Detection of QTLs for Outcrossing-Related Traits in CSSL Population Derived from Primitive Japonica Accession Ludao in the Genetic Background of O. sativa spp. Japonica Restorer C-bao Using RSTEP-LRT Method
Abstract
:1. Introduction
2. Materials and Methods
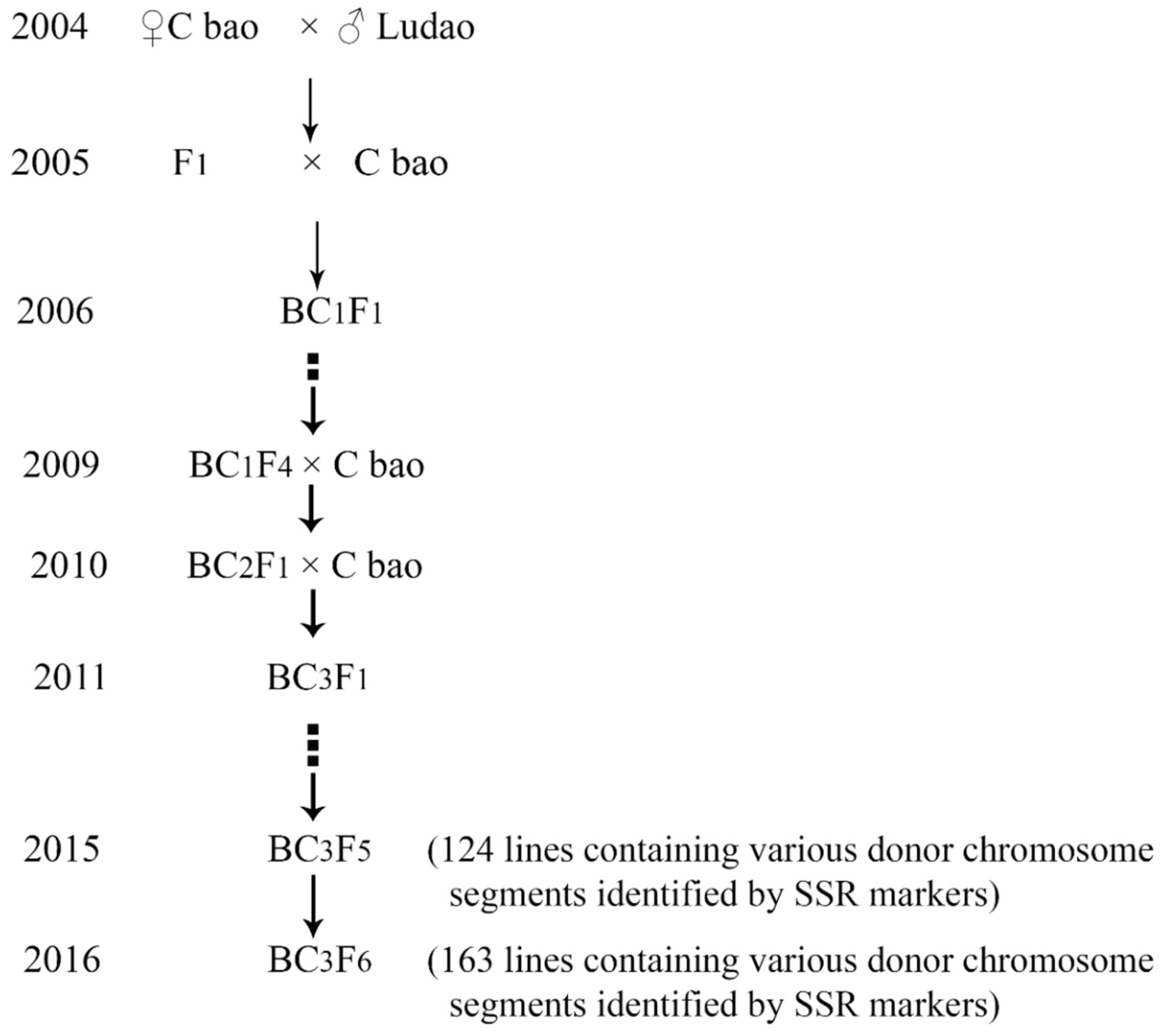
2.1. Plant Materials and Field Planting
2.2. DNA Extraction and SSR Marker Genotyping
2.3. Phenotyping CL-CSSL Population and Their Parents
2.4. Calculation of Heritability in Broad Sense
2.5. Detection of QTLs for the FLL, FLW, FLA, and PNL Traits by the RSTEP-LRT Method
3. Results
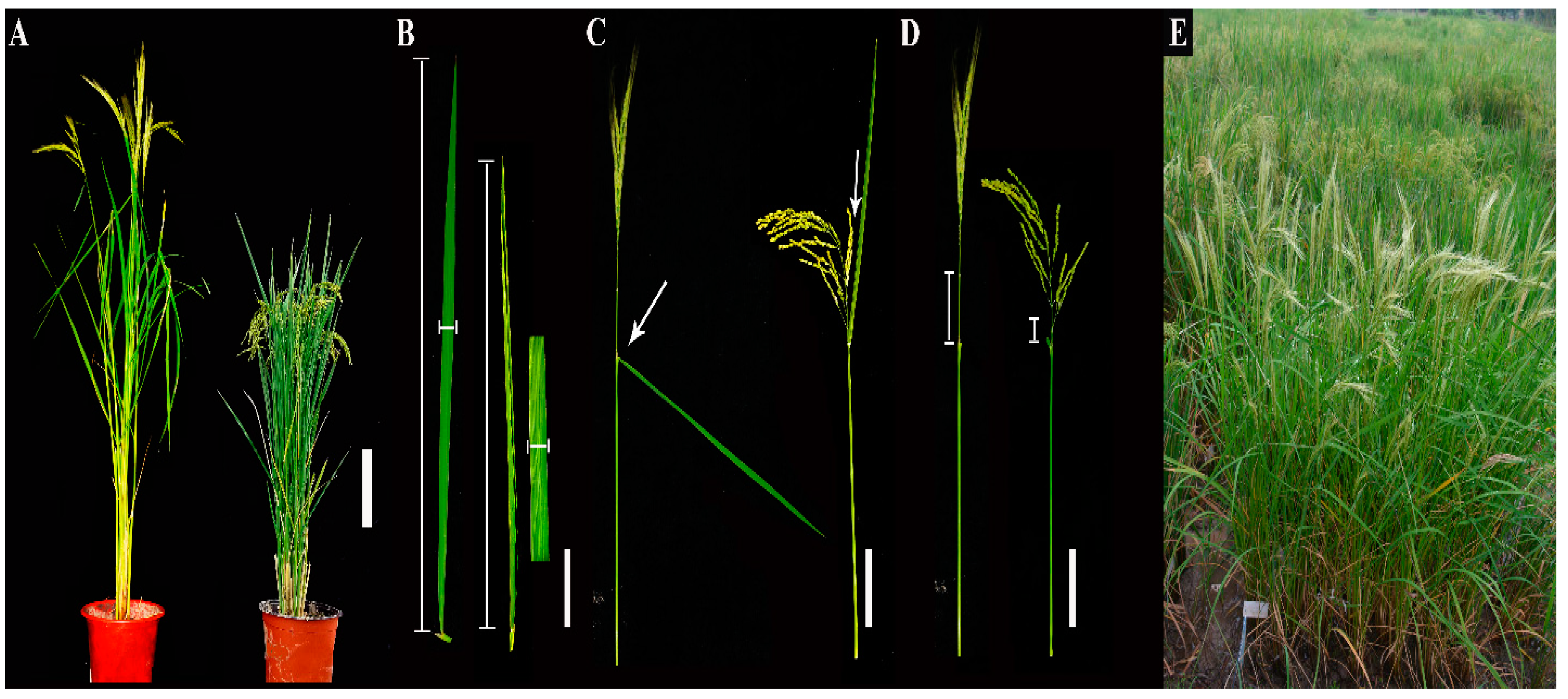
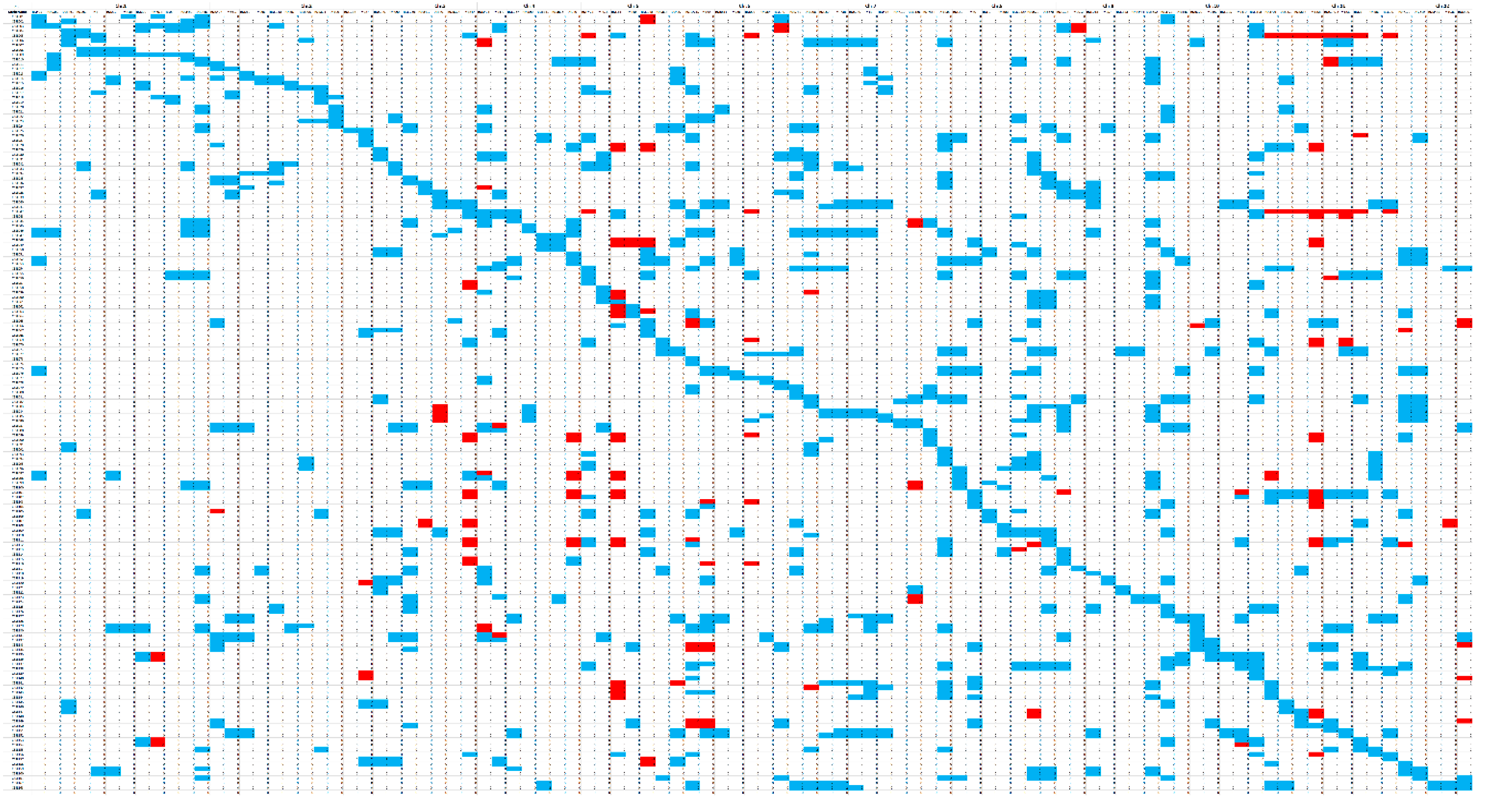
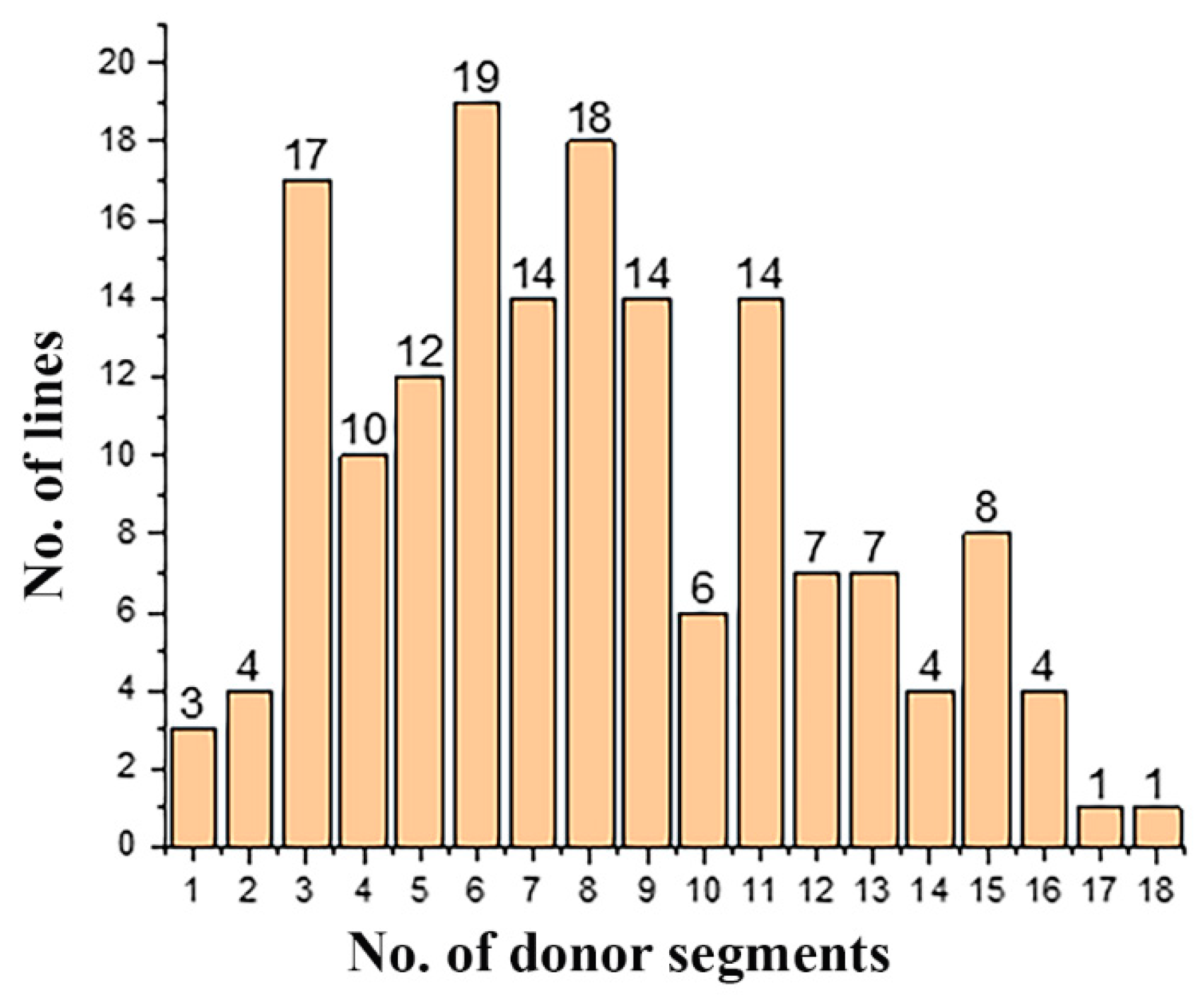
3.1. Construction of the CL-CSSL Population and Its Characteristics of Composition
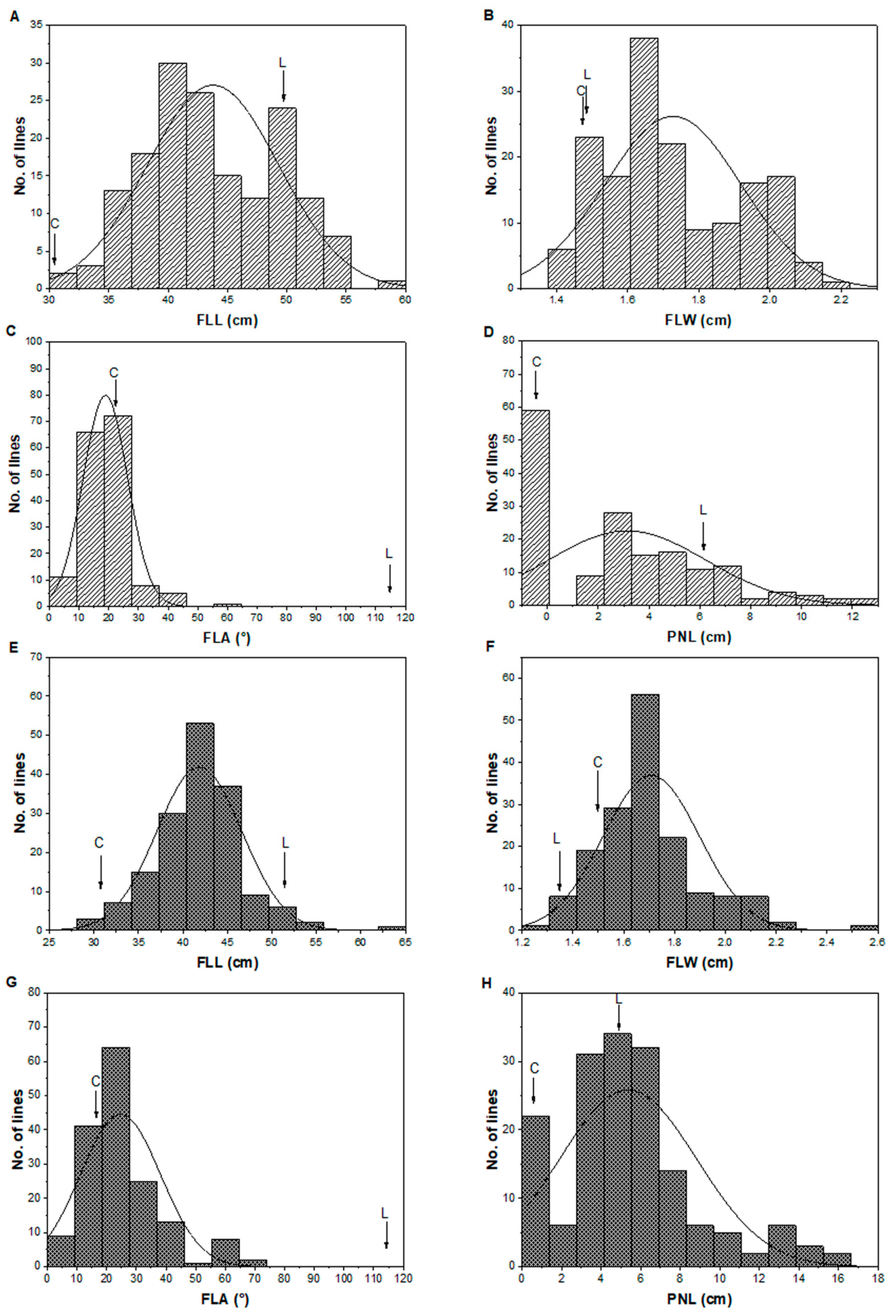
3.2. Phenotypic Variation of the Four Outcrossing Traits in the CL-CSSL Population
3.3. Detection of QTLs for FLL, FLW, FLA, and PNL Traits in the CL-CSSL Population
3.3.1. QTL for Flag Leaf Length
3.3.2. QTL for Flag Leaf Width
3.3.3. QTLs for Flag Leaf Angle
3.3.4. QTLs for Panicle Neck Length
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- GRiSP. Rice Almanac, 4th ed.; Global Rice Science Partnership International Rice Research Institute: Los Baños, Philippines, 2013; pp. 272–275. [Google Scholar]
- Yuan, L.; Virmani, S. Status of hybrid rice research and development. In Hybrid Rice; Smith, W.H., Ed.; International Rice Research Institute: Los Baños, Philippines, 1988; pp. 7–24. [Google Scholar]
- Huang, X.; Han, B. Rice domestication occurred through single origin and multiple introgressions. Nat. Plants 2016, 2, 15207. [Google Scholar] [CrossRef]
- Virmani, S.S.; Aquino, R.C.; Khush, G.S. Heterosis breeding in rice (Oryza sativa L.). Theor. Appl. Genet. 1982, 63, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, R.N.; Pande, K.; Ratho, S.N.; Jachuck, P.J. Heterosis in rice hybrids. Euphytica 1990, 49, 243–247. [Google Scholar]
- Wu, J.; Qi, Y.; Hu, G.; Li, J.; Li, Z.; Zhang, H. Genetic architecture of flag leaf length and width in rice (Oryza sativa L.) revealed by association mapping. Genes Genom. 2017, 39, 341–352. [Google Scholar] [CrossRef]
- Bux, L.; Li, D.; Faheem, M.; Sowadan, O.; Dong, Z.; Liu, E.; Ali, M.; Li, Y.; Sitoe, H.M.; Mirani, A.A.; et al. Detection of QTLs for outcrossing-related traits in rice (Oryza sativa L.) by association mapping and the RSTEP-LRT method. Euphytica 2019, 215, 204. [Google Scholar] [CrossRef]
- Yan, J.; Zhu, J.; He, C.; Benmoussa, M.; Wu, P. Molecular marker-assisted dissection of genotype × environment interaction for plant type traits in rice (Oryza sativa L.). Crop Sci. 1999, 39, 538–544. [Google Scholar] [CrossRef]
- Jiang, D.; Fang, J.; Lou, L.; Zhao, J.; Yuan, S.; Yin, L.; Sun, W.; Peng, L.; Guo, B.; Li, X. Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS ONE 2015, 10, e0118169. [Google Scholar] [CrossRef] [Green Version]
- Fujino, K.; Matsuda, Y.; Ozawa, K.; Nishimura, T.; Koshiba, T.; Fraaije, M.W.; Sekiguchi, H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008, 279, 499–507. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.; Qian, Q.; Bu, Q.; Li, S.; Chen, Q.; Sun, J.; Liang, W.; Zhou, Y.; Chu, C.; Li, X. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008, 147, 1947–1959. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Zhu, L.; Zeng, D.; Gao, Z.; Guo, L.; Fang, Y.; Zhang, G.; Dong, G.; Yan, M.; Liu, J. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 2010, 73, 283–292. [Google Scholar] [CrossRef]
- Li, Z.; Paterson, A.H.; Pinson, S.R.; Stansel, J.W. RFLP facilitated analysis of tiller and leaf angles in rice (Oryza sativa L.). Euphytica 1999, 109, 79–84. [Google Scholar] [CrossRef]
- Kobayashi, S.; Fukuta, Y.; Morita, S.; Sato, T.; Osaki, M.; Khush, G.S. Quantitative trait loci affecting flag leaf development in rice (Oryza sativa L.). Breed. Sci. 2003, 53, 255–262. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Dai, W.; Fan, Y.; Shen, B.; Zheng, K. Genetic dissection of flag leave angle and main panicle yield traits in rice. Chin. Agric. Sci. Bull. 2008, 24, 186–192. [Google Scholar]
- Cai, J. Genetic analysis and QTL mapping of the flag leaf traits in rice (Oryza sativa L.). Master’s Thesis, Chinese Academy of Agricultural Sciences, Hangzhou, China, 2009; pp. 29–42. [Google Scholar]
- Hu, W.; Zhang, H.; Jiang, J. Discovery of a germplasm with large flag leaf angle and its genetic analysis as well as QTL mapping in japonica rice (Oryza sativa L.). Chin. J. Rice Sci. 2012, 26, 34–42. [Google Scholar]
- Bian, J.; He, H.; Shi, H.; Zhu, G.; Li, C.; Zhu, C.; Peng, X.; Yu, Q.; Fu, J.; He, X. Quantitative trait loci mapping for flag leaf traits in rice using a chromosome segment substitution line population. Plant Breed. 2014, 133, 203–209. [Google Scholar] [CrossRef]
- Dong, Z.; Li, D.; Hu, X.; Liang, L.; Wu, G.; Zeng, S.; Liu, E.; Wu, Y.; Wang, H.; Bhanbhro, L.B.; et al. Mining of favorable marker alleles for flag leaf inclination in some rice (Oryza sativa L.) accessions by association mapping. Euphytica 2018, 214, 117. [Google Scholar] [CrossRef]
- Tanaka, A.; Nakagawa, H.; Tomita, C.; Shimatani, Z.; Ohtake, M.; Nomura, T.; Jiang, C.-J.; Dubouzet, J.G.; Kikuchi, S.; Sekimoto, H. BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009, 151, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar] [CrossRef]
- Wan, S.; Wu, J.; Zhang, Z.; Sun, X.; Lv, Y.; Gao, C.; Ning, Y.; Ma, J.; Guo, Y.; Zhang, Q. Activation tagging, an efficient tool for functional analysis of the rice genome. Plant Mol. Biol. 2009, 69, 69–80. [Google Scholar] [CrossRef]
- Liu, J.M.; Park, S.J.; Huang, J.; Lee, E.J.; Xuan, Y.H.; Je, B.I.; Kumar, V.; Priatama, R.A.; Raj, K.V.; Kim, S.H. Loose Plant Architecture1 (LPA1) determines lamina joint bending by suppressing auxin signalling that interacts with C-22-hydroxylated and 6-deoxo brassinosteroids in rice. J. Exp. Bot. 2016, 67, 1883–1895. [Google Scholar] [CrossRef] [Green Version]
- Yin, C.; Gan, L.; Ng, D.; Zhou, X.; Xia, K. Decreased panicle-derived indole-3-acetic acid reduces gibberellin A1 level in the uppermost internode, causing panicle enclosure in male sterile rice Zhenshan 97A. J. Exp. Bot. 2007, 58, 2441–2449. [Google Scholar] [CrossRef] [Green Version]
- Rutger, J.; Carnahan, H. A fourth genetic element to facilitate hybrid cereal production—A recessive tall in rice 1. Crop Sci. 1981, 21, 373–376. [Google Scholar] [CrossRef]
- Yamamoto, T.; Taguchi-Shiobara, F.; Ukai, Y.; Sasaki, T.; Yano, M. Mapping quantitative trait loci for days-to-heading, and culm, panicle and internode lengths in a BC1F3 population using an elite rice variety, Koshihikari, as the recurrent parent. Breed. Sci. 2001, 51, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Hittalmani, S.; Huang, N.; Courtois, B.; Venuprasad, R.; Shashidhar, H.; Zhuang, J.; Zheng, K.; Liu, G.; Wang, G.; Sidhu, J. Identification of QTL for growth-and grain yield-related traits in rice across nine locations of Asia. Theor. Appl. Genet. 2003, 107, 679–690. [Google Scholar] [CrossRef]
- Qiao, B.; Wang, Y.; Zhu, X.; Hong, D. QTL analysis of the uppermost internode length in rice under different growing environments. Yi Chuan Hered. 2007, 29, 1001–1007. [Google Scholar] [CrossRef]
- Xiao, K.; Zuo, H.; Gong, Y.; Zhang, J.; Zhang, Y.; Dong, Y. Location of QTLs controlling panicle exsertion and plant height in rice. Chin. Agric. Sci. Bull. 2008, 24, 95–99. [Google Scholar]
- Yang, D.; Zhu, Z.; Zhang, Y.; Lin, J.; Chen, T.; Zhao, L.; Zhu, W.; Wang, C. Substitution mapping of QTL for panicle exertion using CSSL in rice (Oryza sativa L.). Yi Chuan Hered. 2009, 31, 741–747. [Google Scholar] [CrossRef]
- Guan, H.; Duan, Y.; Liu, H.; Chen, Z.; Zhuo, M.; Zhuang, L.; Qi, W.; Pan, R.; Mao, D.; Zhou, Y. Genetic analysis and fine mapping of an enclosed panicle mutant esp2 in rice (Oryza sativa L.). Chin. Sci. Bull. 2011, 56, 1476–1480. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Kim, H.; Yun, D.; Yoon, U.; Kim, T.; Eun, M.; Lee, G. Characterization and fine mapping of a shortened uppermost internode mutant in rice. Plant Biotechnol. Rep. 2014, 8, 125–134. [Google Scholar] [CrossRef]
- Dang, X.; Fang, B.; Chen, X.; Li, D.; Sowadan, O.; Dong, Z.; Liu, E.; She, D.; Wu, G.; Liang, Y.; et al. Favorable marker alleles for panicle exsertion length in rice (Oryza sativa L.) mined by association mapping and the RSTEP-LRT method. Front. Plant Sci. 2017, 8, 2112. [Google Scholar] [CrossRef] [Green Version]
- Eshed, Y.; Zamir, D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 1995, 141, 1147–1162. [Google Scholar] [PubMed]
- Tanksley, S.; Nelson, J. Advanced backcross QTL analysis: A method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 1996, 92, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, J.H.; Singer, J.B.; Matin, A.; Lander, E.S. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 2000, 24, 221. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wan, X.; Crossa, J.; Crouch, J.; Weng, J.; Zhai, H.; Wan, J. QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet. Res. 2006, 88, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, J.; Yao, J.; Hong, D. Characterization and genetic analysis of grain filling rate of Ludao and restorer line C-Bao in japonica rice. Acta Agron. Sin. 2009, 35, 1229–1235. [Google Scholar] [CrossRef]
- Wei, X.H.; Yang, Z.R.; Dong, L.; Yu, H.Y.; Wang, Y.P.; Yuan, X.P.; Tang, S.X. SSR evidence for taxonomic position of weedy rice ‘Ludao’. Sci. Agric. Sin. 2004, 37, 937–942. [Google Scholar]
- Zhu, S.; Jiang, L.; Wang, C.; Zhai, H.; Li, D.; Wan, J. The origin of weedy rice Ludao in China deduced by genome wide analysis of its hybrid sterility genes. Breed. Sci. 2005, 55, 409–414. [Google Scholar] [CrossRef] [Green Version]
- Three Line Research Group, Crop Institute, Anhui Academy of Agricultural Sciences. Breeding of ‘Dang You C-bao’, a new cultivar in japonica hybrid rice (Oryza sativa L.). Anhui Agric. Sci. 1986, 2, 7–9. [Google Scholar]
- Yang, Z.; Chen, Q.; Chen, R.; Su, Z.; Jia, B.; Tong, J.; Wang, J. The breeding of japonica rice restorer C57. Acta Agron. Sin. 1981, 7, 153–156. [Google Scholar]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [Green Version]
- Temnykh, S.; Park, W.D.; Ayres, N.; Cartinhour, S.; Hauck, N.; Lipovich, L.; Cho, Y.G.; Ishii, T.; McCouch, S.R. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 2000, 100, 697–712. [Google Scholar] [CrossRef]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Thi, T.G.T.; Dong, G.; Wang, H.; Edzesi, W.M.; Hong, D. Genetic diversity and association mapping of seed vigor in rice (Oryza sativa L.). Planta 2014, 239, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. The Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef] [Green Version]
- McCouch, S.R. Gene nomenclature system for rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef] [Green Version]
- Young, N.; Tanksley, S. Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor. Appl. Genet. 1989, 77, 95–101. [Google Scholar] [CrossRef]
- Yue, B.; Xue, W.; Luo, L.; Xing, Y. QTL analysis for flag leaf characteristics and their relationships with yield and yield traits in rice. Acta Genet. Sin. 2006, 33, 824–832. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, X.; Huang, C.; Xing, Y.; Zheng, X.; Xu, Z.; Chen, W. Comparison of genetic linkage map and QTLs controlling flag leaf traits based on F2 and F2:6 populations derived from japonica rice. Chin. J. Rice Sci. 2010, 24, 372–378. [Google Scholar]
- Zeng, D.; Hu, J.; Dong, G.; Liu, J.; Zeng, L.; Zhang, G.; Guo, L.; Zhou, Y.; Qian, Q. Quantitative trait loci mapping of flag-leaf ligule length in rice and alignment with ZmLG1 gene. J. Integr. Plant Biol. 2009, 51, 360–366. [Google Scholar] [CrossRef]
- Farooq, M.; Tagle, A.G.; Santos, R.E.; Ebron, L.A.; Fujita, D.; Kobayashi, N. Quantitative trait loci mapping for leaf length and leaf width in rice cv. IR64 derived lines. J. Integr. Plant Biol. 2010, 52, 578–584. [Google Scholar] [CrossRef]
- Mei, H.; Li, Z.; Shu, Q.; Guo, L.; Wang, Y.; Yu, X.; Ying, C.; Luo, L. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations. Theor. Appl. Genet. 2005, 110, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Qin, R.; Jin, W.; He, G.; Song, Y. Comparative physical mapping of Bph3 with BAC-FISH in Oryza officinalis and O. sativa. Acta Bot. Sin. 2002, 44, 583–587. [Google Scholar]
- Li, C.; Zhou, A.; Sang, T. Rice domestication by reducing shattering. Science 2006, 311, 1936–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toriba, T.; Suzaki, T.; Yamaguchi, T.; Ohmori, Y.; Tsukaya, H.; Hirano, H.-Y. Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 2010, 22, 1452–1462. [Google Scholar] [CrossRef] [Green Version]
- Nagasaki, H.; Itoh, J.-I.; Hayashi, K.; Hibara, K.-I.; Satoh-Nagasawa, N.; Nosaka, M.; Mukouhata, M.; Ashikari, M.; Kitano, H.; Matsuoka, M. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 14867–14871. [Google Scholar] [CrossRef] [Green Version]
| Chromosome | Number of SSR Primers Tested on Parents | Number of Markers | Polymorphic Rate% |
|---|---|---|---|
| 1 | 74 | 12 | 16.2 |
| 2 | 78 | 13 | 16.7 |
| 3 | 69 | 5 | 7.2 |
| 4 | 73 | 7 | 9.6 |
| 5 | 75 | 7 | 9.3 |
| 6 | 66 | 8 | 12.1 |
| 7 | 62 | 9 | 14.5 |
| 8 | 60 | 8 | 13.3 |
| 9 | 76 | 7 | 9.2 |
| 10 | 62 | 7 | 11.3 |
| 11 | 62 | 10 | 16.1 |
| 12 | 74 | 4 | 5.4 |
| Total | 830 | 97 | 11.8 |
| Year | Trait | Ludao | C-bao | Difference between Parents | CL-CSSL Population | CV/% | H2B/% | |
|---|---|---|---|---|---|---|---|---|
| (Mean ± SD) | (Mean ± SD) | (Mean ± SD) | Range | |||||
| 2017 | FLL, cm | 50.6 ± 1.2 | 30.5 ± 0.7 | 20.1 *** | 43.9 ± 6.2 | 31.1~57.8 | 14.1 | 94.0 |
| FLW, cm | 1.5 ± 0.1 | 1.5 ± 0.1 | 0.0 ** | 1.7 ± 0.1 | 1.4~2.1 | 5.9 | 99.0 | |
| FLA, ° | 115.0 ± 2.7 | 22.9 ± 2.5 | 92.1 *** | 18.8 ± 7.5 | 5.0~60.8 | 39.9 | 98.4 | |
| PNL, cm | 6.4 ± 0.7 | 0.0 ± 0.0 | 6.4 *** | 5.1 ± 3.1 | 0.0~12.0 | 60.8 | 99.5 | |
| 2018 | FLL, cm | 52.1 ± 1.7 | 31.3 ± 1.1 | 20.8 *** | 41.8 ± 4.7 | 28.9~63.0 | 11.2 | 91.0 |
| FLW, cm | 1.3 ± 0.3 | 1.5 ± 0.1 | −0.2 * | 1.7 ± 0.1 | 1.2~2.5 | 5.9 | 96.8 | |
| FLA, ° | 115.9 ± 2.7 | 18.7 ± 5.2 | 97.2 *** | 20.7 ± 8.8 | 5.0~65.0 | 42.5 | 98.8 | |
| PNL, cm | 5.5 ± 1.7 | 0.0 ± 0.0 | 5.5 ** | 5.3 ± 3.4 | 0.0~16.5 | 64.1 | 98.2 | |
| Year | FLL | FLW | FLA | PNL | |
|---|---|---|---|---|---|
| FLL | 2017 | 1 | |||
| 2018 | 1 | ||||
| FLW | 2017 | 0.07 | 1 | ||
| 2018 | 0.22 ** | 1 | |||
| FLA | 2017 | −0.02 | −0.25 ** | 1 | |
| 2018 | 0.23 ** | −0.12 | 1 | ||
| PNL | 2017 | 0.09 | −0.36 *** | 0.45 *** | 1 |
| 2018 | 0.34 *** | −0.25 *** | 0.54 *** | 1 |
| Locus | Marker Name | Chr. | 2017 | 2018 | ||||
|---|---|---|---|---|---|---|---|---|
| LOD Value | PVE (%) | Add | LOD Value | PVE (%) | Add | |||
| qFLL-4 | RM349 | 4 | 2.5 | 6.49 | −2.55 | 2.53 | 6.92 | −2.5 |
| qFLW-11 | RM286 | 11 | 5.33 | 13.91 | 0.12 | - | - | - |
| qFLA-1.1 | RM8105 | 1 | 3.67 | 7.87 | 8.71 | 3.65 | 7.84 | 8.67 |
| qFLA-1.2 | RM7075 | 1 | 8.92 | 21.33 | 15.78 | 8.88 | 21.26 | 15.72 |
| qFLA-10 | RM1125 | 10 | - | - | - | 2.87 | 7.74 | 9.77 |
| qPNL-7 | RM11 | 7 | 2.61 | 7.07 | −1.23 | - | - | - |
| qPNL-8 | RM72 | 8 | 3.26 | 8.76 | 2.04 | 3.36 | 8.99 | 2.4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bux, L.; Li, D.; Faheem, M.; Ali, N.; Sirohi, M.H.; Ali, M.; Kumbhar, A.N.; Eltahawy, M.S.; Wu, G.; Liu, E.; et al. Detection of QTLs for Outcrossing-Related Traits in CSSL Population Derived from Primitive Japonica Accession Ludao in the Genetic Background of O. sativa spp. Japonica Restorer C-bao Using RSTEP-LRT Method. Agronomy 2020, 10, 28. https://doi.org/10.3390/agronomy10010028
Bux L, Li D, Faheem M, Ali N, Sirohi MH, Ali M, Kumbhar AN, Eltahawy MS, Wu G, Liu E, et al. Detection of QTLs for Outcrossing-Related Traits in CSSL Population Derived from Primitive Japonica Accession Ludao in the Genetic Background of O. sativa spp. Japonica Restorer C-bao Using RSTEP-LRT Method. Agronomy. 2020; 10(1):28. https://doi.org/10.3390/agronomy10010028
Chicago/Turabian StyleBux, Lal, Dalu Li, Muhammad Faheem, Nour Ali, Muzafar Hussain Sirohi, Mehtab Ali, Ali Nawaz Kumbhar, Moaz Salah Eltahawy, Guocan Wu, Erbao Liu, and et al. 2020. "Detection of QTLs for Outcrossing-Related Traits in CSSL Population Derived from Primitive Japonica Accession Ludao in the Genetic Background of O. sativa spp. Japonica Restorer C-bao Using RSTEP-LRT Method" Agronomy 10, no. 1: 28. https://doi.org/10.3390/agronomy10010028
APA StyleBux, L., Li, D., Faheem, M., Ali, N., Sirohi, M. H., Ali, M., Kumbhar, A. N., Eltahawy, M. S., Wu, G., Liu, E., Dang, X., & Hong, D. (2020). Detection of QTLs for Outcrossing-Related Traits in CSSL Population Derived from Primitive Japonica Accession Ludao in the Genetic Background of O. sativa spp. Japonica Restorer C-bao Using RSTEP-LRT Method. Agronomy, 10(1), 28. https://doi.org/10.3390/agronomy10010028





