NMR Analyses and Statistical Modeling of Biobased Polymer Microstructures—A Selected Review
Abstract
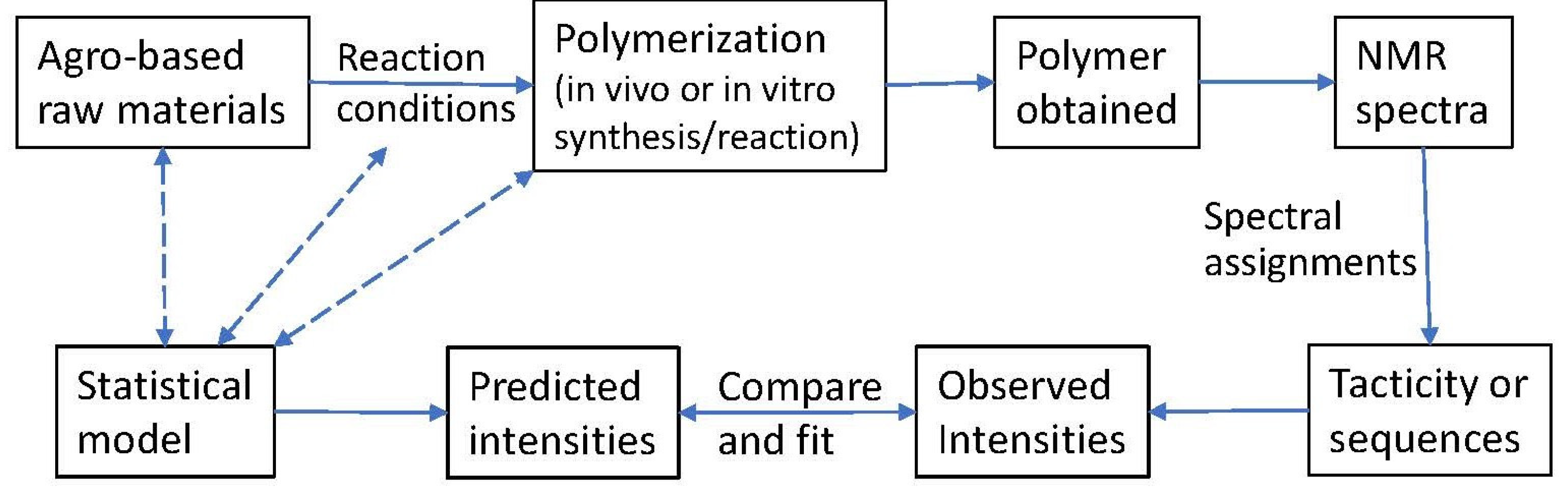
1. Introduction
2. Statistical Models
3. Poly(lactic acid) Tacticity
4. Poly(hydroxyalkanoate) Comonomer Sequences
5. Polysaccharide Sequence Determination
5.1. Alginate Mannuronic/Guluronic Sequence Analysis
5.2. Pectin Galacturonic Acid/Ester Sequence Analysis
5.3. Sequence Analysis of Partially Deacetylated Chitosan
5.4. Comments
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, H.N.; Gross, R.A. (Eds.) Sustainable Green Chemistry in Polymer Research, Vol. 1. Biocatalysis and Biobased Materials; ACS Symposium Series 1450; American Chemical Society: Washington, DC, USA, 2023. [Google Scholar]
- Cheng, H.N.; Gross, R.A. (Eds.) Sustainable Green Chemistry in Polymer Research, Vol. 2. Sustainable Polymers and Applications; ACS Symposium Series 1451; American Chemical Society: Washington, DC, USA, 2023. [Google Scholar]
- Cywar, R.M.; Rorrer, N.A.; Hoyt, C.B.; Beckham, G.T.; Chen, E.Y. Bio-based polymers with performance-advantaged properties. Nat. Rev. Mater. 2022, 7, 83–103. [Google Scholar] [CrossRef]
- Azhar, N.N.H.; Ang, D.T.; Abdullah, R.; Harikrishna, J.A.; Cheng, A. Bio-Based Materials Riding the Wave of Sustainability: Common Misconceptions, Opportunities, Challenges and the Way Forward. Sustainability 2022, 14, 5032. [Google Scholar] [CrossRef]
- Bovey, F.A. High Resolution NMR of Macromolecules; Academic Press: New York, NY, USA, 1972. [Google Scholar]
- Matsuzaki, K.; Uryu, T.; Asakura, T. NMR Spectroscopy and Stereoregularity of Polymers; Japan Scientific Societies Press: Tokyo, Japan, 1996. [Google Scholar]
- Cheng, H.N. Structural Studies of Polymers by Solution NMR; RAPRA Review Reports 125, Vol. 11, Number 5; RAPRA: Shrewsbury, UK, 2001. [Google Scholar]
- Cheng, H.N.; English, A.D. (Eds.) NMR Spectroscopy of Polymers in Solution and in the Solid State; ACS Symposium Series 834; American Chemical Society: Washington, DC, USA, 2003. [Google Scholar]
- Mirau, P.A. A Practical Guide to Understanding the NMR of Polymers; Wiley: Hoboken, NJ, USA, 2004. [Google Scholar]
- Kitayama, T.; Hatada, K. NMR Spectroscopy of Polymers; Springer: New York, NY, USA, 2004. [Google Scholar]
- Cheng, H.N.; Asakura, T.; English, A.D. (Eds.) NMR Spectroscopy of Polymers: Innovative Strategies for Complex Macromolecules; ACS Symposium Series 1077; American Chemical Society: Washington, DC, USA, 2012. [Google Scholar]
- Zhang, R.; Miyoshi, T.; Sun, P. (Eds.) NMR Methods for Characterization of Synthetic and Natural Polymers; Royal Society of Chemistry: London, UK, 2019. [Google Scholar]
- Cheng, H.N. Computer-Assisted NMR Analysis of Polymers in Solution. Polym. News 2000, 25, 114–122. [Google Scholar]
- Cheng, H.N.; Miri, M. Statistical Models and NMR Analysis of Polymer Microstructure. ACS Symp. Ser. 2011, 1077, 371–382. [Google Scholar]
- Odian, G. Principles of Polymerization, 4th ed.; Wiley: Hoboken, NJ, USA, 2004. [Google Scholar]
- Hagiopol, C. Copolymerization: Toward a Systematic Approach; Springer: New York, NY, USA, 1999. [Google Scholar]
- Brar, A.S.; Hekmatyar, S.K. Microstructure determination of the acrylonitrile–styrene–methyl methacrylate terpolymers by NMR spectroscopy. J. Appl. Polym. Sci. 1999, 74, 3026–3032. [Google Scholar] [CrossRef]
- Brar, A.S.; Malhotra, M. Compositional Assignments and Sequence Distribution of Vinylidene Chloride—Methyl Acrylate Copolymers Using One- and Two-Dimensional NMR Spectroscopy. Macromolecules 1996, 29, 7470–7476. [Google Scholar] [CrossRef]
- Coleman, B.D.; Fox, T.G. Multistate mechanism for homogeneous ionic polymerization. I. The diastereosequence distribution. J. Chem. Phys. 1963, 38, 1065–1075. [Google Scholar] [CrossRef]
- Coleman, B.D.; Fox, T.G. General theory of stationary random sequences with applications to the tacticity of polymers. J. Polym. Sci. Part A 1963, 1, 3183–3197. [Google Scholar] [CrossRef]
- Coleman, B.D.; Fox, T.G. A multistate mechanism for homogeneous ionic polymerization. II. The molecular weight distribution. J. Am. Chem. Soc. 1963, 85, 1241–1244. [Google Scholar] [CrossRef]
- Price, F.P. Markov Chains and Monte Carlo Calculations in Polymer Science; Chapter 7; Lowry, G.G., Ed.; Marcel Dekker: New York, NY, USA, 1970. [Google Scholar]
- Shelden, R.A.; Fueno, T.; Tsunetsugu, T.; Furukawa, J. A one-parameter model for isotactic polymerization based on enantiomorphic catalyst sites. J. Polym. Sci. Part B Polym. Lett. 1965, 3, 23–26. [Google Scholar] [CrossRef]
- Inoue, Y.; Itabashi, Y.; Chûjô, R.; Doi, Y. Studies of the stereospecific polymerization mechanism of propylene by a modified Ziegler-Natta catalyst based on 125 MHz 13C n.m.r. spectra. Polymer 1984, 25, 1640–1644. [Google Scholar] [CrossRef]
- Seiner, J.A.; Litt, M. The role of monomer charge-transfer complexes in free radical copolymerization. I. Derivation of terminal complex model equations. Macromolecules 1971, 4, 308–311. [Google Scholar] [CrossRef]
- Hill, D.J.T.; O’Donnell, J.H.; O’Sullivan, P.W. The role of donor-acceptor complexes in polymerization. Prog. Polym. Sci. 1982, 8, 215–275. [Google Scholar] [CrossRef]
- Izu, M.; O’Driscoll, K.F.; Hill, R.J.; Quinn, M.J.; Harwood, H.J. Copolymerization with Depropagation. VII. Coisotacticities in the Free-Radical Copolymerization of α-Methylstyrene and Methyl Methacrylate between their Ceiling Temperatures. Macromolecules 1972, 5, 90–92. [Google Scholar] [CrossRef]
- Cais, R.E.; Hill, D.J.T.; O’Donnell, J.H. Copolymerization of Vinyl Chloride and Sulfur Dioxide. III. Evaluation of the Copolymerization Mechanism. J. Macromol. Sci. Chem. A 1982, 17, 1437–1467. [Google Scholar] [CrossRef]
- Szwarc, M.; Perrin, C.L. General treatment of equilibrium copolymerization of two or more comonomers deduced from the initial state of the system. Macromolecules 1985, 18, 528–533. [Google Scholar] [CrossRef]
- Harwood, H.J. Structures and compositions of copolymers. Makromol. Chem. Makromol. Symp. 1987, 10–11, 331–354. [Google Scholar] [CrossRef]
- Carman, C.J.; Harrington, R.A.; Wilkes, C.E. Monomer Sequence Distribution in Ethylene-Propylene Rubber Measured by 13C NMR. 3. Use of Reaction Probability Model. Macromolecules 1977, 10, 536–544. [Google Scholar] [CrossRef]
- Cheng, H.N. Markovian Statistics and Simplex Algorithm for Carbon-13 NMR Spectra of Ethylene Propylene Copolymers. Anal. Chem. 1982, 54, 1828–1833. [Google Scholar] [CrossRef]
- Cheng, H.N. 13C NMR Analysis of Propylene Butylene Copolymers by a Reaction Probability Model. J. Polym. Sci. Polym. Phys. Ed. 1983, 21, 573–581. [Google Scholar] [CrossRef]
- Cheng, H.N. Analysis of NMR Triad Distribution in Ethylene/1 Butene Copolymers. Polym. Bull. 1990, 23, 589–596. [Google Scholar] [CrossRef]
- Cheng, H.N.; Lee, G.H. Characterization of Ethylene Copolymers with 1H NMR Techniques and Reaction Probability Models. Macromolecules 1988, 21, 3164–3170. [Google Scholar] [CrossRef]
- Cheng, H.N. Computerized Model Fitting Approach for the NMR Analysis of Polymers. J. Chem. Inf. Comput. Sci. 1987, 27, 8–13. [Google Scholar] [CrossRef]
- Cheng, H.N. 13C NMR Sequence Determination for Multicomponent Polymer Mixtures. J. Appl. Polym. Sci. 1988, 35, 1639–1650. [Google Scholar] [CrossRef]
- Cheng, H.N. Perturbed Markovian Probability Models. Macromolecules 1992, 25, 2351–2358. [Google Scholar] [CrossRef]
- Cheng, H.N. NMR Characterization of Copolymers that Exhibit Non-Symmetric Compositional Heterogeneity. Macromolecules 1997, 30, 4117–4125. [Google Scholar] [CrossRef]
- Koenig, J.L. Chemical Microstructure of Polymer Chains; Wiley: New York, NY, USA, 1980. [Google Scholar]
- Cheng, H.N. Polymerization and Statistical Models. In Encyclopedia of NMR; Grant, D.M., Harris, R.K., Eds.; Wiley: New York, NY, USA, 1996; pp. 3713–3721. [Google Scholar]
- Cheng, H.N. Cheng. Stereochemistry of Vinyl Polymers and NMR Characterization. J. Appl. Polym. Sci. 1988, 36, 229–241. [Google Scholar] [CrossRef]
- Cheng, H.N.; Babu, G.N.; Newmark, R.A.; Chien, J.C.W. Consecutive Two State Statistical Models for Vinyl Polymerization. Macromolecules 1992, 25, 6980–6987. [Google Scholar] [CrossRef]
- Cheng, H.N. NMR analysis of multicomponent polyolefins. In New Advances in Polyolefins; Chung, T.C., Ed.; Plenum: New York, NY, USA, 1993; pp. 159–174. [Google Scholar]
- Roland, M.T.; Cheng, H.N. Reaction Probability Model for Four Component Copolymerization. Macromolecules 1991, 24, 2015–2018. [Google Scholar] [CrossRef]
- Cheng, H.N.; Kasehagen, L.J. NMR Analysis of Copolymers through Kinetic Modeling. ACS Polym. Prepr. 2003, 44, 381–382. [Google Scholar]
- Cheng, H.N.; Kasehagen, L.J. NMR Studies and Simulation of Polymerization. ACS Polym. Prepr. 1997, 38, 863–864. [Google Scholar]
- Lasprilla, A.J.R.; Martinez, G.A.R.; Lunelli, B.H.; Jardini, A.L.; Filho, R.M. Poly-lactic acid synthesis for application in biomedical devices—A review. Biotechnol. Adv. 2012, 30, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Bergsma, J.E.; Bos, R.R.M.; Rozema, F.R.; Jong, W.D.; Boering, G.J. Biocompatibility of intraosseously implanted predegraded poly(lactide): An animal study. J. Mater. Sci. Mater. Med. 1996, 7, 1–7. [Google Scholar] [CrossRef]
- Fambri, L.; Pergoretti, A.; Fenner, R.; Incardona, S.D.; Migliarisi, C. Biodegradable fibres of poly(L-lactic acid) produced by melt spinning. Polymer 1997, 38, 79–85. [Google Scholar] [CrossRef]
- Masutani, K.; Kimura, Y. PLA synthesis, from the monomer to the polymer. In Poly(Lactic Acid) Science and Technology: Processing, Properties, Additives and Applications; Jiemenz, A., Peltzer, M., Ruseckeite, R., Eds.; RSC Publishing: Cambridge, UK, 2015; pp. 3–36. [Google Scholar]
- Fukushima, K.; Kimura, Y. A novel synthetic approach to stereo-block poly(lactic acid). Macromol. Symp. 2005, 224, 133–143. [Google Scholar] [CrossRef]
- Zell, M.T.; Padden, B.E.; Paterick, A.J.; Thakur, K.A.M.; Kean, R.T.; Hillmyer, M.A.; Munson, E.J. Unambiguous determination of the 13C and 1H NMR stereosequence assignments of polylactide using high resolution solution NMR spectroscopy. Macromolecules 2002, 35, 7700–7707. [Google Scholar] [CrossRef]
- Thakur, K.A.M.; Kean, R.T.; Hall, E.S.; Kolstad, J.J.; Lindgren, T.A.; Doscotch, M.A.; Siepmann, J.I.; Munson, E.J. High-resolution 13C and 1H solution NMR study of poly(lactide). Macromolecules 1997, 30, 2422–2428. [Google Scholar] [CrossRef]
- Suganuma, K.; Horiuchi, K.; Matsuda, H.; Cheng, H.N.; Aoki, A.; Asakura, T. Stereoregularity of poly(lactic acid) and their model compounds as studied by NMR and quantum chemical calculations. Macromolecules 2011, 44, 9247–9253. [Google Scholar] [CrossRef]
- Suganuma, K.; Horiuchi, K.; Matsuda, H.; Cheng, H.N.; Aoki, A.; Asakura, T. NMR Analysis and chemical shift calculations of poly(lactic acid) dimer model compounds with different tacticities. Polym. J. 2012, 44, 838–844. [Google Scholar] [CrossRef]
- Suganuma, K.; Matsuda, H.; Cheng, H.N.; Iwai, M.; Nonokawa, R.; Asakura, T. Tacticity determination of 1H and 13C NMR spectra of poly(lactic acid) in C5D5N. Polym. Test. 2014, 38, 35–39. [Google Scholar] [CrossRef]
- Suganuma, K.; Asakura, T.; Oshimura, M.; Hirano, T.; Ute, K.; Cheng, H.N. NMR analysis of poly(lactic acid) via statistical models. Polymers 2019, 11, 725. [Google Scholar] [CrossRef]
- Kumar, M.; Rathour, R.; Singh, R.; Sun, Y.; Pandey, A.; Gnansounou, E.; Lin, K.A.; Tsang, D.C.W.; Thakur, I.S. Bacterial polyhydroxyalkanoates: Opportunities, challenges, and prospects. J. Clean. Prod. 2020, 263, 121500. [Google Scholar] [CrossRef]
- Anjum, A.; Zuber, M.; Zia, K.M.; Noreen, A.; Anjum, M.N.; Tabasum, S. Microbial production of polyhydroxyalkanoates (PHAs) and its copolymers: A review of recent advancements. Int. J. Biol. Macromol. 2016, 89, 161–174. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, J.; Chen, G.-Q. Polyhydroxyalkanoates, challenges and opportunities. Curr. Opin. Biotechnol. 2014, 30, 59–65. [Google Scholar] [CrossRef]
- Westlie, A.H.; Quinn, E.C.; Parker, C.R.; Chen, E.Y. Synthetic biodegradable polyhydroxyalkanoates (PHAs): Recent advances and future challenges. Prog. Polym. Sci. 2022, 134, 101608. [Google Scholar] [CrossRef]
- Adamus, G.; Domiński, A.; Kowalczuk, M.; Kurcok, P.; Radecka, I. From Anionic Ring-Opening Polymerization of β-Butyrolactone to Biodegradable Poly(hydroxyalkanoate)s: Our Contributions in This Field. Polymers 2021, 13, 4365. [Google Scholar] [CrossRef]
- Cherpinski, A.; Torres-Giner, S.; Vartiainen, J.; Peresin, M.S.; Lahtinen, P.; Lagaron, J.M. Improving the water resistance of nanocellulose-based films with polyhydroxyalkanoates processed by the electrospinning coating technique. Cellulose 2018, 25, 1291–1307. [Google Scholar] [CrossRef]
- Gross, R.A.; Demello, A.C.; Lenz, R.W.; Brandl, H.; Fuller, R.C. The biosynthesis and characterization of poly(β-hydroxyalkanoates) produced by Pseudomonas oleovorans. Macromolecules 1989, 22, 1106–1115. [Google Scholar] [CrossRef]
- Noda, I.; Green, P.R.; Satkowski, M.M.; Schechtman, L.A. Preparation and properties of a novel class of polyhydroxyalkanote copolymers. Biomacromolecules 2005, 6, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, G.; Zhou, X.R.; Chen, G.Q. Biosynthesis of poly(3-hydroxydecanoate) and 3-hydroxydodecanoate dominating polyhydroxyalkanoates by β-oxidation pathway inhibited Pseudomonas putida. Metab. Eng. 2011, 13, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Matsusaki, H.; Abe, H.; Taguchi, K.; Fukui, T.; Doi, Y. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) by recombinant bacteria expressing the PHA synthase gene phaC1 from Pseudomonas sp. 61-3. Appl. Microbiol. Biotechnol. 2000, 53, 401–409. [Google Scholar] [CrossRef]
- Phithakrotchanakoon, C.; Champreda, V.; Aiba, S.; Pootanakit, K.; Tanapongpipat, S. Production of polyhydroxyalkanoates from crude glycerol using recombinant Escherichia coli. J. Polym. Environ. 2015, 23, 38–44. [Google Scholar] [CrossRef]
- Cheng, H.N.; Biswas, A.; Vermillion, K.; Melendez-Rodriguez, B.; Lagaron, S.M. NMR Analysis and Triad Sequence Distributions of Poly(3-hydroxybutyrate-co-3-hydroxyvalerate). Polym. Test. 2020, 90, 106754. [Google Scholar] [CrossRef]
- Abe, M.M.; Martins, J.R.; Sanvezzo, P.B.; Macedo, J.V.; Branciforti, M.C.; Halley, P.; Botaro, V.R.; Brienzo, M. Advantages and Disadvantages of Bioplastics Production from Starch and Lignocellulosic Components. Polymers 2021, 13, 2484. [Google Scholar] [CrossRef]
- Lim, C.; Yusoff, S.; Ng, C.G.; Lim, P.E.; Ching, Y.C. Bioplastic made from seaweed polysaccharides with green production methods. J. Environ. Chem. Eng. 2021, 9, 105895. [Google Scholar] [CrossRef]
- Whistler, R.L.; BeMiller, J.N. (Eds.) Industrial Gums: Polysaccharides and their Derivatives, 3rd ed.; Academic Press: San Diego, CA, USA, 1993. [Google Scholar]
- Shaghaleh, H.; Xu, X.; Wang, S. Current progress in production of biopolymeric materials based on cellulose, cellulose nanofibers, and cellulose derivatives. RSC Adv. 2018, 8, 825–842. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.N.; Biswas, A.; Furtado, R.F.; Alves, C.A. Hydrophobic Modifications of Agro-based Polymers—A Selected Review. ACS Symp. Ser. 2023, 1450, 249–258. [Google Scholar]
- Cheng, H.N. Use of green chemistry for process development and improvement. In Green Chemistry: Research and Connections to Climate Change; Benvenuto, M.A., Welch, L., Eds.; De Gruyter: Berlin, Germany; Boston, MA, USA, 2023; pp. 1–12. [Google Scholar]
- Cheng, H.N.; Neiss, T.G. NMR Spectroscopy of Food Polysaccharides in Solution. Polym. Rev. 2012, 52, 81–114. [Google Scholar] [CrossRef]
- Yao, H.Y.; Wang, J.; Yin, J.; Nie, S.; Xie, M. A review of NMR analysis in polysaccharide structure and conformation: Progress, challenge and perspective. Food Res. Int. 2021, 143, 110290. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, L. Chemical structural and chain conformational characterization of some bioactive polysaccharides isolated from natural sources. Carbohydr. Polym. 2009, 76, 349–361. [Google Scholar] [CrossRef]
- Ren, Y.; Bai, Y.; Zhang, Z.; Cai, W.; Flores, A.D.R. The Preparation and Structure Analysis Methods of Natural Polysaccharides of Plants and Fungi: A Review of Recent Development. Molecules 2019, 24, 3122. [Google Scholar] [CrossRef] [PubMed]
- Pettolino, F.A.; Walsh, C.; Fincher, G.B.; Bacic, A. Determining the polysaccharide composition of plant cell walls. Nat. Protoc. 2012, 7, 1590–1607. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.N.; Edgar, K.J. Alginate derivatization: A review of chemistry, properties and applications. Biomaterials 2012, 33, 3279–3305. [Google Scholar] [CrossRef] [PubMed]
- Grasdalen, H.; Larsen, B.; Smidsrod, O. 13C-n.m.r. studies of monomeric composition and sequence in alginate. Carbohydr. Res. 1981, 89, 179–191. [Google Scholar] [CrossRef]
- Grasdalen, H. High-field ‘H-n.m.r. spectroscopy of alginate: Sequential structure and linkage conformations. Carbohydr. Res. 1983, 118, 255–260. [Google Scholar] [CrossRef]
- Cheng, H.N. Compositional Heterogeneity of Alginates through NMR Analysis. Polym. Bull. 1999, 43, 247. [Google Scholar] [CrossRef]
- Neiss, T.G.; Cheng, H.N. Analysis of Polysaccharides by NMR and Separation Techniques. ACS Polym. Prepr. 2001, 42, 76–77. [Google Scholar]
- Neiss, T.G.; Cheng, H.N. Coupled SEC-NMR Analysis of Alginates. ACS Symp. Ser. 2002, 834, 382–395. [Google Scholar]
- Kawarada, H.; Hirai, A.; Odani, H.; Lida, T.; Nakajima, A. Structure characterization of alginate and conformational behaviors of various alkali-metal alginates in solution. Polym. Bull. 1990, 24, 551–557. [Google Scholar] [CrossRef]
- Cheng, H.N. (USDA-ARS-SRRC, New Orleans, LA, USA). Personal communication.
- Freitas, C.M.P.; Coimbra, J.S.R.; Souza, V.G.; Sousa, R.C.S. Structure and Applications of Pectin in Food, Biomedical, and Pharmaceutical Industry: A Review. Coatings 2021, 11, 922. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Neiss, T.G.; Cheng, H.N.; Daas, P.J.H.; Schols, H.A. NMR and statistical analysis of the galacturonic acid and methyl ester distributions in pectic polysaccharides. ACS Polym. Prepr. 1998, 39, 688–689. [Google Scholar]
- Neiss, T.G.; Cheng, H.N.; Daas, P.J.H.; Schols, H.A. Compositional Heterogeneity in Pectic Polysaccharides: NMR and Statistical Analysis. Macromol. Symp. 1999, 140, 165–178. [Google Scholar] [CrossRef]
- Kou, S.G.; Peters, L.M.; Mucalo, M.R. Chitosan: A review of sources and preparation methods. Int. J. Biol. Macromol. 2021, 169, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Ahmed, S. A review on chitosan and its nanocomposites in drug delivery. Int. J. Biol. Macromol. 2018, 109, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Varum, K.M.; Anthonsen, M.W.; Grasdalen, H.; Smidsrod, O. Determination of the degree of N-acetylation and the distribution of N-acetyl groups in partially N-deacetylated chitins (chitosans) by high-field n.m.r. spectroscopy. Carbohydr. Res. 1991, 211, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Varum, K.M.; Anthonsen, M.W.; Grasdalen, H.; Smidsrod, O. 13C-N.m.r. studies of the acetylation sequences in partially N-deacetylated chitins (chitosans). Carbohydr. Res. 1991, 217, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.N. Compositional Heterogeneity in NMR Polymer Analysis. ACS Polym. Prepr. 2003, 44, 261–262. [Google Scholar]
- Moser, E.; Laistler, E.; Schmitt, F.; Kontaxis, G. Ultra-High Field NMR and MRI—The Role of Magnet Technology to Increase Sensitivity and Specificity. Front. Phys. 2017, 5, 33, Corrigendum Front. Phys. 2017, 5, 41. [Google Scholar] [CrossRef]
- Wren, E. Evidence that NMR Chemical Shifts Depend on Magnetic Field Strength. Chemistry World. 29 April 2020. Available online: https://www.chemistryworld.com/news/evidence-that-nmr-chemical-shifts-depend-on-magnetic-field-strength/4011613.article (accessed on 6 January 2024).


| NMR Information | Statistical Models | References |
|---|---|---|
| Homopolymer tacticity and copolymer sequence | 1. One-component models (discrete) a. Chain-end control: B, M1, M2 b. Catalytic-site control: E model c. Both end and site control: EM1, EM2 | [5,40,41,42] |
| 2. Two-component models (discrete) a. Consecutive B/B, B/E, E/E b. Concurrent B/B, B/E, E/E | [43] | |
| 3. Multi-component models (discrete) a. Consecutive multisite models b. Concurrent multisite models | [37,44] | |
| 4. Perturbed models (continuous) a. Symmetric B and M1 models b. Non-symmetric B and M1 models c. General-case models | [38,39] | |
| Terpolymers and Tetrapolymers | Higher-copolymerization models | [45] |
| Branched polymers and more complex polymers | Kinetic models | [46,47] |
| Tetrad | Model 1 a | Model 2 b | Model 3 c | Obsd. % | Calc. % Mod. 1 | Calc. % Mod. 3 |
|---|---|---|---|---|---|---|
| mmm | (p22 + q22 + p23 + q23)/2 | p14 + q14 | f2[(p22 + q22 + p23 + q23)/2] + f1[p14 + q14] | 39.9 | 40.1 | 39.9 |
| mrm | p2q2 | 2p12q12 | f2 p2q2 + f1 [2p12q12] | 21.4 | 24.0 | 21.3 |
| mmr | p2q2/2 | p13q1 + p1q13 | f2 p2q2/2 + f1 [p13q1 + p1q13] | 11.3 | 12.0 | 11.6 |
| rmm | p2q2/2 | p13q1 + p1q13 | f2 p2q2/2 + f1 [p13q1 + p1q13] | 10.9 | 12.0 | 11.6 |
| rmr | p2q2/2 | 2p12q12 | f2 p2q2/2 + f1 [2p12q12] | 12.0 | 12.0 | 11.3 |
| rrm | 0 | p13q1 + p1q13 | f1 [p13q1 + p1q13] | 2.5 | 0 | 1.5 |
| mrr | 0 | p13q1 + p1q13 | f1 [p13q1 + p1q13] | 1.5 | 0 | 1.5 |
| rrr | 0 | 2p12q12 | f1 [2p12q12] | 0.7 | 0 | 1.2 |
| MD | 1.2 | 0.4 | ||||
| Reaction probabilities | p2 = 0.6 | p1 = 0.66 f1 = 0.12 p2 = 0.65 f2 = 0.88 |
| Triad | Obsd % | M1 Model Expressions * | Calc. % | Two-Component B/B Model Expressions ** | Calc. % |
|---|---|---|---|---|---|
| VVV | 4.7 | kPBVPVV2 | 4.7 | w1PV13 + w2PV23 | 4.7 |
| BVV + VVB | 8.3 | 2kPBVPVVPVB | 11.6 | 2w1PV12(1 − Pv1) + 2w2PV22(1 − Pv2) | 8.3 |
| BVB | 9.7 | kPVB2PBV | 7.1 | w1PV1(1 − Pv1)2 + w2PV2(1 − Pv2)2 | 9.9 |
| VBV | 3.9 | kPBV2PVB | 2.2 | w1PV12(1 − Pv1) + w2PV22(1 − Pv2) | 4.1 |
| BBV + VBB | 20.3 | 2kPVBPBBPBV | 21.4 | 2w1PV1(1 − Pv1)2 + 2w2PV2(1 − Pv2)2 | 19.9 |
| BBB | 53.1 | kPVBPBB2 | 53.1 | w1(1 − PV1)3 + w2(1 − PV2)3 | 53.1 |
| mean deviation | 1.5 | mean deviation | 0.2 | ||
| PBV | 0.168 | PV1 | 0.134 | ||
| PVB | 0.551 | w1 | 0.802 | ||
| r1 r2 | 4.03 | PV2 | 0.610 | ||
| w2 | 0.198 |
| NMR Triad | Obsd % | Discrete Models | Continuous Model | |
|---|---|---|---|---|
| Calc % (for B) | Calc % (for B/B) | Calc % (for Perturbed B) | ||
| MMM | 39 | 39 | 39 | 39 |
| MMG | 17 | 29 | 19 | 18 |
| GMG | 8 | 5 | 7 | 7 |
| MGM | 10 | 14 | 9 | 9 |
| GGM | 14 | 11 | 14 | 15 |
| GGG | 12 | 2 | 12 | 12 |
| Mean dev. | 5.4 | 0.6 | 0.6 | |
| Reaction probabilities | PM = 0.731 | Component 1: w1 = 0.592 PM = 0.858 Component 2: w2 = 0.408 PM = 0.338 | PM = 0.648 σ = 0.253 τ = −0.004 | |
| NMR Triad | Obsd. % | Discrete Models | Continuous Model | |
|---|---|---|---|---|
| Calc % (for B) | Calc % (for B/B) | Calc % (for Perturbed B) | ||
| EEE | 32.4 | 32.4 | 32.5 | 32.4 |
| EEG | 28.0 | 29.5 | 28.0 | 27.5 |
| GEG | 6.4 | 6.7 | 7.1 | 6.7 |
| EGE | 13.0 | 14.8 | 14.0 | 13.8 |
| GGE | 14.0 | 13.5 | 14.0 | 13.4 |
| GGG | 6.2 | 3.1 | 4.4 | 6.2 |
| Mean dev. | 1.2 | 0.6 | 0.4 | |
| Reaction probabilities | PE = 0.687 | Component 1: w1 = 0.793 PE = 0.724 Component 2: w2 = 0.207 PE = 0.489 | PE = 0.675 σ = 0.118 τ = −0.008 | |
| NMR Triad | Obsd. % | Discrete Models | Continuous Model | |
|---|---|---|---|---|
| Calc % (for B) | Calc % (for B/B) | Calc % (for Perturbed B) | ||
| EEE | 2.1 | 1.6 | 2.4 | 2.5 |
| EEG | 10.6 | 9.6 | 10.6 | 10.6 |
| GEG | 13.4 | 14.1 | 13.3 | 13.3 |
| EGE | 5.5 | 4.8 | 5.3 | 5.3 |
| GGE | 26.7 | 28.2 | 26.7 | 26.6 |
| GGG | 41.7 | 41.7 | 41.7 | 41.7 |
| Mean dev. | 0.8 | 0.1 | 0.1 | |
| Reaction probabilities | PE = 0.253 | Component 1: w1 = 0.337 PE = 0.586 Component 2: w2 = 0.159 PE = 0.414 | PE = 0.264 σ = 0.0928 τ = 0.0003 | |
| NMR Triad | Obsd. % | Discrete Models | Continuous Model | |
|---|---|---|---|---|
| Calc % (for B) | Calc % (for B/B) | Calc % (for Perturbed B) | ||
| AAA | 15 | 15 | 16 | 15 |
| AAD | 28 | 27 | 25 | 28 |
| DAD | 10 | 12 | 10 | 9 |
| ADA | 14 | 13 | 13 | 14 |
| DDA | 16 | 23 | 20 | 17 |
| DDD | 17 | 10 | 17 | 17 |
| Mean dev. | 3.0 | 1.5 | 0.4 | |
| Reaction probabilities | PA = 0.531 | Component 1: w1 = 0.906 PA = 0.357 Component 2: w2 = 0.094 PA = 0.009 | PA = 0.547 σ = 0.083 τ = −0.031 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, H.N.; Asakura, T.; Suganuma, K.; Lagaron, J.M.; Melendez-Rodriguez, B.; Biswas, A. NMR Analyses and Statistical Modeling of Biobased Polymer Microstructures—A Selected Review. Polymers 2024, 16, 620. https://doi.org/10.3390/polym16050620
Cheng HN, Asakura T, Suganuma K, Lagaron JM, Melendez-Rodriguez B, Biswas A. NMR Analyses and Statistical Modeling of Biobased Polymer Microstructures—A Selected Review. Polymers. 2024; 16(5):620. https://doi.org/10.3390/polym16050620
Chicago/Turabian StyleCheng, Huai N., Tetsuo Asakura, Koto Suganuma, Jose M. Lagaron, Beatriz Melendez-Rodriguez, and Atanu Biswas. 2024. "NMR Analyses and Statistical Modeling of Biobased Polymer Microstructures—A Selected Review" Polymers 16, no. 5: 620. https://doi.org/10.3390/polym16050620
APA StyleCheng, H. N., Asakura, T., Suganuma, K., Lagaron, J. M., Melendez-Rodriguez, B., & Biswas, A. (2024). NMR Analyses and Statistical Modeling of Biobased Polymer Microstructures—A Selected Review. Polymers, 16(5), 620. https://doi.org/10.3390/polym16050620








