHB) Production. Philipp. J. Sci. 2023, 152, 381–395. [Google Scholar] [CrossRef]
- Saratale, G.D.; Saratale, R.G.; Varjani, S.; Cho, S.-K.; Ghodake, G.S.; Kadam, A.; Mulla, S.I.; Bharagava, R.N.; Kim, D.-S.; Shin, H.S. Development of ultrasound aided chemical pretreatment methods to enrich saccharification of wheat waste biomass for polyhydroxybutyrate production and its characterization. Ind. Crops Prod. 2020, 150, 112425. [Google Scholar] [CrossRef]
- Zou, Y.; Yang, M.; Tao, Q.; Zhu, K.; Liu, X.; Wan, C.; Harder, M.K.; Yan, Q.; Liang, B.; Ntaikou, I.; et al. Recovery of polyhydroxyalkanoates (PHAs) polymers from mixed microbial culture through combined ultrasonic disruption and alkaline digestion. J. Environ. Manag. 2023, 326, 116786. [Google Scholar] [CrossRef]
- Silambarasan, S.; Logeswari, P.; Sivaramakrishnan, R.; Pugazhendhi, A.; Kamaraj, B.; Ruiz, A.; Ramadoss, G.; Cornejo, P. Polyhydroxybutyrate production from ultrasound-aided alkaline pretreated finger millet straw using Bacillus megaterium strain CAM12. Bioresour. Technol. 2021, 325, 124632. [Google Scholar] [CrossRef]
- Huang, T.-Y.; Duan, K.-J.; Huang, S.-Y.; Chen, C.W. Production of polyhydroxyalkanoates from inexpensive extruded rice bran and starch by Haloferax mediteranei. J. Ind. Microbiol. Biotechnol. 2006, 33, 701–706. [Google Scholar] [CrossRef]
- Kumar, M.; Singhal, A.; Verma, P.K.; Thakur, I.S. Production and Characterization of Polyhydroxyalkanoate from Lignin Derivatives by Pandoraea sp. ISTKB. ACS Omega 2017, 2, 9156–9163. [Google Scholar] [CrossRef] [PubMed]
- de Souza, L.; Manasa, Y.; Shivakumar, S. Bioconversion of lignocellulosic substrates for the production of polyhydroxyalkanoates. Biocat. Agric. Biotechnol. 2020, 28, 101754. [Google Scholar] [CrossRef]
- Martínez-Avila, O.; Llimós, J.; Ponsá, S. Integrated solid-state enzymatic hydrolysis and solid-state fermentation for producing sustainable polyhydroxyalkanoates from low-cost agro-industrial residues. Food Bioprod. Process 2021, 126, 334–344. [Google Scholar] [CrossRef]
- Elain, A.; Le Grand, A.; Corre, Y.-M.; Le Fellic, M.; Hachet, N.; Le Tilly, V.; Loulergue, P.; Audic, J.-L.; Bruzaud, S. Valorisation of local agro-industrial processing waters as growth media for polyhydroxyalkanoates (PHA) production. Ind. Crops Prod. 2016, 80, 1–5. [Google Scholar] [CrossRef]
- Sarmiento-Vásquez, Z.; de Souza Vandenberghe, L.P.; Karp, S.G.; Soccol, C.R. Production of Polyhydroxyalkanoates through Soybean Hull and Waste Glycerol Valorization: Subsequent Alkaline Pretreatment and Enzymatic Hydrolysis. Fermentation 2022, 8, 433. [Google Scholar] [CrossRef]
- Tello-Cruzado, B.K.; Azanedo-Vargas, M.; Quinones-Cerna, C.E.; Fuentes-Olivera, A.; Rodríguez-Soto, J.C.; Quezada-Alvarez, M.A.; Cruz-Monzon, J.A. Use of Enzymatic Hydrolysate from Agroindustrial Asparagus Waste as Substrate for the Production of Polyhydroxyalkanoate by Bacilluy thuringiensis. Environ. Res. Eng. Manag. 2024, 80, 8–16. [Google Scholar] [CrossRef]
- Elgharbawy, A.A.; Alam, M.Z.; Moniruzzaman, M.; Goto, M. Ionic liquid pretreatment as emerging approaches for enhanced enzymatic hydrolysis of lignocellulosic biomass. Biochem. Eng. J. 2016, 109, 252–267. [Google Scholar] [CrossRef]
- Didion, Y.P.; Alvan Vargas, M.V.G.; Tjaslma, T.G.; Woodley, J.; Nikel, P.I.; Malankowska, M.; Su, Z.; Pinelo, M. A novel strategy for extraction of intracellular poly(3-hydroxybutyrate) from engineered Pseudomonas putida using deep eutectic solvents: Comparison with traditional biobased organic solvents. Sep. Purif. Technol. 2024, 338, 126465. [Google Scholar] [CrossRef]
- Mondal, S.; Syed, U.T.; Gil, C.; Hilliou, L.; Duque, A.F.; Reis, M.A.M.; Brazinha, C. A novel sustainable PHA downstream method. Green Chem. 2023, 25, 1137–1149. [Google Scholar] [CrossRef]
- Escobar, E.L.N.; da Silva, T.A.; Pirich, C.L.; Corazza, M.L.; Ramos, L.P. Supercritical Fluids: A Promising Technique for Biomass Pretreatment and Fractination. Front. Bioeng. Biotechnol. 2020, 8, 252. [Google Scholar] [CrossRef]
- Awogbemi, O.; Von Kallon, D.V. Pretreatment techniques for agricultural waste. Cast Stud. Chem. Environ. Eng. 2022, 6, 100229. [Google Scholar] [CrossRef]
- Kumar, V.; Ahluwalia, V.; Saran, S.; Kumar, J.; Patel, A.K.; Singhania, R.R. Recent developments on solid-state fermentation for production of microbial secondary metabolites, challenges and solutions. Bioresour. Technol. 2021, 323, 124566. [Google Scholar] [CrossRef] [PubMed]
- Rebocho, A.T.; Pereira, J.R.; Freitas, F.; Neves, L.A.; Alves, V.D.; Sevrin, C.; Grandfils, C.; Reis, M.A.M. Production of Medium-Chain Length Polyhydroxyalkanoates by Pseudomonas citronellolis Grown in Apple Pulp Waste. Appl. Food Biotechnol. 2019, 6, 71–82. [Google Scholar]
- Kurt-Kızıldoğan, A.; Türe, E.; Okay, S.; Otur, Ç. Improved production of poly(3-hydroxybutyrate) by extremely halophilic archaeon Haloarcula sp. TG1 by utilization of rCKT3eng-treated sugar beet pulp. Biom. Conv. Bioref. 2023, 13, 10911–10921. [Google Scholar] [CrossRef]
- Sirohi, R. Sustainable utilization of food waste: Production and characterization of polyhydroxybutyrate (PHB) from damaged wheat grains. Environ. Technol. Inn. 2021, 23, 101715. [Google Scholar] [CrossRef]
- Alva Munoz, L.E.; Riley, M.R. Utilization of Cellulosic Waste From Tequila Bagasse and Production of Polyhydroxyalkanoate (PHA) Bioplastics by Saccharophagus degradans. Biotechnol. Bioeng. 2008, 100, 882–888. [Google Scholar] [CrossRef]
- Obruca, S.; Benesova, P.; Petrik, S.; Oborna, J.; Prikryl, R.; Marova, I. Production of polyhydroxyalkanoates using hydrolysate of spent coffe grounds. Process Biochem. 2014, 49, 1409–1414. [Google Scholar] [CrossRef]
- López-Cuellar, M.R.; Alba-Flores, J.; Garcida Rodríguez, J.N.; Pérez-Guevara, F. Production of polyhydroxyalkanoates (PHAs) with canola oil as carbon source. Int. J. Biol. Macromol. 2011, 48, 74–80. [Google Scholar] [CrossRef]
- Reis, M.; Albuquerque, M.; Villano, M.; Majone, M. Mixed Culture Processes for Polyhydroxyalkanoate Production from Agro-Industrial Surplus/Wastes as Feedstocks. Compreh. Biotechnol. 2011, 5, 669–683. [Google Scholar]
- Catherine, M.-C.; Guwy, A.; Massanet-Nicolau, J. Effect of acetate concentration, temperature, pH and nutrient concentration on polyhydroxyalkanoates (PHA) production by glycogen accumulating organisms. Bioresour. Technol. Rep. 2022, 20, 101226. [Google Scholar] [CrossRef]
- Villano, M.; Beccari, M.; Dionisi, D.; Lampis, S.; Miccheli, A.; Vallini, G.; Majone, M. Effect of pH on the production of bacterial polyhydroxyalkanoates by mixedcultures enriched under periodic feeding. Process Biochem. 2010, 45, 714–723. [Google Scholar] [CrossRef]
- Lopez-Vazquez, C.M.; Hooijmans, C.M.; Brdjanovic, D.; Gijzen, H.J.; van Loosdrecht, M.C.M. Temperature effects on glycogen accumulating organisms. Water Res. 2009, 43, 2852–2864. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hua, F.; Tsang, Y. Synthesis of PHA from waste under various C:N ratios. Bioresour. Technol. 2007, 98, 1690–1693. [Google Scholar] [CrossRef] [PubMed]
- Koller, M. A Review on Established and Emerging Fermentation Schemes for Microbial Production of Polyhydroxyalkanoate (PHA) Biopolyesters. Fermentation 2018, 4, 30. [Google Scholar] [CrossRef]
- Lizardi-Jiménez, M.A.; Hernández-Martínez, R. Solid state fermentation (SSF): Diversity of applications to valorize waste and biomass. 3 Biotech 2017, 7, 44. [Google Scholar] [CrossRef]
- Yafetto, L. Application of solid-state fermentation by microbial biotechnology for bioprocessing of agro-industrial wastes from 1970 to 2020: A review and bibliometric analysis. Heliyon 2022, 8, e09173. [Google Scholar] [CrossRef]
- Kolaković, A. Influence of Corn Silage Addition on Laccase Activity During Submerge Cultivation of Trametes versicolor. Master’s Thesis, University Josip Juraj Strossmayer in Osijek, Osijek, Croatia, 17 July 2015. [Google Scholar]
- Behl, M.; Thakar, S.; Ghai, H.; Sakhuja, D.; Bhatt, A.K. Fundamentals of fermentation technology. In Basic Biotechniques for Bioprocess and Bioentrepreneurship, 1st ed.; Bhatt, A.K., Bhatia, R.K., Bhalla, T.C., Eds.; Elsevier: London, UK, 2023; Volume 1, pp. 313–328. [Google Scholar]
- Ćurković, I. Design of Bioreactor for Microorganisms Cultivation in Semi-Solid and Solid Substrates. Master’s Thesis, University Josip Juraj Strossmayer in Osijek, Osijek, Croatia, 13 June 2013. [Google Scholar]
- Mohapatra, S.; Samantaray, D.P.; Samantaray, S.M. Impact of process recovery on PHA production by Bacillus thuringiensis RKD-12. Poll Res. 2015, 34, 395–400. [Google Scholar]
- Montiel-Jarillo, G.; Morales-Urrea, D.A.; Contreras, E.M.; López-Córdoba, A.; Gómez-Pachón, E.Y.; Carrera, J.; Suárez-Ojeda, M.E. Improvement of the Polyhydroxyalkanoates Recovery from Mixed Microbial Cultures Using Sodium Hypochlorite Pre-treatment Coupled with Solvent Extraction. Polymers 2022, 14, 3938. [Google Scholar] [CrossRef]
- Heinrich, D.; Madkour, M.H.; Al-Ghamdi, M.A.; Shabbaj, I.I.; Steinbüchel, A. Large scale extraction of poly(3-hydroxybutyrate from Ralstonia eutropha H16 using sodium hypochlorite. AMB Exress 2012, 2, 59. [Google Scholar] [CrossRef]
- Obruca, S.; Marova, I.; Stankova, M.; Mravcova, L.; Svoboda, Z. Effect of ethanol and hydrogen peroxide on poly(3-hydroxybutyrate) biosynthetic pathway in Cupriavidus necator H16. World J. Microbiol. Biotechnol. 2010, 26, 1261–1267. [Google Scholar] [CrossRef]
- Younas, T.; Ali, I.; Jamil, N. Polyhydroxyalkanoates production using canola oil by bacteria isolated from paper pulp industry. Kuwait J. Sci. 2015, 42, 236–249. [Google Scholar]
- Park, S.L.; Cho, J.Y.; Choi, T.-R.; Song, H.-S.; Bhatia, S.K.; Gurav, R.; Park, S.-H.; Park, K.; Joo, J.C.; Hwang, S.Y.; et al. Improvement of polyhydroxybutyrate (PHB) plate-based screening method for PHB degrading bacteria using cell-grown amorphous PHB and recovered by sodium dodecyl sulfate (SDS). Int. J. Biol. Macromol. 2021, 177, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Neves, A.; Müller, J. Use of enzymes in extraction of polyhydroxyalkanoates produced by Cupriavidus necator. Biotechnol. Prog. 2012, 28, 1575–1580. [Google Scholar] [CrossRef] [PubMed]
- Kunasundari, B.; Sudesh, K. Isolation and recovery of microbial polyhydroxyalkanoates. eXPRESS Polym. Lett. 2011, 5, 620–634. [Google Scholar] [CrossRef]
- Marudkla, J.; Patjawit, A.; Chuensangjun, C.; Sirisansaneeyakul, S. Optimization of poly(3-hydroxybutyrate) extraction from Cupriavidus necator DSM 545 using sodium dodecyl sulfate and sodium hypochlorite. Agric. Nat. Resour. 2018, 52, 266–273. [Google Scholar] [CrossRef]
- Song, J.-H.; Jeon, C.-O.; Choi, M.-H.; Yoon, S.-C.; Park, W.-J. Polyhydroxyalkanoate (PHA) production Using Waste Vegetable Oil by Pseudomonas sp. Strain DR2. J. Microbiol. Biotechnol. 2008, 18, 1408–1415. [Google Scholar]
- Fernández, D.; Rodríguez, E.; Bassas, M.; Vinas, M.; Solanas, A.M.; Llorens, J.; Marqués, A.M.; Manresa, A. Agro-industrial oily wastes as substrates for PHA production by the new strain Pseudomonas aeruginosa NCIB 40045: Effect of culture conditions. Biochem. Eng. J. 2005, 26, 159–167. [Google Scholar] [CrossRef]
- Yang, Y.-H.; Jeon, J.-M.; Yi, D.H.; Kim, J.-H.; Seo, H.-M.; Rha, C.K.; Sinskey, A.J.; Brigham, C.J. Application of a Non-halogenated Solvent, Methyl Ethyl Ketone (MEK) for Recovery of Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) [P(HB-co-HV)] from Bacterial Cells. Biotechnol. Bioprocess Eng. 2015, 20, 291–297. [Google Scholar] [CrossRef]
- Nayir, T.Y.; Konuk, S.; Kara, S. Extraction of polyhydroxyalkanoate from activated sludge using supercritical carbon dioxide process and biopolymer characterization. J. Biotechnol. 2023, 364, 50–57. [Google Scholar] [CrossRef]
- Hejazi, P.; Vasheghani-Farahani, E.; Yamini, Y. Supercritical Fluid Disruption of Ralstonia eutropha for Poly(β-hydroxybutyrate) Recovery. Biotechnol. Prog. 2008, 19, 1519–1523. [Google Scholar] [CrossRef]
- Hand, S.; Gill, J.; Chu, K.-H. Phage-based extraction of polyhydroxybutyrate (PHB) produced from synthetic crude glycerol. Sci. Total Environ. 2016, 557, 317–321. [Google Scholar] [CrossRef] [PubMed]
- ISO 18134-3; Solid biofuels—Determination of moisture content—Oven dry method—Part 3: Moisture in General Analysis Sample. ISO Publishing: Geneva, Switzerland, 2023.
- ISO 18123:2023; Solid Biofuels—Determination of Volatile Matter. ISO Publishing: Geneva, Switzerland, 2023.
- Önorm, S. Analytical Methods and Quality Control for Waste Compost; Austrian Standardization Institute: Vienna, Austria, 2023. [Google Scholar]
- ASTM D982-16; Standard Test Method for Organic Nitrogen in Paper and Paperboard. ASTM International: West Conshohocken, PA, USA, 2016.
- Marsden, W.; Gray, P.P.; Nippard, G.J.; Quinlan, M.R. Evaluation of the DNS method for analysing lignocellulosic hydrolysates. J. Chem. Technol. Biotechnol. 1982, 32, 1016–1022. [Google Scholar] [CrossRef]
- ISO 21268-3:2019; Soil Quality—Leaching Procedures for Subsequent Chemical and Ecotoxicological Testing of Soil and Soil-like Materials. Part 3: Up-Flow Percolation Test. ISO: Geneva, Switxerland, 2019.
- Briški, F.; Kopčić, N.; Ćosić, I.; Kučić, D.; Vuković, M. Biodegradation of tobacco waste by composting: Genetic identification of nicotine-degrading bacteria and kinetic analysis of transformations in leachate. Chem. Pap. 2012, 66, 1103–1110. [Google Scholar] [CrossRef]
- Kumar, A.; Shah, S. Production and characterization of polyhydroxyalkanoates from industrial waste using soil bacterial isolates. Braz. J. Microbiol. 2021, 52, 715–726. [Google Scholar]
- Benson, H.J. Microbiological Applications. In A Laboratory Manual in General Microbiology, 8th ed.; The McGraw-Hill Companies: New York, NY, USA, 2001; pp. 154–156, 160, 168, 169. [Google Scholar]
- Law, J.H.; Slepecky, R.A. A rapid spectrophotometric assay of alpha, betaunsaturated acid and beta-hydroxy acids. Anal. Chem. 1960, 32, 1697–1699. [Google Scholar]
- Law, J.H.; Slepecky, R.A. Assay of poly-b-hydroxybutyric acid. J. Bacteriol. 1961, 82, 33.e36. [Google Scholar] [CrossRef]
- Porras, M.A.; Villar, M.A.; Cubitto, M.A. Improved intracellular PHA determinations with novel spectrophotometric quantification methodologies based on Sudan black dye. J. Microbiol. Meth. 2018, 148, 1–11. [Google Scholar] [CrossRef]
- Takayoshi, T. Structural insights into starch-metabolizing enzymes and their applications. Biosci. Biotechnol. Biochem. 2024, 88, 864–871. [Google Scholar]
- Gamage, A.; Thiviya, P.; Mani, S.; Ponnusamy, P.G.; Manamperi, A.; Evon, P.; Merah, O.; Madhujith, T. Environmental Properties and Applications of Biodegradable Starch-Based Nanocomposites. Polymers 2022, 14, 4578. [Google Scholar] [CrossRef]
- Talaie, A.; Alaee, S.; Mohanadoss, P. Guidelines for quick application of biochemical tests to identify unknown bacteria. Acc. Biotechnol. Res. 2015, 2, 65–82. [Google Scholar]
- Chen, C.-L.; Ge, L.; Li, L.; Li, Y.; Mo, Y.; Razaad, I.M.N.; Tan, G.-Y.; Wang, J.-Y.; Wang, L.; Zhao, L. Start a research on biopolymer polyhydroxyalkanoate (PHA): A review. Polymers 2014, 6, 706–754. [Google Scholar] [CrossRef]
- Adebajo, S.O.; Bankole, P.O.; Ojo, A.E.; Akintokun, P.O. Screening and optimization of polyhydroxybutyrate (PHB) by Lysinibacillus fusiformis from diverse environmental sources. Microbe 2024, 2, 100043. [Google Scholar] [CrossRef]
- Hamdy, S.M.; Danial, A.W.; Gad El-Rab, S.M.F.; Shoreit, A.A.M.; Hesham, A.E.-L. Production and optimization of bioplastic (Polyhydroxybutyrate) from Bacillus cereus strain SH-02 using response surface methodology. BMC Microbiol. 2022, 22, 183. [Google Scholar] [CrossRef] [PubMed]
- Cibichakravarthy, B.; Venkatachalam, S.; Prabagaran, S.R. Chapter 9—Unleashing Extremophilic Metabolites and Its Industrial Perspectives. In New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Secondary Metabolites Biochemistry and Applications, 1st ed.; Gupta, V.K., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 1, pp. 119–130. [Google Scholar]
- de Oliviera Schmidt, V.K.; dos Santos, E.F.; de Oliveira, D.; Ayub, M.A.Z.; Cesca, K.; Cortivo, P.R.D.; de Andrade, C.J.; Hickert, L.R. Production of Polyhydroxyalkanoates by Bacillus megaterium: Prospecting on Rice Hull and Residual Glycerol Potential. Biomass 2022, 2, 412–425. [Google Scholar] [CrossRef]
- Vu, D.H.; Wainaina, S.; Taherzadeh, M.J.; Akesson, D.; Ferreira, J.A. Production of polyhydroxyalkanoates (PHAs) by Bacillus megaterium using food waste acidogenic fermentation-derived volatile fatty acids. Bioengineered 2021, 12, 2480–2498. [Google Scholar] [CrossRef]
- Trakunjae, C.; Boondaeng, A.; Apiwatanapiwat, W.; Kosugi, A.; Arai, T.; Sudesh, K.; Vaithanomsat, P. Enhanced polyhydroxybutyrate (PHB) production by newly isolated rare actinomycetes Rhodococcus sp. Strain BSRT1-1 using response surface methodology. Sci. Rep. 2021, 11, 1896. [Google Scholar] [CrossRef]
- Maurya, A.C.; Bhattacharya, A.; Khare, S.K. Biodegradation of terephthalic acid using Rhodococcus erythropolis MTCC 3951: Insights into the degradation process, applications in wastewater treatment and polyhydroxyalkanoate production. Environ. Sci. Pollut. Res. Int. 2024, 31, 57376–57385. [Google Scholar] [CrossRef]
- Wong, A.L.; Chua, H.; Hoi Fu Yu, P. Microbial Production of Polyhydroxyalkanoates by Bacteria Isolated from Oil Wastes. Appl. Biochem. Biotechnol. 2000, 85, 843–858. [Google Scholar] [CrossRef]
- Panchal, H.K. Polyhydroxybutyrate (PHB) Production and Optimization using Staphylococcus Gallinarum and One More Soil Isolate. Int. J. Pharm. Biol. Sci. 2019, 2, 52–57. [Google Scholar]
- Jin, C.; Li, J.; Huang, Z.; Han, X.; Bao, J. Engineering Corynebacterium glutamicum for synthesis of poly(3-hydroxybutyrate) from lignocellulose biomass. Biotechnol. Bioeng. 2022, 119, 1598–1613. [Google Scholar] [CrossRef]
- Matias, F.; Bonatto, D.; Padilla, G.; de Andrade Rodrigues, M.F.; Henriques, J.A.P. Polyhydroxyalkanoates production by Actinobacteria isolated from soil. Can. J. Microbiol. 2009, 55, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Mitra, R.; Xu, T.; Chen, G.-Q.; Xiang, H.; Han, J. An updated overview on the regulatory circuits of polyhydroxyalkanoates synthesis. Microb. Biotechnol. 2021, 15, 1446–1470. [Google Scholar] [CrossRef] [PubMed]
- Powers, E.M. Efficacy of the Ryu nonstaining KOH technique for rapidly determining gram reactions of food-borne and waterborne bacteria and yeasts. Appl. Environ. Microbiol. 1995, 61, 3756–3758. [Google Scholar] [CrossRef]
- Microbe Notes. Available online: https://microbenotes.com/biochemical-test-of-bacteria/ (accessed on 9 October 2024).
- VetBact. Available online: https://www.vetbact.org/biochemtest/1 (accessed on 9 October 2024).
- Microbe Notes. Available online: https://microbenotes.com/nitrate-reduction-test-objectives-principle-procedure-and-results/ (accessed on 9 October 2024).
- Maria, W.; Johan, B.; Sabine, L. Comparison of Auxacolor with API 20 C Aux in yeast identification. Clin. Microbiol. Infect. 1997, 3, 369–375. [Google Scholar]
- Boyd, M.A.; Antonio, M.A.D.; Hillier, S.L. Comparison of API 50 CH strips to whole—Chromosomal DNA probes for identification of Lactobacillus species. J. Clin. Microbiol. 2005, 43, 5309–5311. [Google Scholar] [CrossRef]
- Dance, D.A.; Wuthiekanun, V.; Naigowit, P.; White, N.J. Identification of Pseudomonas pseudomallei in clinical practice: Use of simple screening tests and API 20 NE. J. Clin. Pathol. 1989, 42, 645–648. [Google Scholar] [CrossRef]
- Almeida, R.J.; Jorgensen, J.H. Identification of coagulase-negative staphylococci with the API STAPH-IDENT system. J. Clin. Microbiol. 1983, 18, 254–257. [Google Scholar] [CrossRef]
- Groothuis, D.G.; Elzenaar, C.P.; van Silfhout, A. An Evaluation of the API-20 Strep System (Rapid-Strep System). System. Appl. Microbiol. 1986, 8, 137–141. [Google Scholar] [CrossRef]
- Edinger, R.C.; Migneault, P.C.; Nolte, F.S. Supplementary Rapid Biochemical Test Panel for the API 20E Bacterial Identification System. J. Clin. Microbiol. 1985, 22, 1063–1065. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: And emerging technology for microbial identification for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Hrubanova, K.; Kalina, M.; Koller, M.; Kourilova, X.; Kovalcik, A.; Krzyzanek, V.; Masilko, J.; Nebesarova, J.; Novackova, I.; Obruca, S.; et al. Introducing the Newly Isolated Bacterium Aneurinibacillus sp. H1 as an Auspicious Thermophilic Producer of Various Polyhydroxyalkanoates (PHA) Copolymers-1. Isolation and Characterization of the Bacterium. Polymers 2020, 12, 1235. [Google Scholar] [CrossRef] [PubMed]
- Bosco, F.; Cirrincione, S.; Carletto, R.; Marmo, L.; Chiesa, F.; Mazzoli, R.; Pessione, E. PHA Production from Cheese Whey and “Scotta”: Comparison between a Consortium and a Pure Culture of Leuconostoc mesenteroides. Microorganisms 2021, 9, 2426. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.; Onions, A.H.S.; Allsopp, D.; Eggins, H.O.W. Smith’s Introduction to Industrial Mycology, 7th ed.; Edward Arnold: London, UK, 1981; pp. 1–398. [Google Scholar]
- Aslim, B.; Beyatli, Y.; Mercan, N.; Şafak, S. A study on the production of poly-beta-hydroxybutyrate by some eukaryotic microorganisms. Turk. Elec. J. Biotechnol. 2002, 11–17. [Google Scholar]
- Banu, J.R.; Ilamathi, P.M.K.; Khanna, Y.; Murugesan, K.; Selvam, A.; Udayakumar, M.; Wong, J. Food waste properties, In Current Developments in Biotechnology and Bioengineering: Sustainable Food Waste Management: Resource Recovery and Treatment, 1st ed.; Kaur, G., Lasaridi, K., Pandey, A., Taherzadeh, M., Wong, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 11–41. [Google Scholar]
- Raghavarao, K.S.M.S.; Ranganathan, T.V.; Karanth, N.G. Some engineering aspects of solid-state fermentation. Biochem. Eng. J. 2003, 13, 127–135. [Google Scholar] [CrossRef]
- Bigelis, R. Solid-phase fermentation: Aerobic and anaerobic. In Manual of Industrial Microbiology and Biotechnology, 3rd ed.; Bull, A.T., Junker, B., Katz, L., Lynd, L.R., Masurekar, P., Reeves, C.D., Zhao, H., Eds.; John Wiley & Sons: New York, NY, USA, 2014; Volume 1, pp. 235–252. [Google Scholar]
- Llenas, L.; Martínez, O.; Ponsá, S. Sustainable polyhydroxyalkanoates production via solid-state fermentation: Influence of the operational parameters and scaling up of the process. Food Bioprod. Process 2021, 132, 13–22. [Google Scholar]
- Blunt, W.; Shah, P.; Vasquez, V.; Ye, M.; Doyle, C.; Liu, Y.; Saeidlou, S.; Monteil-Rivera, F. Biosynthesis and properties of polyhydroxyalkanoates synthesized from mixed C5 and C6 sugars obtained from hardwood hydrolysis. New Biotechnol. 2023, 77, 40–49. [Google Scholar] [CrossRef]
- Londono-Hernandez, L.; de Jesús García-Gómez, M.; Huerta-Ochoa, S.; Polanía-Rivera, A.M.; Aguilar, C.N.; Prado-Barragán, L.A. Effect of Glucose Concentration on the Production of Proteolytic Extract by Different Strains of Aspergillus under Solid-State Fermentation. Fermentation 2024, 10, 97. [Google Scholar] [CrossRef]
- Yaverino-Gutierrez, M.A.; Ramos, L.; Ascencio, J.J.; Chandel, A.K. Enhanced Production of Clean Fermentable Sugars by Acid Pretreatment and Enzymatic Saccharification of Sugarcane Bagasse. Processes 2024, 12, 978. [Google Scholar] [CrossRef]
- Marakana, P.G.; Dey, A.; Saini, B. Isolation of nanocellulose from lignocellulosic biomass: Synthesis, characterization, modification and potential applications. J. Environ. Chem. Eng. 2021, 9, 106606. [Google Scholar] [CrossRef]
- de Souza, G.A.; Reis, G.A.; Michels, M.H.A.; Fajardo, G.L.; Lamot, I.; de Best, J.H. Optimization of Green Extraction and Purification of PHA Produced by Mixed Microbial Cultures from Sludge. Water 2020, 12, 1185. [Google Scholar] [CrossRef]
- Getino, L.; Martín, J.L.; Chamizo-Ampudia, A. A Review of Polyhydroxyalkanoates: Characterization, Production, and Application from Waste. Microorganisms 2024, 12, 2028. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Oehmen, A.; Carvalho, G.; Reis, M.A.M. Community profile governs substrate competition in polyhydroxyalkanoate (PHA)-producing mixed cultures. New Biotechnol. 2020, 58, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Oehmen, A.; Freitas, E.B.; Carvalho, G.; Reis, M.A.M. The link of feast—Phase dissolved oxygen (DO) with substrate competition and microbial selection in PHA production. Water Res. 2017, 112, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Zhu, C.; Singh, R.P.; Chen, W. Chickpea: Its Origin, Distribution, Nutrition, Benefits, Breeding and Symbiotic Relationship with Mesorhizobium Species. Plants 2024, 13, 429. [Google Scholar] [CrossRef]
- Gautam, S.; Gautam, A.; Pawaday, J.; Kanzariya, R.K.; Yao, Z. Current Status and Challenges in the Commercial Production of Polyhydroxyalkanoate-Based Bioplastic: A Review. Processes 2024, 12, 1720. [Google Scholar] [CrossRef]
- Yang, J.; Gao, C.; Yang, X.; Su, Y.; Shi, S.; Han, L. Effect of combined wet alkaline mechanical pretreatment on enzymatic hydrolysis of corn stover and its mechanism. Biotechnol. Biofuels Bioprod. 2022, 15, 31. [Google Scholar] [CrossRef]
- Ramezani, M.; Amoozegar, M.A.; Ventosa, A. Screening and comparative assay of poly-hydroxyalkanoates produced by bacteria isolated from the Gavkhooni Wetland in Iran and evaluation of poly-β-hydroxybutyrate production by halotolerant bacterium Oceanimonas sp. GK1. Ann. Microbiol. 2015, 65, 517–526. [Google Scholar] [CrossRef]
- Divyashree, M.S.; Davis, R.; Kumari, K.S.; Raj, B.; Shamala, T.R.; Vijayendra, S.V. Production and characterization of bacterial polyhydroxyalkanoate copolymers and evaluation of their blends by Fourier transform infrared spectroscopy and scanning electron microscopy. Indian J. Microbiol. 2009, 49, 251–258. [Google Scholar]
- Bhati, R.; Mallick, N.; Sankhla, I.S.; Singh, A.K. Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) co-polymer production from a local isolate, Brevibacillus invocatus MTCC 9039. Bioresour. Technol. 2010, 101, 1947–1953. [Google Scholar]
- Dias, M.L.; Freire, D.M.; Oliveira, F.C.; Castilho, L.R. Characterization of poly(3-hydroxybutyrate) produced by Cupriavidus necator in solid-state fermentation. Bioresour. Technol. 2007, 98, 633–638. [Google Scholar]
- Akbari, V.; Cochez, M.; Langlois, V.; Michely, L.; Moradkhani, G.; Renard, E.; Saeb, M.R.; Vagner, C. Thermal stability and flammability behavior of poly(3-hydroxybutyrate) (PHB) based composites. Materials 2019, 12, 2239. [Google Scholar] [CrossRef]
- Dikshit, P.K.; Pradhan, S. Production, ultrasonic extraction, and characterization of poly(3-hydroxybutyrate) (PHB) using Bacillus megaterium and Cupriavidus necator. Polym. Adv. Technol. 2018, 29, 2392–2400. [Google Scholar]


| Agro-Industrial Waste | Pretreatment Method | Microorganism | PHA Yield | Ref. |
|---|---|---|---|---|
| Apple pulp waste | n.a. | Pseudomonas citronellolis NRRL B-2504 | 30 ± 1.7% | [76] |
| Sugar beetroot pulp | Enzymatic–recombinant endoglucanase (rCKT3eng), chemical hydrolysis | Haloarcula sp. TG1 | 45.6%/17.8% | [77] |
| Wheat grains | Acidic hydrolysis (hydrochloric acid) | Bacillus sp. NII2 | 1.413 mg/L | [78] |
| Tequila bagasse | Bacterial pretreatment (Saccharophagus degradans ATCC 43961) | Saccharophagus degradans ATCC 43961 | 1.5 mg/L | [79] |
| Coffee ground | n-hexane oil extraction, acidic hydrolysis (sulphuric acid), and enzymatic digestion | Burkholderia cepacia ATCC 17759 | 2.69 ± 0.07 g/L | [80] |
| Canola oil | n.a. | Wautersia eutropha ATCC 17699 | 18.27 g/L | [81] |
| Sugarcane molasse | n.a. | Bacillus cereus SPV | 61.07% | [15] |
| Substrate | w(MC)/% | w(DM)/% | w(VM)/% | pH/− | ω(RS)/mg gDM−1 | CFUbacteria/gDM | CFUfungi/gDM | w(C)/% | w(N)/% |
|---|---|---|---|---|---|---|---|---|---|
| Chickpea 1 | 58.06 | 41.94 | 97.69 | 5.545 | 45.47 | 9.8 × 109 | 1.6 × 107 | 54.27 | 3.93 |
| Chickpea 2 | 59.86 | 40.14 | 97.72 | 5.011 | 56.78 | 1.2 × 109 | 1.5 × 108 | 54.28 | 4.02 |
| Starch | 44.77 | 55.23 | 99.36 | 4.453 | 33.54 | 1.5 × 108 | 7.4 × 106 | 55.20 | 0.02 |
| Substrate | Identified Microorganism | Morphology |
|---|---|---|
| Chickpea 1 | Brevibacillus sp. | Transparent with flat elevation, and regular round configuration, rod shaped |
| Empedobacter brevis | Orange with flat elevation, and regular round configuration, rod shaped | |
| Aneurinibacillus aneurinilyticus | Brownish with raised elevation, and regular round configuration, rod shaped | |
| Chickpea 2 | Micrococcus spp. | Orange with flat elevation, and regular round configuration, round shaped (cocci) |
| Trichosporon asahii | White and cracked in the middle with smooth and shiny edges | |
| Starch | Leuconostoc sp. | White with flat elevation, and regular round configuration, cocci/coccobacilli |
| Bacillus licheniformis | White with raised elevation, wavy and smooth edges, rod shaped | |
| Staphylococcus lentus | Transparent with raised elevation, and regular round configuration, round shaped (cocci) | |
| Citrobacter freundii | Transparent with raised elevation, irregular shape with twisted edges, rod shaped | |
| Cryptococcus humicola | Yellowish with raised elevation, round shape with jagged edges | |
| Geotrichum klebahnii | White with flat elevation, filamentous shape with jagged edges | |
| Candida krusei | White with raised elevation, and regular round configuration |
| Identified Bacteria | Gram Staining | KOH Test | Oxidase | Catalase | Nitrate-Reductase |
|---|---|---|---|---|---|
| Brevibacillus sp. | +ve | + | + | + | + |
| Empedobacter brevis | −ve | + | +/− | + | − |
| Aneurinibacillus aneurinilyticus | +ve | + | + | + | − |
| Micrococcus spp. | +ve | + | +/− | + | +/− |
| Leuconostoc sp. | +ve | − | − | - | − |
| Bacillus licheniformis | +ve | − | − | + | + |
| Staphylococcus lentus | +ve | − | − | + | + |
| Citrobacter freundii | −ve | − | − | + | + |
| Substrate | w(MC)/% | w(DM)/% | w(VM)/% | pH/- | ω(RS)/mg gDM−1 | CFUbacteria/gDM | CFUfungi/gDM | w(C)/% | w(N)/% |
|---|---|---|---|---|---|---|---|---|---|
| Chickpea 1 | 73.66 | 26.34 | 93.99 | 5.557 | 30.34 | 3.8 × 1010 | 2.4 × 108 | 52.21 | 1.73 |
| Chickpea 2 | 74.99 | 25.01 | 91.90 | 5.219 | 12.36 | 4.7 × 1010 | 2.1 × 108 | 51.05 | 1.10 |
| Starch | 45.90 | 54.10 | 96.32 | 7.219 | 21.04 | 3.4 × 109 | 1.3 × 108 | 53.51 | 0.07 |
| Substrate | PHA Accumulation/% |
|---|---|
| Chickpea 1 | 5.42 ± 0.062 |
| Chickpea 2 | 13.81 ± 0.048 |
| Starch | 5.29 ± 0.086 |
| Sample | Tg/°C | Tm/°C | ΔHm/J g−1 | Tc/°C | ΔHc/J g−1 |
|---|---|---|---|---|---|
| Chickpea 1 | / | 128.16 ± 0.007 | −8.46 ± 0.002 | 99.10 ± 0.001 | 16.20 ± 0.009 |
| Chickpea 2 | 1.81 ± 0.007 | 124.52 ± 0.010 | −6.28 ± 0.017 | 95.24 ± 0.001 | 11.33 ± 0.007 |
| Starch | / | 119.53 ± 0.007 | −0.46 ± 0.021 | 85.37 ± 0.006 | 1.96 ± 0.010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grgurević, K.; Bramberger, D.; Miloloža, M.; Stublić, K.; Ocelić Bulatović, V.; Ranilović, J.; Ukić, Š.; Bolanča, T.; Cvetnić, M.; Markić, M.; et al. Producing and Characterizing Polyhydroxyalkanoates from Starch and Chickpea Waste Using Mixed Microbial Cultures in Solid-State Fermentation. Polymers 2024, 16, 3407. https://doi.org/10.3390/polym16233407
Grgurević K, Bramberger D, Miloloža M, Stublić K, Ocelić Bulatović V, Ranilović J, Ukić Š, Bolanča T, Cvetnić M, Markić M, et al. Producing and Characterizing Polyhydroxyalkanoates from Starch and Chickpea Waste Using Mixed Microbial Cultures in Solid-State Fermentation. Polymers. 2024; 16(23):3407. https://doi.org/10.3390/polym16233407
Chicago/Turabian StyleGrgurević, Karlo, Dora Bramberger, Martina Miloloža, Krešimir Stublić, Vesna Ocelić Bulatović, Jasmina Ranilović, Šime Ukić, Tomislav Bolanča, Matija Cvetnić, Marinko Markić, and et al. 2024. "Producing and Characterizing Polyhydroxyalkanoates from Starch and Chickpea Waste Using Mixed Microbial Cultures in Solid-State Fermentation" Polymers 16, no. 23: 3407. https://doi.org/10.3390/polym16233407
APA StyleGrgurević, K., Bramberger, D., Miloloža, M., Stublić, K., Ocelić Bulatović, V., Ranilović, J., Ukić, Š., Bolanča, T., Cvetnić, M., Markić, M., & Kučić Grgić, D. (2024). Producing and Characterizing Polyhydroxyalkanoates from Starch and Chickpea Waste Using Mixed Microbial Cultures in Solid-State Fermentation. Polymers, 16(23), 3407. https://doi.org/10.3390/polym16233407









