Purification and Characterization of Transglutaminase Isolated from Sardine (Sardina pilchardus) Flesh Waste
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Transglutaminase Extraction and Purification
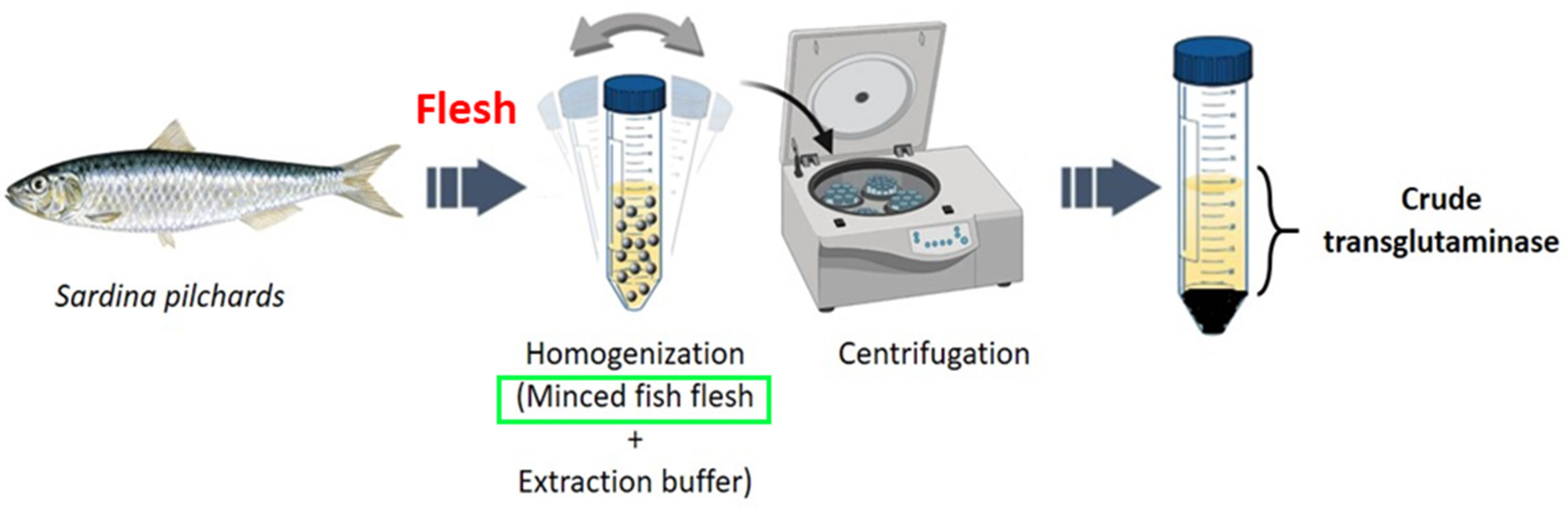
2.2.1. Preparation of the Crude Transglutaminase Extract
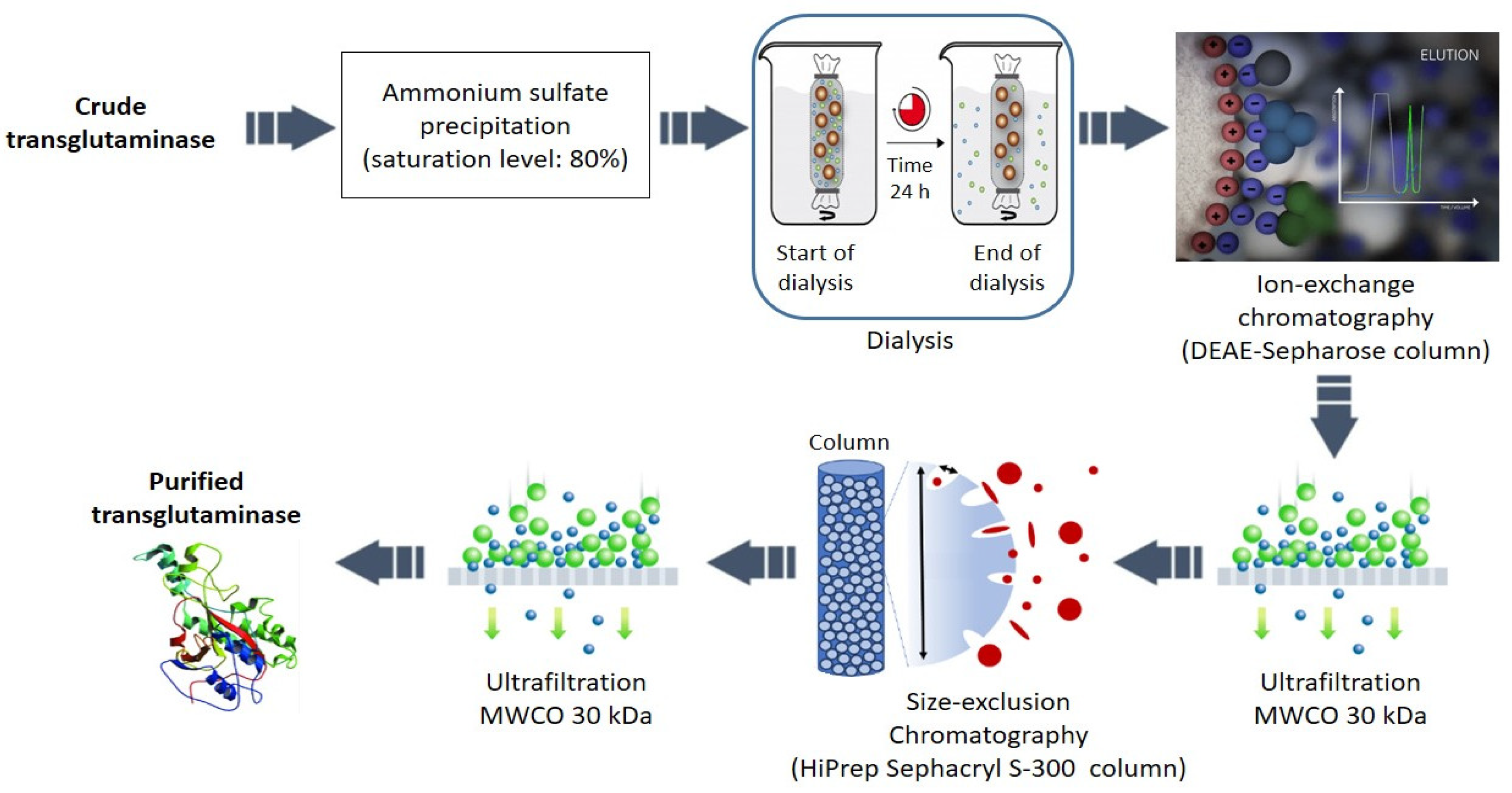
2.2.2. Purification of Sardine Transglutaminase
2.3. Characterization of Sardine Transglutaminase
2.3.1. Transglutaminase Activity Assay
2.3.2. Purification Fold and Yield of Transglutaminase
2.3.3. Protein Content
2.3.4. SDS–PAGE Electrophoresis and Molecular Weight
2.3.5. Optimal pH
2.3.6. Optimal Temperature
2.3.7. Thermal Stability
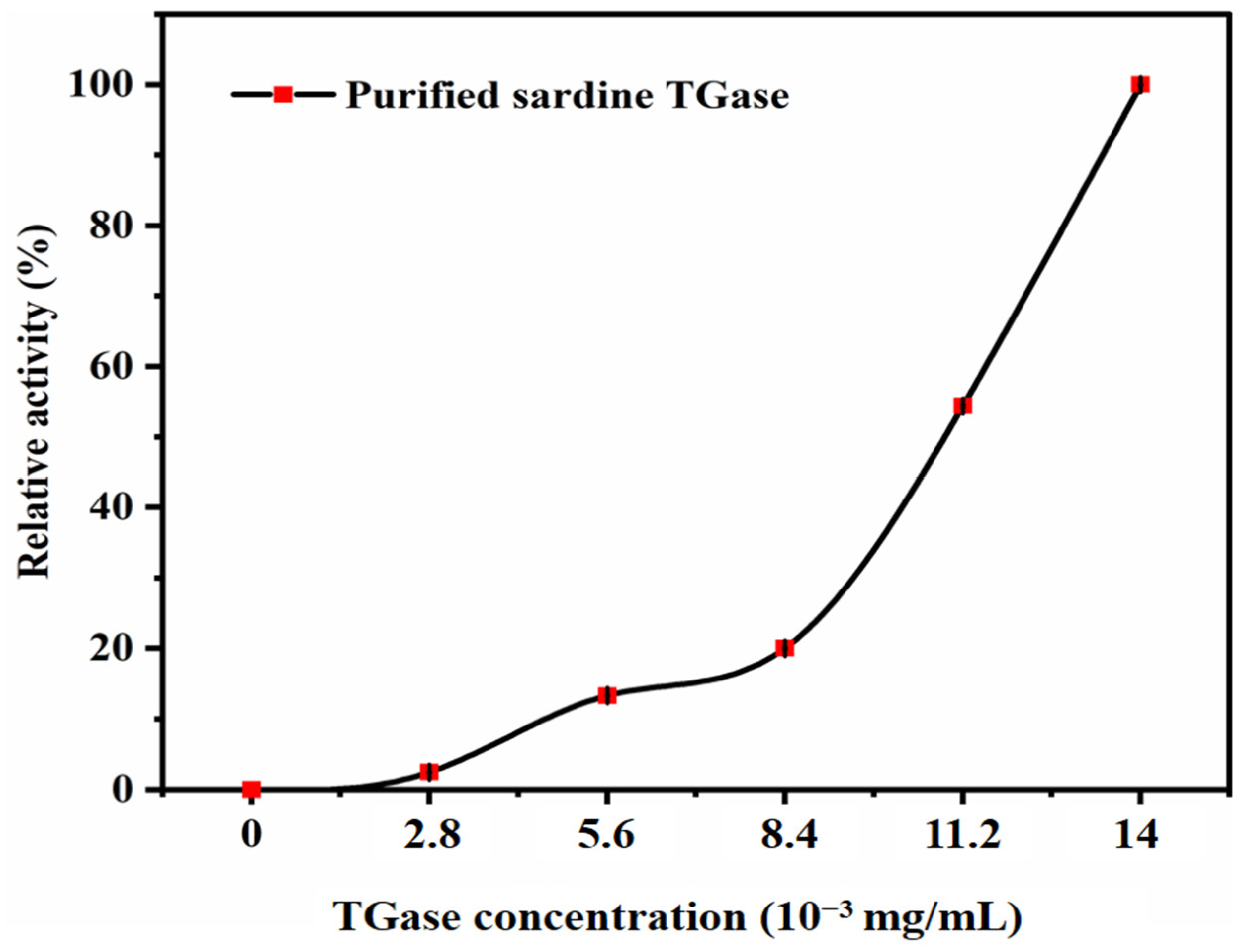
2.3.8. Effect of Enzyme Concentration on Transglutaminase Activity
2.3.9. Effect of Salts, Chelating Agents, and Metal Ions on Transglutaminase Activity
2.4. Effect of Added Sardine Transglutaminase on the Natural Actomyosin (NAM) Cross-Linking
2.4.1. Incubation of NAM with TGase
2.4.2. Protein Solubility
2.4.3. Surface Hydrophobicity
2.5. Statistical Analysis
3. Results and Discussion
3.1. Transglutaminase Extraction and Purification
3.2. Transglutaminase Characterization
3.2.1. Purity and Molecular Weight
3.2.2. Optimal pH
3.2.3. Optimal Temperature
3.2.4. Thermal Stability
3.2.5. Effect of Enzyme Concentration on Transglutaminase Activity
3.2.6. Effect of Chemical Additives on Enzyme Activity
Effect of Calcium Chloride (CaCl2)
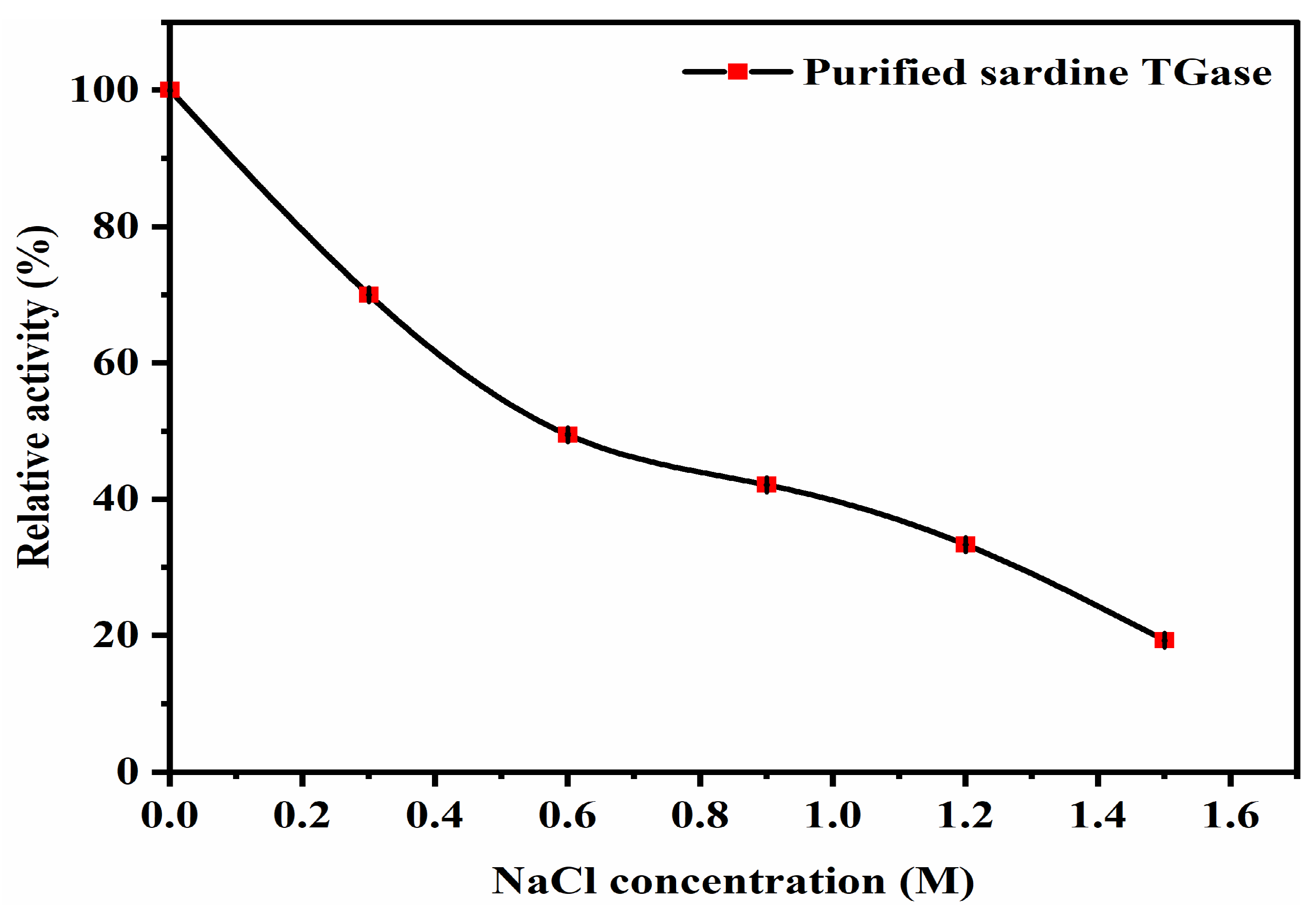
Effect of Sodium Chloride (NaCl)
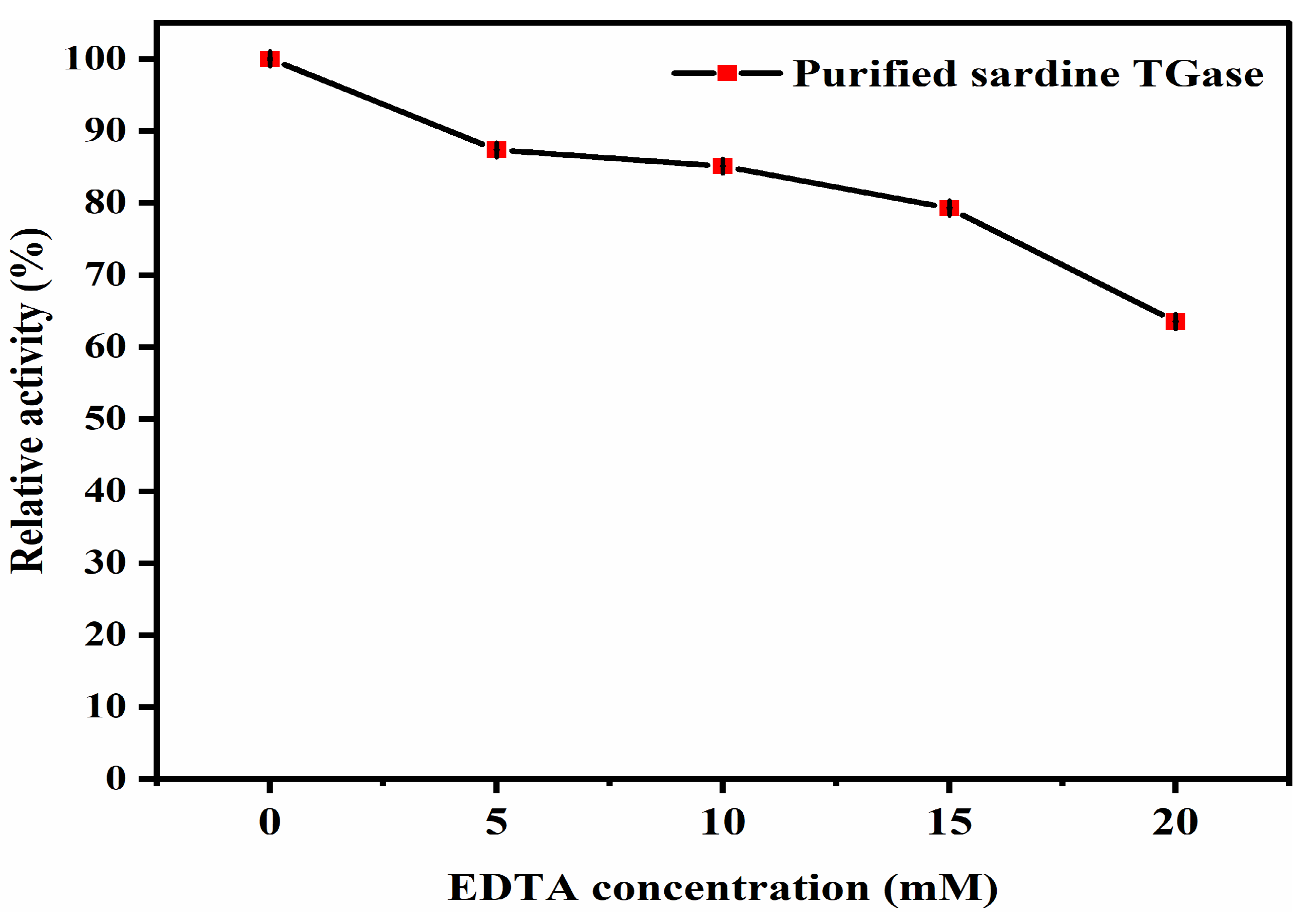
Effect of Ethylenediaminetetraacetic Acid (EDTA)
Effect of Metal Ions on Transglutaminase Activity
3.3. Effect of Added Sardine Transglutaminase on the Natural Actomyosin (NAM) Cross-Linking
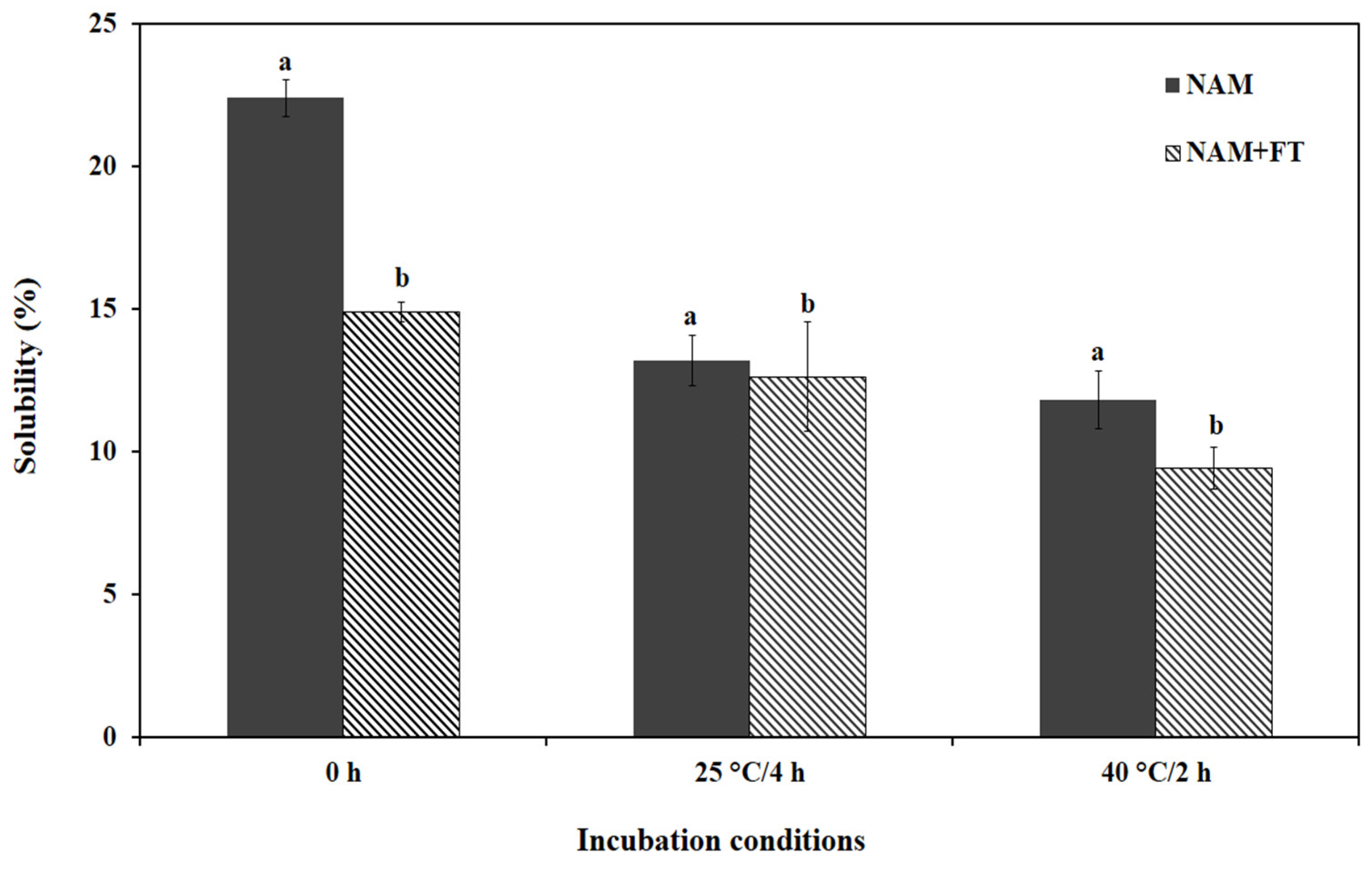
3.3.1. Protein Solubility
3.3.2. Surface Hydrophobicity
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Long, M.; Zheng, N.; Deng, Y.; Wang, Q.; Osire, T.; Xia, T. Microstructural, physicochemical properties, and interaction mechanism of hydrogel nanoparticles modified by high catalytic activity transglutaminase crosslinking. Food Hydrocoll. 2024, 147, 109384. [Google Scholar] [CrossRef]
- Alhasani, H.A.W.; Al-Younis, Z.K. Extraction, purification and characterization of transglutaminase from some plants. In Proceedings of the 4th International Conference for Agricultural and Sustainability Sciences, IOP Conference Series: Earth Environmental Science, Online, 4–5 October 2021; Volume 910, p. 012061. [Google Scholar]
- Sulaiman, N.S.; Sintang, M.D.; Zaini, H.M.; Munsu, E.; Matanjun, P.; Pindi, W. Applications of protein crosslinking in food products. Int. Food Res. J. 2022, 29, 723–739. [Google Scholar] [CrossRef]
- Binsi, P.K.; Shamasundar, B.A. Purification and characterisation of transglutaminase from four fish species: Effect of added transglutaminase on the viscoelastic behaviour of fish mince. Food Chem. 2012, 132, 1922–1929. [Google Scholar] [CrossRef]
- Gaspar, A.L.C.; de Góes-Favoni, S.P. Action of microbial transglutaminase (MTGase) in the modification of food proteins: A review. Food Chem. 2015, 171, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Amirdivani, S.; Khorshidian, N.; Fidelis, M.; Granato, D.; Koushki, M.R.; Mohammadi, M.; Khoshtinat, K.; Mortazavian, A.M. Effects of transglutaminase on health properties of food products. Curr. Opin. Food Sci. 2018, 22, 74–80. [Google Scholar] [CrossRef]
- El-Hofi, M.; Ismail, A.; Nour, M.; Ibrahim, O. Isolation, purification and characterisation of transglutaminase from rosemary (Rosmarinus officinalis L.) leaves. Acta Sci. Pol. Technol. Aliment. 2014, 13, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, S.; Simpson, B.K. A cold active transglutaminase from Antarctic krill (Euphausia superba): Purification, characterization and application in the modification of cold-set gelatin gel. Food Chem. 2017, 232, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Icekson, I.; Apelbaum, A. Evidence for transglutaminase activity in plant tissue. Plant Physiol. 1987, 84, 972–974. [Google Scholar] [CrossRef] [PubMed]
- Signorini, M.; Beninati, S.; Bergamini, D. Identification of transglutaminase activity in the leaves of silver beet (Beta vulgaris L.). J. Plant Physiol. 1991, 137, 547–552. [Google Scholar] [CrossRef]
- Kang, H.; Cho, Y.D. Purification and properties of transglutaminase from soybean (Glycine max) leaves. Biochem. Biophys. Res. Commun. 1996, 223, 288–292. [Google Scholar] [CrossRef]
- Worratao, A.; Yongsawatdigul, J. Purification and characterization of transglutaminase from Tropical tilapia (Oreochromis niloticus). Food Chem. 2005, 93, 651–658. [Google Scholar] [CrossRef]
- Batista, I.; Salteiro, A.T.; Mateus, M.J. Preliminary Characterization of European Sardine Transglutaminase. J. Aquat. Food Prod. Technol. 2002, 11, 57–64. [Google Scholar] [CrossRef]
- Nozawa, H.; Mamegoshi, S.; Seki, N. Partial purification and characterization of six transglutaminases from ordinary muscles of various fishes and marine invertebrates. Comp. Biochem. Physiol. B 1997, 118, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Kishi, H.; Nozawa, H.; Seki, N. Reactivity of muscle transglutaminase on carp myofibrils and myosin B. Nippon Suisan Gakk. 1991, 57, 1203–1210. [Google Scholar] [CrossRef]
- Vasić, K.; Knez, Ž.; Leitgeb, M. Transglutaminase in Foods and Biotechnology. Int J Mol Sci. 2023, 24, 12402. [Google Scholar] [CrossRef] [PubMed]
- Imsiridou, A.; Karnezi, S.; Minos, G.; Exadactylos, A. Comparison of European sardine (Sardina pilchardus, Walbaum 1792) Greek halptypes with those found in the global distribution of the species. Appl. Ecol. Env. Res. 2021, 19, 4025–4035. [Google Scholar] [CrossRef]
- IMARC Group. Sardine Market Report by Species, Type, Distribution Channel, and Region 2024–2032; Report ID: 5936388; Research and Markets: Dublin, Ireland, 2024; p. 146. Available online: https://www.researchandmarkets.com/report/sardines (accessed on 27 January 2025).
- Pérez Roda, M.A.; Gilman, E.; Huntington, T.; Kennelly, S.J.; Suuronen, P.; Chaloupka, M.; Medley, P. A Third Assessment of Global Marine Fisheries Discards. In ICES Document No. 63; ICES: Copenhagen, Denmark, 2019; p. 78. [Google Scholar]
- Manni, L.; Zouine, N.; Ouahidi, I.; Bouraqqadi, M.; Ananou, S. Purification of Trypsin from Sardine (Sardina pilchardus) Viscera and Its Application in Preparation of Antioxidative Fish Protein Hydrolysates. Trop. J. Nat. Prod. Res. 2024, 8, 5881–5888. [Google Scholar] [CrossRef]
- Ferraro, V.; Carvalho, A.P.; Piccirillo, C.; Santos, M.M.; Castro, P.M.; Pintado, M.E. Extraction of high added value biological compounds from sardine, sardinetype fish and mackerel canning residues—A review. Mater. Sci. Eng. C. 2013, 33, 3111–3120. [Google Scholar] [CrossRef] [PubMed]
- Tsukamasa, Y.; Miyake, Y.; Ando, M.; Makinodan, Y. Total activity of transglutaminase at various temperatures in several fish meats. Fish. Sci. 2002, 68, 929–933. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitve method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1979, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 15, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hemung, B.; Eunice, C.Y.; Li-Chan, B.; Yongsawatdigul, J. Thermal stability of fish natural actomyosin affects reactivity to cross-linking by microbial and fish transglutaminases. Food Chem. 2008, 111, 439–446. [Google Scholar] [CrossRef]
- Wang, J.Y.; Yang, Y.L.; Tang, X.Z.; Ni, W.X.; Zhou, L. Effects of pulsed ultrasound on rheological and structural properties of chicken myofibrillar protein. Ultrason. Sonochem. 2017, 38, 225–233. [Google Scholar] [CrossRef]
- Benjakul, S.; Seymour, T.A.; Morrissey, M.T.; An, H. Physicochemical changes in Pacific whiting muscle proteins during iced storage. J. Food Sci. 1997, 62, 729–733. [Google Scholar] [CrossRef]
- Confortin, T.C.; Spannemberg, S.S.; Todero, I.; Luft, L.; Brun, T.; Alves, E.A.; Kuhn, R.C.; Marcio, A.; Mazutti, M.A. Microbial Enzymes as Control Agents of Diseases and Pests in Organic Agriculture. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 321–332. ISBN 9780444635044. [Google Scholar]
- Westphal, A.H.; van Berkel, W.J.H. Techniques for Enzyme Purification. In Biocatalysis for Practitioners: Techniques, Reactions and Applications, 1st ed.; de Gonzalo, G., Lavandera, I., Eds.; WILEY-VCH GmbH: Weinheim, Germany, 2021. [Google Scholar]
- Gendeh, G.; Decrop, W.; Olivo, M.J.; Sneekes, E.J.; Swart, R. Exploration of pH-Gradient Ion-Exchange Chromatography for High-Resolution Protein Separations in Biotechnology and Proteomics; Thermo Fisher Scientific Note: Waltham, MA, USA, 2012; pp. 1–7. Available online: https://tools.thermofisher.com/content/sfs/posters/110535-PN-LC-pH-Gradient-Proteins-12Mar2012-PO70013.pdf (accessed on 1 November 2024).
- Laksono, U.T.; Suprihatin, S.; Nurhayati, T.; Romli, M. Purification of endogenous transglutaminase from daggertooth pike conger fish (Muraenesox cinerus) meat. Int. Food Res. J. 2021, 28, 1030–1037. [Google Scholar] [CrossRef]
- Hemung, B.O.; Yongsatwadigul, J. Partial purification and characterisation of transglutaminase from threadfin bream (Nemipterus sp.) liver. J. Food Biochem. 2008, 32, 182–200. [Google Scholar] [CrossRef]
- Kumazawa, Y.; Sano, K.I.; Seguro, K.; Yasueda, H.; Nio, N.; Motoki, M. Purification and characterization of transglutaminase from Japanese oyster (Crassostrea gigas). J. Agric. Food Chem. 1997, 45, 604–610. [Google Scholar] [CrossRef]
- Kumazawa, Y.; Nakanishi, K.; Yasueda, H.; Motoki, M. Purification and characterization of transglutaminase from walleye pollack liver. Fish. Sci. 1996, 62, 959–964. [Google Scholar] [CrossRef]
- Hemker, A.K.; Nguyen, L.T.; Salvi, D. Effect of high-pressure technologies on enzyme activity and stability. In Effect of High-Pressure Technologies on Enzymes: Science and Applications, 1st ed.; Júnior, B.R.C.L., Tribst, A.A.L., Eds.; Elsevier Inc. Academic Press: Amsterdam, The Netherlands, 2023; pp. 49–75. [Google Scholar]
- Klesk, K.; Yongsawatdigul, J.; Park, J.W.; Viratchakul, S.; Virulhakul, P. Gel forming ability of tropical tilapia surimi as compared with Alaska pollock and Pacific whiting surimi. J. Aquat. Food Prod. Technol. 2000, 9, 91–104. [Google Scholar] [CrossRef]
- Evstatiev, R.; Cervenka, A.; Austerlitz, T.; Deim, G.; Baumgartner, M.; Beer, A.; Krnjic, A.; Gmainer, C.; Lang, M.; Frick, A.; et al. The food additive EDTA aggravates colitis and colon carcinogenesis in mouse models. Sci. Rep. 2021, 11, 5188. [Google Scholar] [CrossRef]
- Oviedo, C.; Rodríguez, J. EDTA: The chelating agent under environmental scrutiny. Quím. Nova 2003, 26, 901–905. [Google Scholar] [CrossRef]
- de Cassia Pereira, J.; Giese, E.C.; de Souza Moretti, M.M.; dos Santos Gomes, A.C.; Perrone, O.M.; Boscolo, M.; da Silva, R.; Gomes, E.; Martins, D.A.B. Effect of Metal Ions, Chemical Agents and Organic Compounds on Lignocellulolytic Enzymes Activities. In Enzyme Inhibitors and Activators; Senturk, M., Ed.; IntechOpen: London, UK, 2017. [Google Scholar]
- Sun, Q.; Kong, B.; Zheng, O.; Liu, S.; Dong, X. Effect of protein structure changes during different power ultrasound thawing on emulsification properties of common carp (Cyprinus carpio) myofibrillar protein. Ultrason. Sonochem. 2023, 101, 106719. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.P.; Xiong, Y.L. Microbial transglutaminase-induced structural and rheological changes of cationic and anionic myofibrillar proteins. Meat Sci. 2012, 91, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.A.; Ahmed, S.H.; Mohamed, E.A.; Ahmed, I.A.M.; Babiker, E.E. Effect of transglutaminase cross linking on the functional properties as a function of NaCl concentration of legumes protein isolate. Int. J. Biol. Life Sci. 2010, 6, 8–13. [Google Scholar]
- Li, T.; Wang, C.; Li, T.; Ma, L.; Sun, D.; Hou, J.; Jiang, Z. Surface hydrophobicity and functional properties of citric acid cross-linked whey protein isolate: The impact of pH and concentration of citric acid. Molecules 2018, 23, 2383. [Google Scholar] [CrossRef] [PubMed]







| Purification Step | Total Protein * (mg/mL) | Total Activity (U/mL) | Specific Activity (U/mg Protein) | Purification Fold | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 862.29 | 1683.00 | 1.95 | 1.00 | 100.00 |
| Dialyzed fraction | 227.84 | 156.00 | 0.68 | 0.35 | 98.11 |
| DEAE-Sepharose | 0.84 | 136.08 | 162.16 | 83.15 | 87.23 |
| Hiprep Sephacryl S-300 | 0.14 | 50.00 | 357.14 | 183.15 | 36.74 |
| Common Name | Species | Specific Activity (U/mg) | Yield (%) | References |
|---|---|---|---|---|
| Sardine | Sardina pilchardus | 357.14 | 36.74 | This study |
| Daggertooth pike conger | Muraenesox cinerus | 3.65 | 17.14 | Lakonso et al. [31] |
| Bigeye snapper | Priacanthus hamrur | 47.44 | 29.54 | Binsi and Shamasundar [4] |
| Oil sardine | Sardinella longiceps | 66.35 | 24.85 | |
| Tilapia | Oreochromis mossambicus | 69.14 | 22.66 | |
| Common carp | Cyprinus carpio | 49.33 | 33.01 | |
| Tropical tilapia | Oreochromis niloticus | 197.00 | 12.90 | Warratao and Yongsawatdigul [12] |
| Carp | Cyprinus carpio | 8.60 | 24.00 | Nozawa et al. [14] |
| Rainbow trout | Oncorhynchus mykiss | 1.17 | 21.00 | |
| Atka mackerel | Pleurogrammus azonus | 1.10 | 2.00 |
| Reagent | Concentration (mM) | Relative Activity (%) |
|---|---|---|
| Control: CaCl2 | 10 | 100 |
| Ba2+ | 10 | 24.91 ± 1.6 |
| Mg2+ | 10 | 55.2 ± 1.58 |
| Sr2+ | 10 | 0 |
| Zn2+ | 10 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaghbib, I.; Abdullah, J.A.A.; Hassouna, M.; Romero, A. Purification and Characterization of Transglutaminase Isolated from Sardine (Sardina pilchardus) Flesh Waste. Polymers 2025, 17, 510. https://doi.org/10.3390/polym17040510
Zaghbib I, Abdullah JAA, Hassouna M, Romero A. Purification and Characterization of Transglutaminase Isolated from Sardine (Sardina pilchardus) Flesh Waste. Polymers. 2025; 17(4):510. https://doi.org/10.3390/polym17040510
Chicago/Turabian StyleZaghbib, Imen, Johar Amin Ahmed Abdullah, Mnasser Hassouna, and Alberto Romero. 2025. "Purification and Characterization of Transglutaminase Isolated from Sardine (Sardina pilchardus) Flesh Waste" Polymers 17, no. 4: 510. https://doi.org/10.3390/polym17040510
APA StyleZaghbib, I., Abdullah, J. A. A., Hassouna, M., & Romero, A. (2025). Purification and Characterization of Transglutaminase Isolated from Sardine (Sardina pilchardus) Flesh Waste. Polymers, 17(4), 510. https://doi.org/10.3390/polym17040510








