Comprehensive In Silico Characterization and Expression Profiling of Nine Gene Families Associated with Calcium Transport in Soybean
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Ca2+ Transport Proteins in the Soybean Genome
2.2. Phylogenetic Analysis and Chromosomal Mapping
2.3. Gene Duplication and Calculation of Ka/Ks Values
2.4. Identification of Conserved Domains and Cis-Acting Regulatory Elements
2.5. RNA-Seq Datasets
3. Results
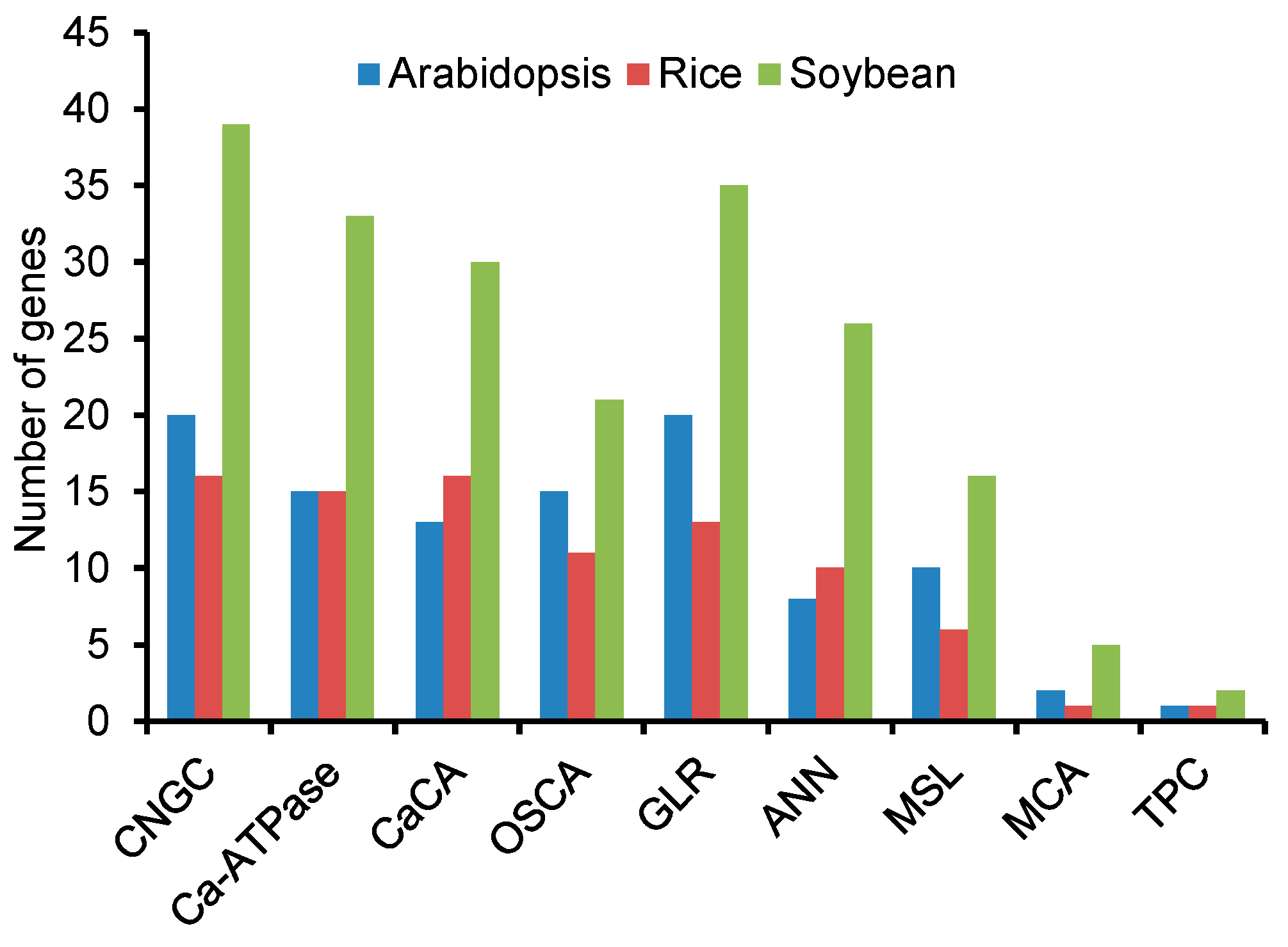
3.1. Identification of Putative Ca2+ Transport Proteins in the Soybean Genome
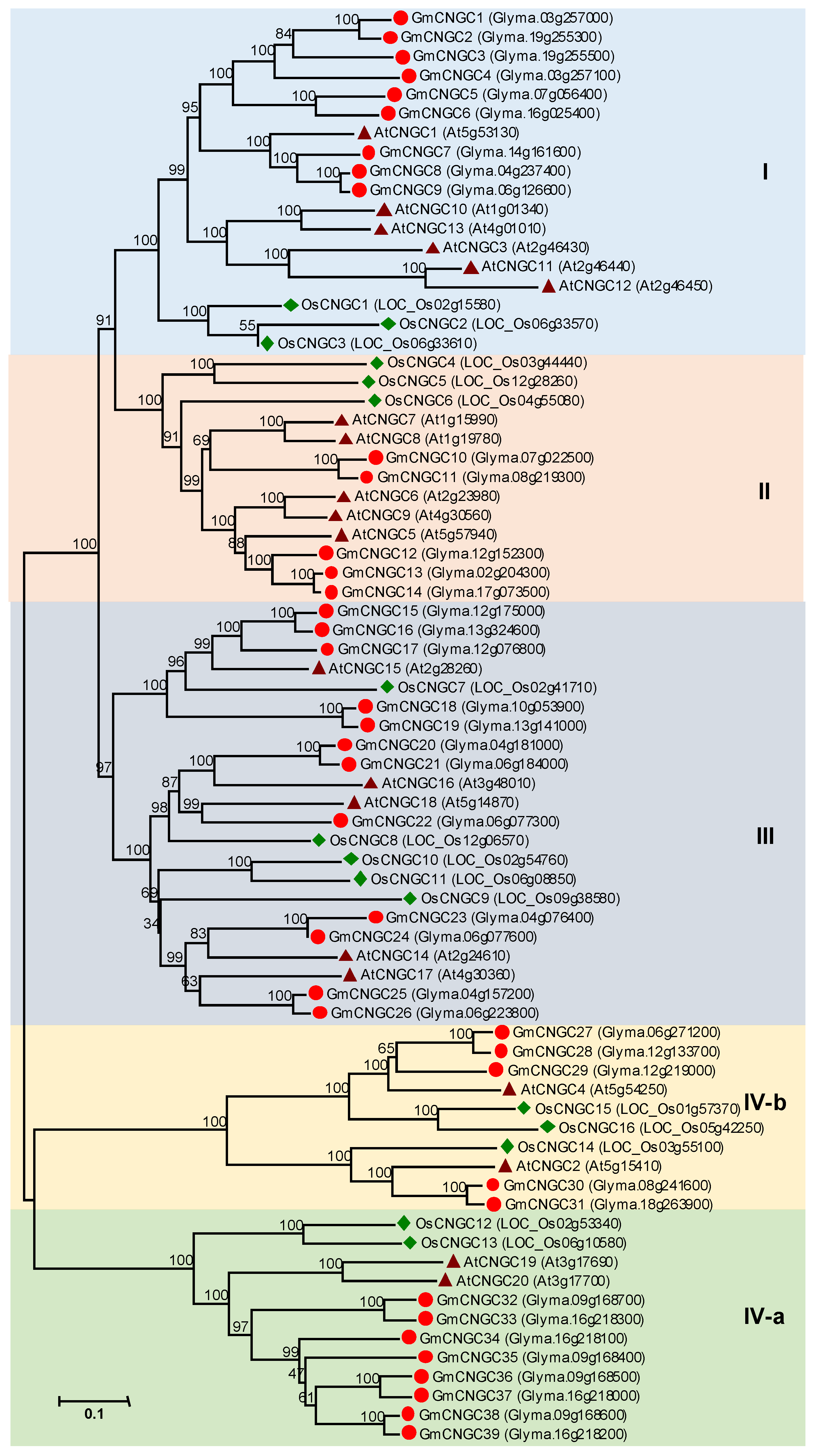
3.2. CNGC Gene Family in Soybean
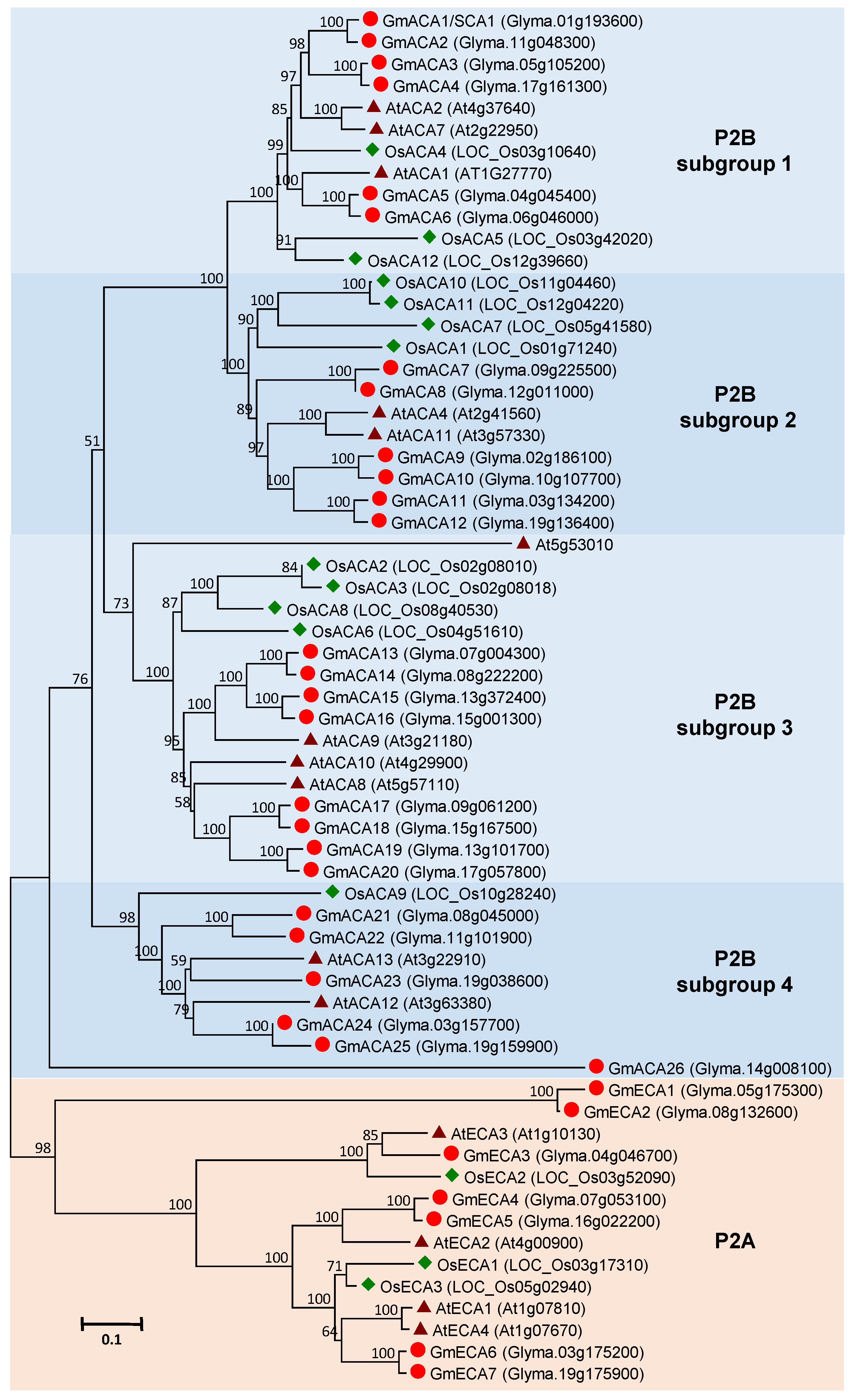
3.3. Ca2+-ATPase Gene Family in Soybean
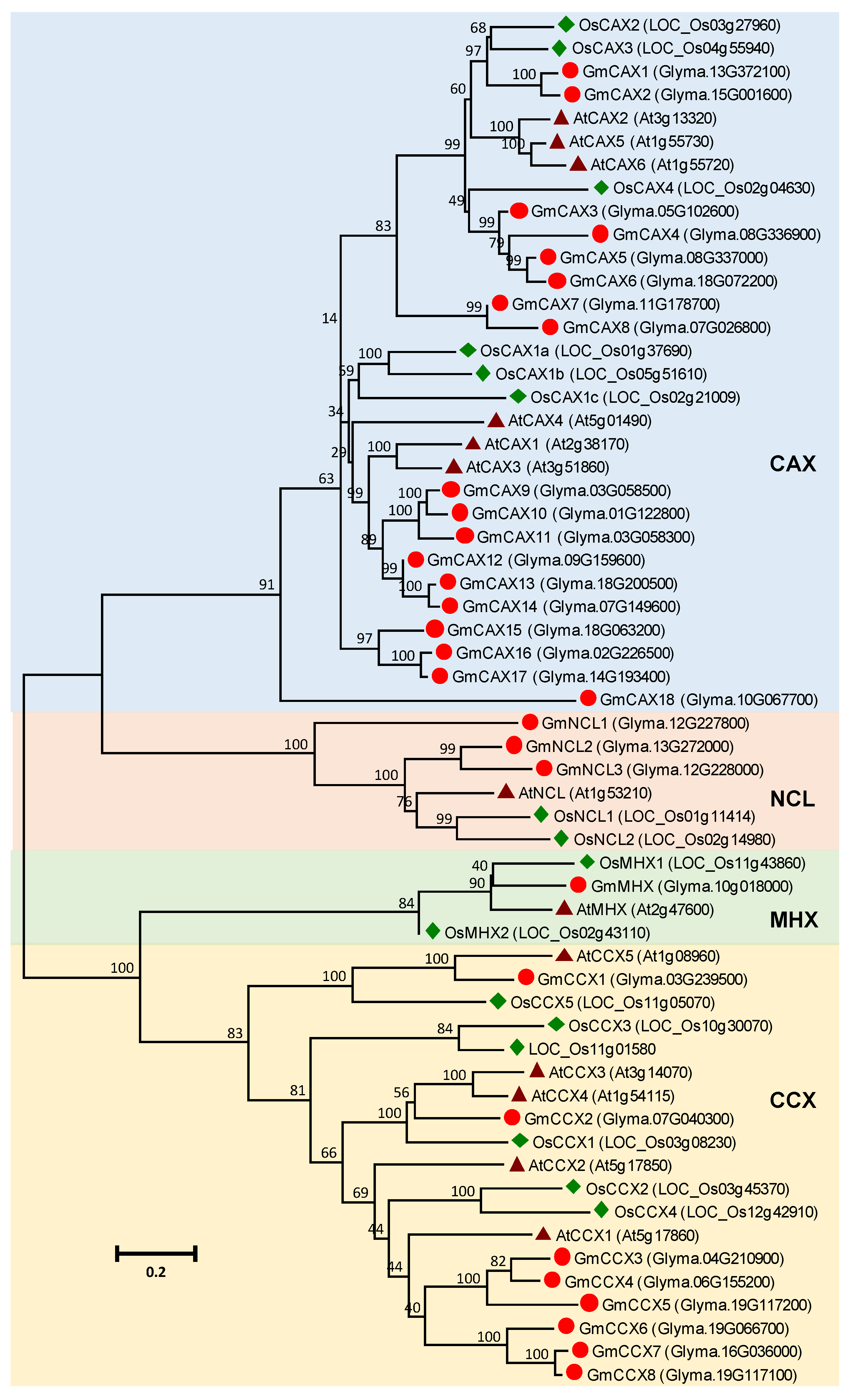
3.4. CaCA Gene Family in Soybean
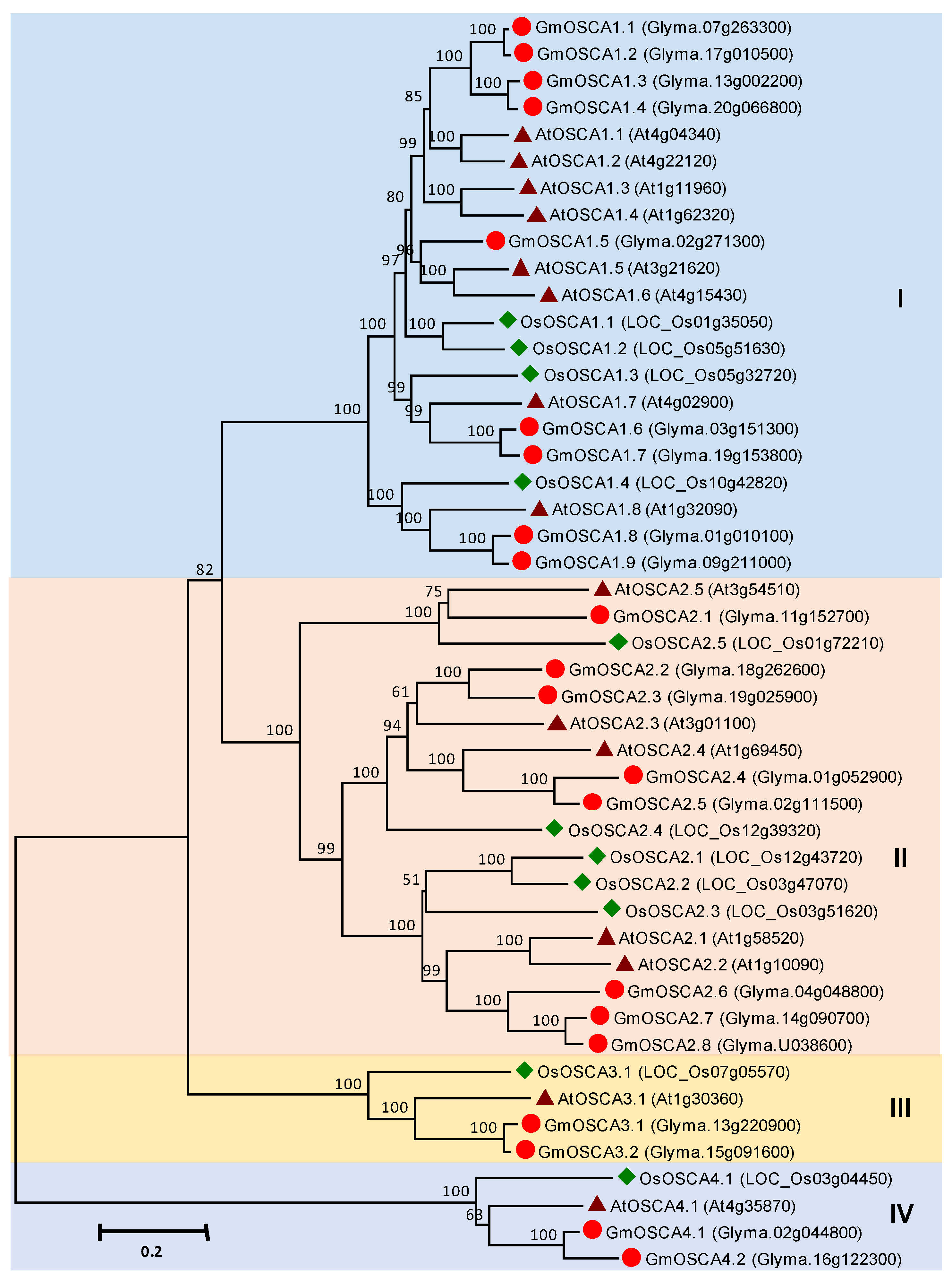
3.5. OSCA Gene Family in Soybean
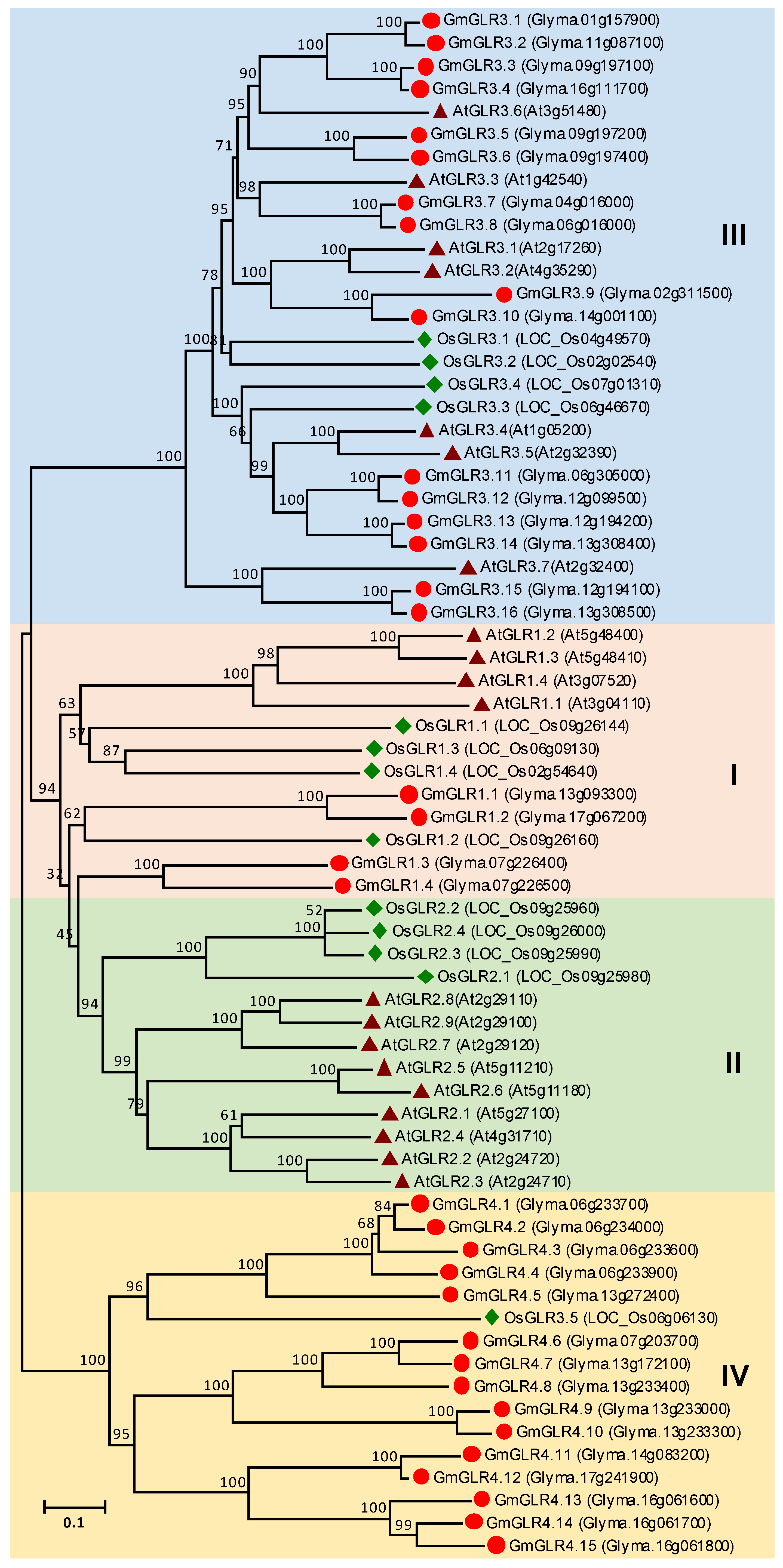
3.6. GLR Gene Family in Soybean
3.7. ANN Gene Family in Soybean
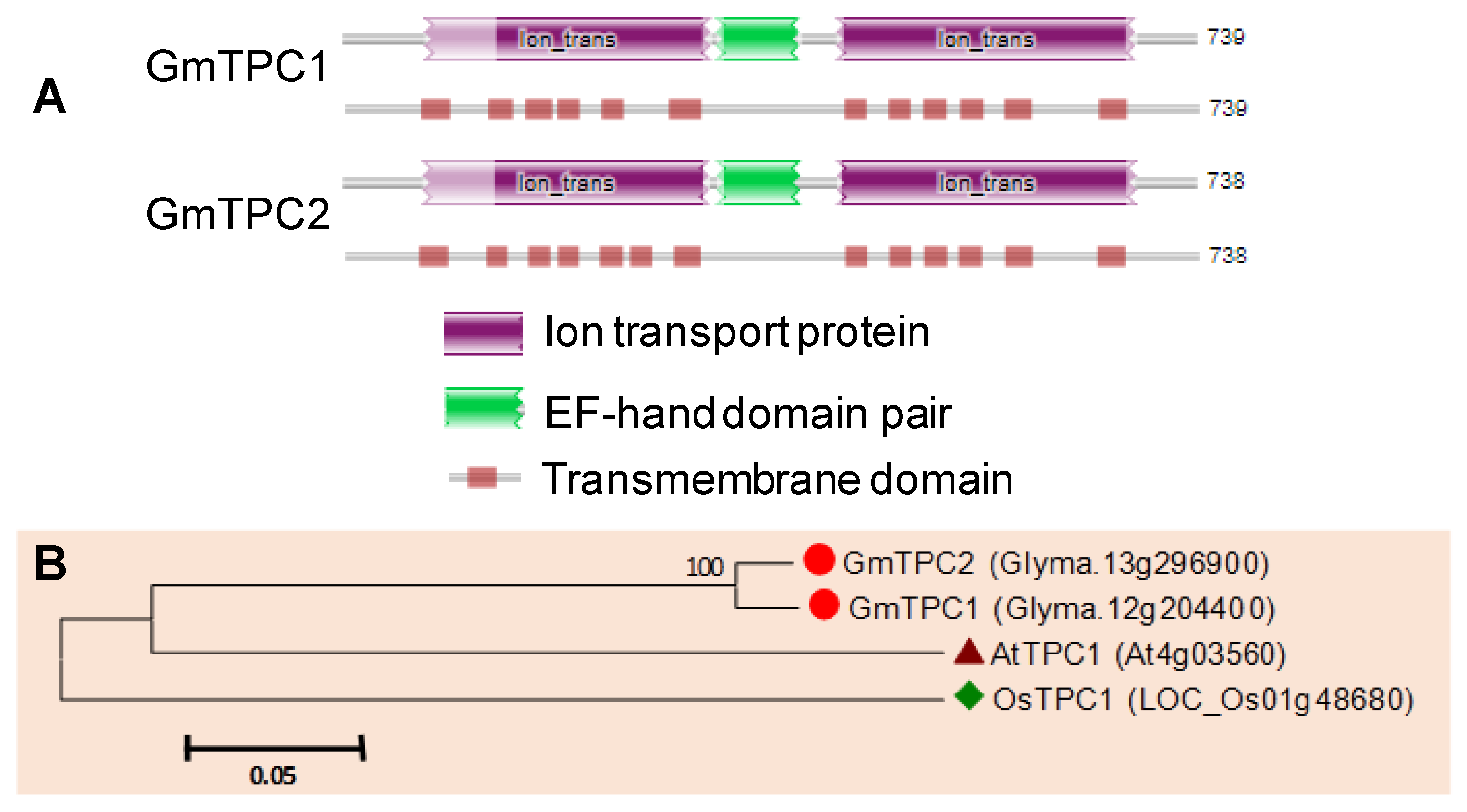
3.8. TPC Proteins in Soybean
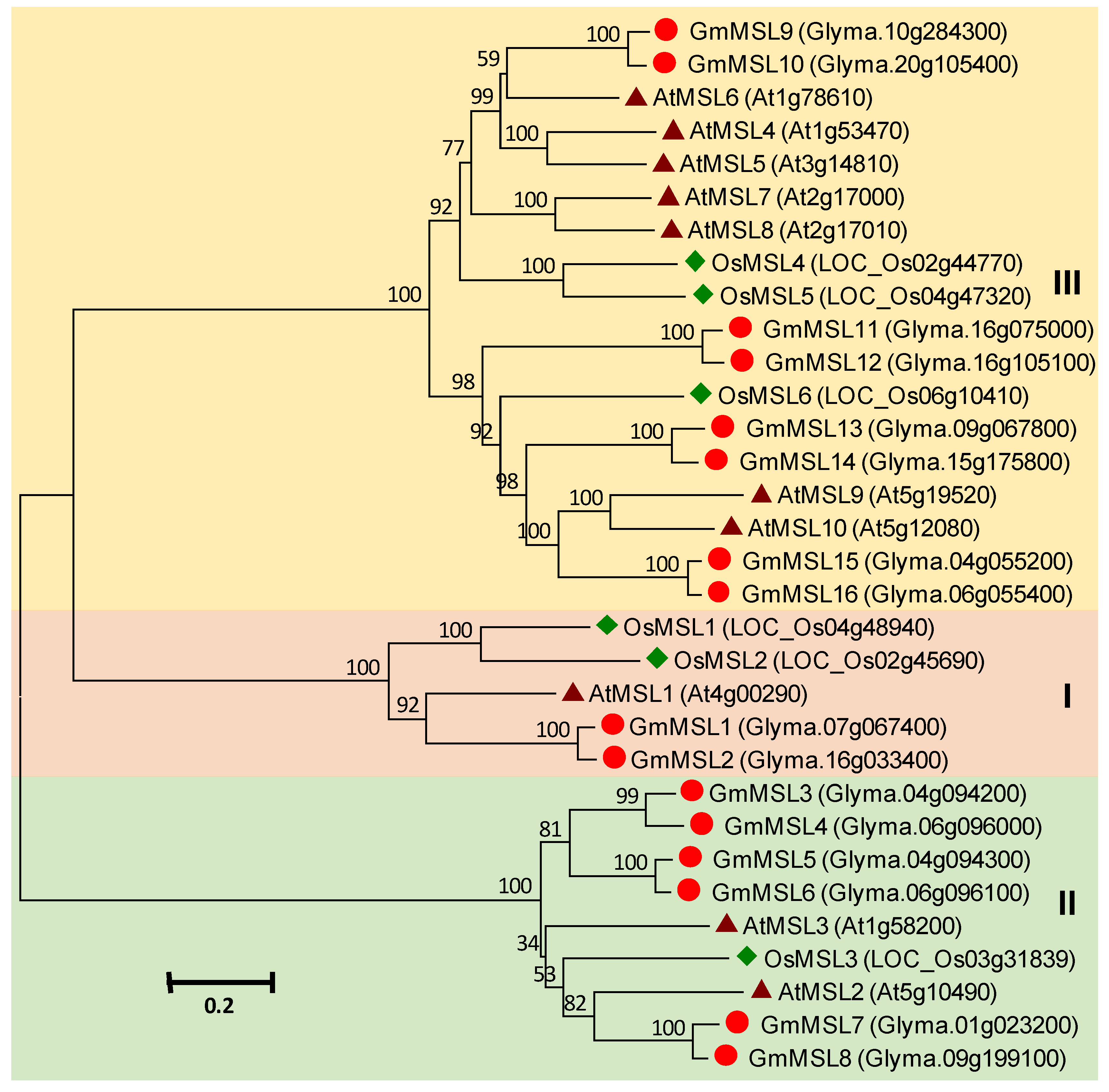
3.9. MSL Gene Family in Soybean
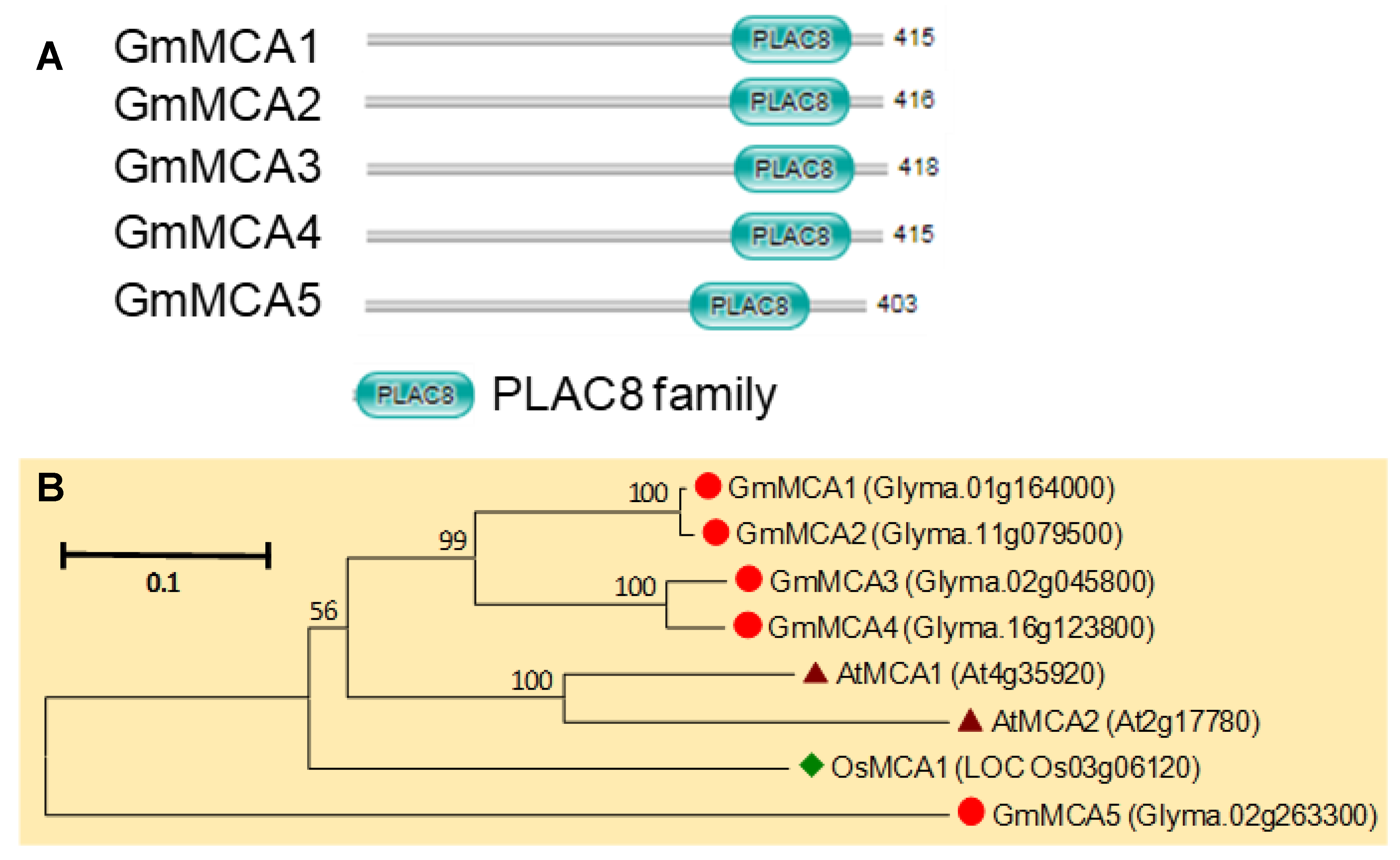
3.10. MCA Gene Family in Soybean
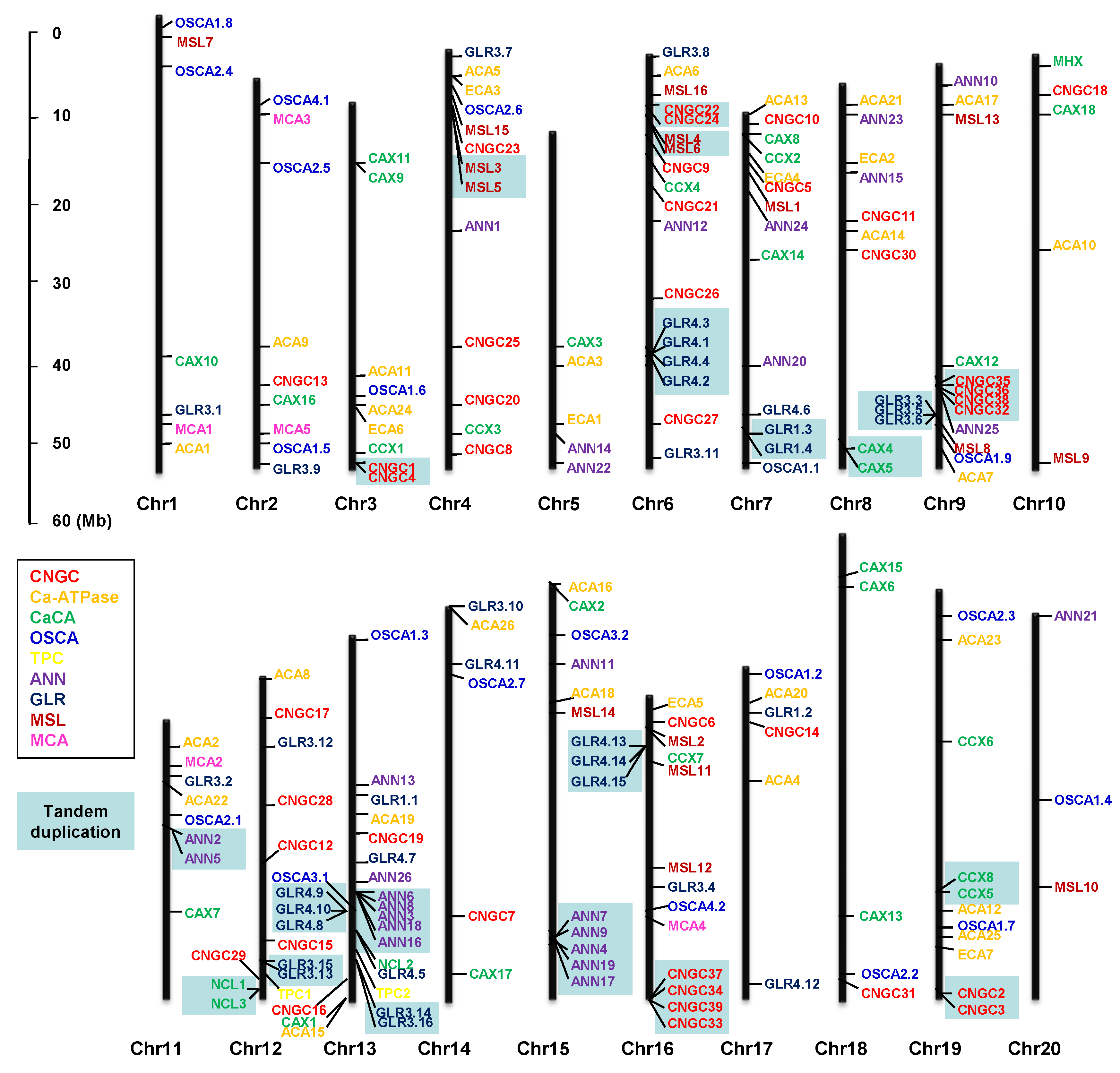
3.11. Chromosomal Distribution and Gene Duplication
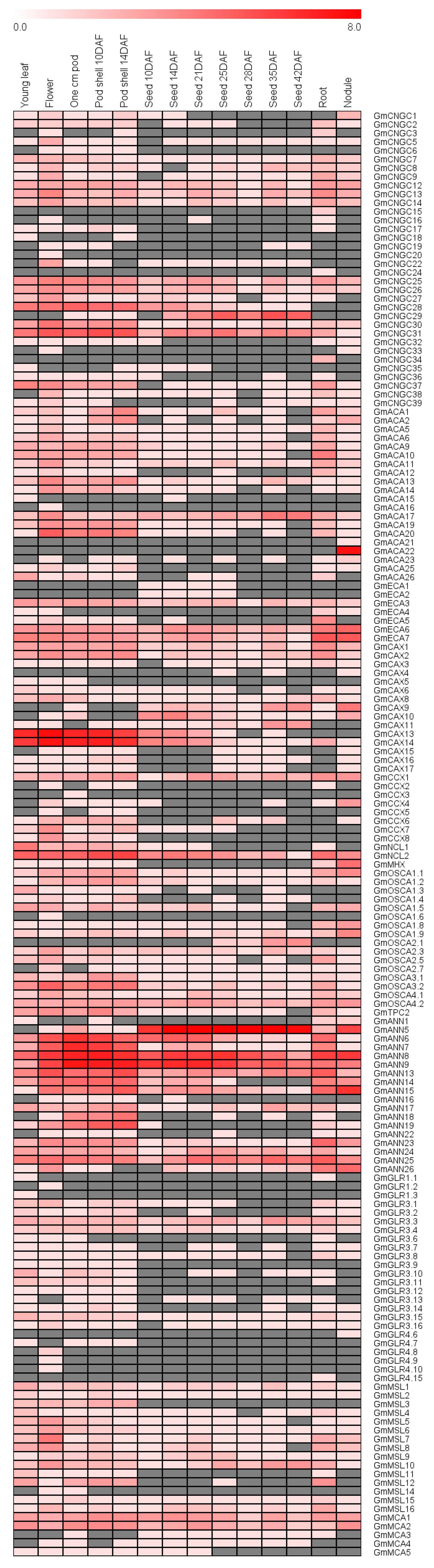
3.12. Expression Profile of Genes Encoding Putative Ca2+ Transport Proteins during Development
3.13. Responses of Genes Encoding Putative Ca2+ Transport Proteins to Stresses and Rhizobia Infection
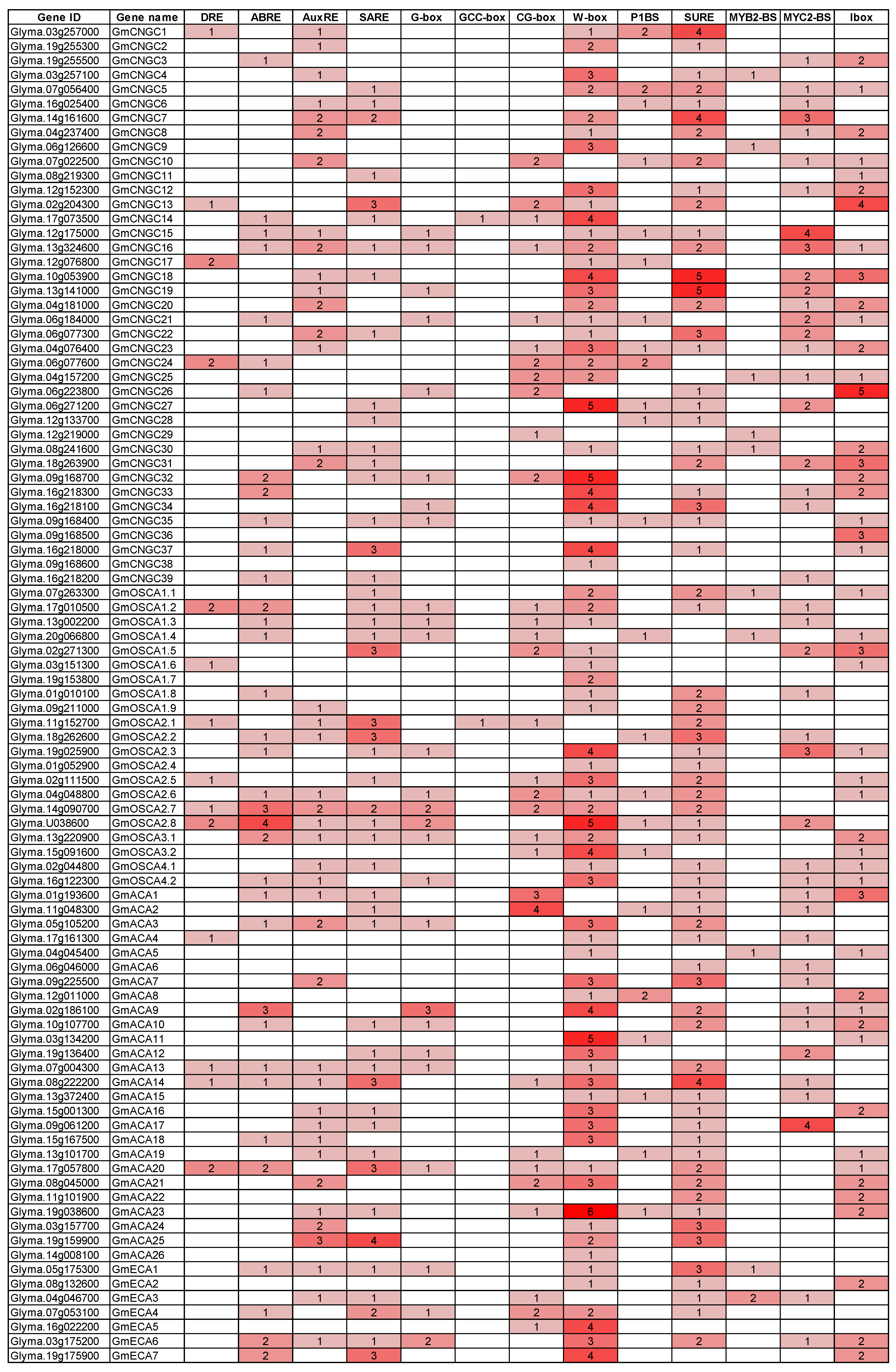
3.14. Hormone and Stress-Related Cis-Elements in the Promoters
3.15. Analysis of Potential MicroRNA Target Sites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hawkesford, M.; Horst, W.; Kichey, T.; Lambers, H.; Schjoerring, J.; Møller, I.S.; White, P. Functions of macronutrients. In Marschner’s Mineral Nutrition of Higher Plants; Elsevier: Amsterdam, The Netherlands, 2012; pp. 135–189. [Google Scholar]
- White, P.J.; Broadley, M.R. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Shabala, S.; Isayenkov, S.; Cuin, T.A.; Pottosin, I. Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 2018, 220, 49–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edel, K.H.; Kudla, J. Increasing complexity and versatility: How the calcium signaling toolkit was shaped during plant land colonization. Cell Calcium 2015, 57, 231–246. [Google Scholar] [CrossRef]
- Reddy, A.S. Calcium: Silver bullet in signaling. Plant Sci. 2001, 160, 381–404. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- Yuan, P.; Jauregui, E.; Du, L.; Tanaka, K.; Poovaiah, B.W. Calcium signatures and signaling events orchestrate plant–microbe interactions. Curr. Opin. Plant Biol. 2017, 38, 173–183. [Google Scholar] [CrossRef]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.; Wang, C.; Gao, Q.; Li, L.; Luan, S. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 2020, 6, 750–759. [Google Scholar] [CrossRef]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca(2+) influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Hilleary, R.; Paez-Valencia, J.; Vens, C.; Toyota, M.; Palmgren, M.; Gilroy, S. Tonoplast-localized Ca(2+) pumps regulate Ca(2+) signals during pattern-triggered immunity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2020, 117, 18849–18857. [Google Scholar] [CrossRef] [PubMed]
- McAinsh, M.R.; Pittman, J.K. Shaping the calcium signature. New Phytol. 2009, 181, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Jammes, F.; Hu, H.C.; Villiers, F.; Bouten, R.; Kwak, J.M. Calcium-permeable channels in plant cells. FEBS J. 2011, 278, 4262–4276. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Peiter, E.; Maathuis, F.J.; Mills, L.N.; Knight, H.; Pelloux, J.; Hetherington, A.M.; Sanders, D. The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 2005, 434, 404–408. [Google Scholar] [CrossRef]
- Dietrich, P.; Moeder, W.; Yoshioka, K. Plant cyclic nucleotide-gated channels: New insights on their functions and regulation. Plant Physiol. 2020, 184, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.F.; Gu, L.L.; Wang, H.Q.; Fei, C.F.; Fang, X.; Hussain, J.; Sun, S.J.; Dong, J.Y.; Liu, H.; Wang, Y.F. Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 3096–3101. [Google Scholar] [CrossRef] [Green Version]
- Nawaz, Z.; Kakar, K.U.; Saand, M.A.; Shu, Q.-Y. Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genom. 2014, 15, 853. [Google Scholar] [CrossRef] [Green Version]
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.M.; Sanders, D.; et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646. [Google Scholar] [CrossRef] [Green Version]
- Thor, K.; Jiang, S.; Michard, E.; George, J.; Scherzer, S.; Huang, S.; Dindas, J.; Derbyshire, P.; Leitao, N.; DeFalco, T.A.; et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 2020, 585, 569–573. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; et al. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol. 2015, 15, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurusu, T.; Sakurai, Y.; Miyao, A.; Hirochika, H.; Kuchitsu, K. Identification of a putative voltage-gated Ca2+-permeable channel (OsTPC1) involved in Ca2+ influx and regulation of growth and development in rice. Plant Cell Physiol. 2004, 45, 693–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davenport, R. Glutamate receptors in plants. Ann. Bot. 2002, 90, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Vincill, E.D.; Bieck, A.M.; Spalding, E.P. Ca(2+) conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol. 2012, 159, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Michard, E.; Lima, P.T.; Borges, F.; Silva, A.C.; Portes, M.T.; Carvalho, J.E.; Gilliham, M.; Liu, L.H.; Obermeyer, G.; Feijo, J.A. Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 2011, 332, 434–437. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, J.; Ma, C.; Zhao, Y.; Wang, Y.; Hasi, A.; Qi, Z. Glutamate receptor-like channel3.3 is involved in mediating glutathione-triggered cytosolic calcium transients, transcriptional changes, and innate immunity responses in Arabidopsis. Plant Physiol. 2013, 162, 1497. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Yu, Z.; Du, G.; Zhang, Y.; Taylor, J.L.; Shen, C.; Xu, J.; Liu, X.; Wang, Y.; Wu, Y. Heterologous expression and functional analysis of rice glutamate receptor-like family indicates its role in glutamate triggered calcium flux in rice roots. Rice 2016, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Hamill, O.P.; Martinac, B. Molecular basis of mechanotransduction in living cells. Physiol. Rev. 2001, 81, 685–740. [Google Scholar] [CrossRef]
- Hamilton, E.S.; Schlegel, A.M.; Haswell, E.S. United in diversity: Mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 2015, 66, 113–137. [Google Scholar] [CrossRef] [Green Version]
- Pivetti, C.D.; Yen, M.R.; Miller, S.; Busch, W.; Tseng, Y.H.; Booth, I.R.; Saier, M.H., Jr. Two families of mechanosensitive channel proteins. Microbiol. Mol. Biol. Rev. MMBR 2003, 67, 66–85. [Google Scholar] [CrossRef] [Green Version]
- Saddhe, A.A.; Kumar, K. In silico identification and expression analysis of MscS like gene family in rice. Plant Gene 2015, 1, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, Y.; Katagiri, T.; Shinozaki, K.; Qi, Z.; Tatsumi, H.; Furuichi, T.; Kishigami, A.; Sokabe, M.; Kojima, I.; Sato, S.; et al. Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. USA 2007, 104, 3639–3644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurusu, T.; Nishikawa, D.; Yamazaki, Y.; Gotoh, M.; Nakano, M.; Hamada, H.; Yamanaka, T.; Iida, K.; Nakagawa, Y.; Saji, H.; et al. Plasma membrane protein OsMCA1 is involved in regulation of hypo-osmotic shock-induced Ca2+ influx and modulates generation of reactive oxygen species in cultured rice cells. BMC Plant Biol. 2012, 12, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, T.; Nakagawa, Y.; Mori, K.; Nakano, M.; Imamura, T.; Kataoka, H.; Terashima, A.; Iida, K.; Kojima, I.; Katagiri, T.; et al. MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol. 2010, 152, 1284–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laohavisit, A.; Davies, J.M. Annexins. New Phytol. 2011, 189, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.M. Annexin-mediated calcium signalling in plants. Plants 2014, 3, 128–140. [Google Scholar] [CrossRef] [Green Version]
- Laohavisit, A.; Richards, S.; Shabala, L.; Chen, C.; Colaco, R.; Swarbreck, S.; Shaw, E.; Dark, A.; Shabala, S.; Shang, Z.; et al. Salinity-induced calcium signaling and root adaptation in Arabidopsis thaliana require the calcium regulatory protein annexin1. Plant Physiol. 2013, 163, 253–262. [Google Scholar] [CrossRef] [Green Version]
- Richards, S.L.; Laohavisit, A.; Mortimer, J.C.; Shabala, L.; Swarbreck, S.M.; Shabala, S.; Davies, J.M. Annexin 1 regulates the H2O2-induced calcium signature in Arabidopsis thaliana roots. Plant J. 2014, 77, 136–145. [Google Scholar] [CrossRef]
- Laohavisit, A.; Mortimer, J.C.; Demidchik, V.; Coxon, K.M.; Stancombe, M.A.; Macpherson, N.; Brownlee, C.; Hofmann, A.; Webb, A.A.; Miedema, H.; et al. Zea mays annexins modulate cytosolic free Ca2+ and generate a Ca2+-permeable conductance. Plant Cell 2009, 21, 479–493. [Google Scholar] [CrossRef] [Green Version]
- Clark, G.B.; Morgan, R.O.; Fernandez, M.P.; Roux, S.J. Evolutionary adaptation of plant annexins has diversified their molecular structures, interactions and functional roles. New Phytol. 2012, 196, 695–712. [Google Scholar] [CrossRef]
- Jami, S.K.; Clark, G.B.; Ayele, B.T.; Roux, S.J.; Kirti, P.B. Identification and characterization of annexin gene family in rice. Plant Cell Rep. 2012, 31, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Tang, Y.; Gao, S.; Su, S.; Hong, L.; Wang, W.; Fang, Z.; Li, X.; Ma, J.; Quan, W.; et al. Comprehensive analyses of the annexin gene family in wheat. BMC Genom. 2016, 17, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonza, M.C.; De Michelis, M.I. The plant Ca2+-ATPase repertoire: Biochemical features and physiological functions. Plant Biol. 2011, 13, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.S.; Lee, S.H.; Kim, J.C.; Heo, W.D.; Kim, M.C.; Park, C.Y.; Park, H.C.; Lim, C.O.; Kim, W.B.; Harper, J.F.; et al. Identification of a calmodulin-regulated soybean Ca(2+)-ATPase (SCA1) that is located in the plasma membrane. Plant Cell 2000, 12, 1393–1407. [Google Scholar] [CrossRef]
- Huda, K.M.K.; Banu, M.S.A.; Tuteja, R.; Tuteja, N. Global calcium transducer P-type Ca2+-ATPases open new avenues for agriculture by regulating stress signalling. J. Exp. Bot. 2013, 64, 3099–3109. [Google Scholar] [CrossRef]
- Baxter, I.; Tchieu, J.; Sussman, M.R.; Boutry, M.; Palmgren, M.G.; Gribskov, M.; Harper, J.F.; Axelsen, K.B. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003, 132, 618–628. [Google Scholar] [CrossRef] [Green Version]
- Kamrul Huda, K.M.; Yadav, S.; Akhter Banu, M.S.; Trivedi, D.K.; Tuteja, N. Genome-wide analysis of plant-type II Ca2+ ATPases gene family from rice and Arabidopsis: Potential role in abiotic stresses. Plant Physiol. Biochem. 2013, 65, 32–47. [Google Scholar] [CrossRef]
- Pittman, J.K.; Hirschi, K.D. Phylogenetic analysis and protein structure modelling identifies distinct Ca2+/Cation antiporters and conservation of gene family structure within Arabidopsis and rice species. Rice 2016, 9, 3. [Google Scholar] [CrossRef] [Green Version]
- Emery, L.; Whelan, S.; Hirschi, K.; Pittman, J. Protein phylogenetic analysis of Ca2+/cation antiporters and insights into their evolution in plants. Front. Plant Sci. 2012, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Deshmukh, R.; Sonah, H.; Patil, G.; Chen, W.; Prince, S.; Mutava, R.; Vuong, T.; Valliyodan, B.; Nguyen, H.T. Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 2014, 5, 244. [Google Scholar] [CrossRef]
- Zeng, H.; Zhang, X.; Zhang, X.; Pi, E.; Xiao, L.; Zhu, Y. Early Transcriptomic response to phosphate deprivation in soybean leaves as revealed by RNA-sequencing. Int. J. Mol. Sci. 2018, 19, 2145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanuzzaman, M.; Nahar, K.; Rahman, A.; Mahmud, J.A.; Hossain, M.S.; Fujita, M. 4-soybean production and environmental stresses. In Environmental Stresses in Soybean Production; Miransari, M., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 61–102. [Google Scholar]
- Frietsch, S.; Wang, Y.-F.; Sladek, C.; Poulsen, L.R.; Romanowsky, S.M.; Schroeder, J.I.; Harper, J.F. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. USA 2007, 104, 14531–14536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Kanwar, P.; Yadav, A.K.; Mishra, M.; Jha, S.K.; Baranwal, V.; Pandey, A.; Kapoor, S.; Tyagi, A.K.; Pandey, G.K. Genome-wide expressional and functional analysis of calcium transport elements during abiotic stress and development in rice. FEBS J. 2014, 281, 894–915. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37 (Suppl. S1), D211–D215. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Zhang, Y.; Zhang, X.; Pi, E.; Zhu, Y. Analysis of EF-Hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front. Plant Sci. 2017, 8, 877. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.-H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Medina-Rivera, A.; Defrance, M.; Sand, O.; Herrmann, C.; Castro-Mondragon, J.A.; Delerce, J.; Jaeger, S.; Blanchet, C.; Vincens, P.; Caron, C.; et al. RSAT 2015: Regulatory sequence analysis tools. Nucleic Acids Res. 2015, 43, W50–W56. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [Green Version]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhang, X.; Ding, M.; Zhang, X.; Zhu, Y. Transcriptome profiles of soybean leaves and roots in response to zinc deficiency. Physiol. Plant 2019, 167, 330–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.Q.; Wang, G.P.; Zhang, Y.Q.; Hu, X.Y.; Pi, E.X.; Zhu, Y.Y.; Wang, H.Z.; Du, L.Q. Genome-wide identification of phosphate-deficiency-responsive genes in soybean roots by high-throughput sequencing. Plant Soil 2016, 398, 207–227. [Google Scholar] [CrossRef]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 2014, 15, 950. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L.; et al. Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanubile, A.; Muppirala, U.K.; Severin, A.J.; Marocco, A.; Munkvold, G.P. Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genom. 2015, 16, 1089. [Google Scholar] [CrossRef] [Green Version]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010, 152, 541–552. [Google Scholar] [CrossRef] [Green Version]
- Zelman, A.K.; Dawe, A.; Gehring, C.; Berkowitz, G.A. Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front. Plant Sci. 2012, 3, 95. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Kumar, R.; Tripathi, A.K.; Gupta, B.K.; Pareek, A.; Singla-Pareek, S.L. Genome-wide investigation and expression analysis of Sodium/Calcium exchanger gene family in rice and Arabidopsis. Rice 2015, 8, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.H.; Gojobori, T.; Nei, M. Pseudogenes as a paradigm of neutral evolution. Nature 1981, 292, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Wang, G.; Hu, X.; Wang, H.; Du, L.; Zhu, Y. Role of microRNAs in plant responses to nutrient stress. Plant Soil 2014, 374, 1005–1021. [Google Scholar] [CrossRef]
- Li, S.; Castillo-Gonzalez, C.; Yu, B.; Zhang, X. The functions of plant small RNAs in development and in stress responses. Plant J. 2017, 90, 654–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [Green Version]
- Huda, K.M.; Banu, M.; Akhter, S.; Garg, B.; Tula, S.; Tuteja, R.; Tuteja, N. OsACA6, a P-type IIB Ca2+ ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing stress-responsive genes. Plant J. 2013, 76, 997–1015. [Google Scholar] [CrossRef]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.-Q.; Yang, Y.; Zhang, A.; Fei, C.-F.; Gu, L.-L.; Sun, S.-J.; Xu, W.; Wang, L.; Liu, H.; Wang, Y.-F. Three CNGC family members, CNGC5, CNGC6, and CNGC9, are required for constitutive growth of Arabidopsis root hairs as Ca2+-permeable channels. Plant Commun. 2020, 1, 100001. [Google Scholar] [CrossRef]
- Pan, Y.; Chai, X.; Gao, Q.; Zhou, L.; Zhang, S.; Li, L.; Luan, S. Dynamic interactions of plant CNGC subunits and calmodulins drive oscillatory Ca2+ channel activities. Dev. Cell 2019, 48, 710–725. [Google Scholar] [CrossRef] [Green Version]
- Vincill, E.D.; Clarin, A.E.; Molenda, J.N.; Spalding, E.P. Interacting glutamate receptor-like proteins in phloem regulate lateral root initiation in Arabidopsis. Plant Cell 2013, 25, 1304–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Yan, J.; Du, X.; Hua, J. Overlapping and differential roles of plasma membrane calcium ATPases in Arabidopsis growth and environmental responses. J. Exp. Bot. 2018, 69, 2693–2703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, L.; Romanowsky, S.M.; Harper, J.F.; Sharrock, R.A. The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol. 2008, 146, 716–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charpentier, M.; Sun, J.; Vaz Martins, T.; Radhakrishnan, G.V.; Findlay, K.; Soumpourou, E.; Thouin, J.; Very, A.A.; Sanders, D.; Morris, R.J.; et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 2016, 352, 1102–1105. [Google Scholar] [CrossRef]
- Capoen, W.; Sun, J.; Wysham, D.; Otegui, M.S.; Venkateshwaran, M.; Hirsch, S.; Miwa, H.; Downie, J.A.; Morris, R.J.; Ané, J.-M.; et al. Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 2011, 108, 14348–14353. [Google Scholar] [CrossRef] [Green Version]
- Perez-Prat, E.; Narasimhan, M.L.; Binzel, M.L.; Botella, M.A.; Chen, Z.; Valpuesta, V.; Bressan, R.A.; Hasegawa, P.M. Induction of a putative Ca-ATPase mRNA in NaCl-adapted cells. Plant Physiol. 1992, 100, 1471–1478. [Google Scholar] [CrossRef] [Green Version]
- Qudeimat, E.; Faltusz, A.M.C.; Wheeler, G.; Lang, D.; Holtorf, H.; Brownlee, C.; Reski, R.; Frank, W. A PIIB-type Ca2+-ATPase is essential for stress adaptation in Physcomitrella patens. Proc. Natl. Acad. Sci. USA 2008, 105, 19555–19560. [Google Scholar] [CrossRef] [Green Version]
- Geisler, M.; Frangne, N.; Gomès, E.; Martinoia, E.; Palmgren, M.G. The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast. Plant Physiol. 2000, 124, 1814–1827. [Google Scholar] [CrossRef] [Green Version]
- Anil, V.S.; Rajkumar, P.; Kumar, P.; Mathew, M.K. A plant Ca2+ pump, ACA2, relieves salt hypersensitivity in yeast. Modulation of cytosolic calcium signature and activation of adaptive Na+ homeostasis. J. Biol. Chem. 2008, 283, 3497–3506. [Google Scholar] [CrossRef] [Green Version]
- Konopka-Postupolska, D.; Clark, G.; Goch, G.; Debski, J.; Floras, K.; Cantero, A.; Fijolek, B.; Roux, S.; Hennig, J. The role of annexin 1 in drought stress in Arabidopsis. Plant Physiol. 2009, 150, 1394–1410. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Mehta, S.; Turano, F.J. The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 2004, 45, 1380–1389. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, H.; Zhao, B.; Wu, H.; Zhu, Y.; Chen, H. Comprehensive In Silico Characterization and Expression Profiling of Nine Gene Families Associated with Calcium Transport in Soybean. Agronomy 2020, 10, 1539. https://doi.org/10.3390/agronomy10101539
Zeng H, Zhao B, Wu H, Zhu Y, Chen H. Comprehensive In Silico Characterization and Expression Profiling of Nine Gene Families Associated with Calcium Transport in Soybean. Agronomy. 2020; 10(10):1539. https://doi.org/10.3390/agronomy10101539
Chicago/Turabian StyleZeng, Houqing, Bingqian Zhao, Haicheng Wu, Yiyong Zhu, and Huatao Chen. 2020. "Comprehensive In Silico Characterization and Expression Profiling of Nine Gene Families Associated with Calcium Transport in Soybean" Agronomy 10, no. 10: 1539. https://doi.org/10.3390/agronomy10101539
APA StyleZeng, H., Zhao, B., Wu, H., Zhu, Y., & Chen, H. (2020). Comprehensive In Silico Characterization and Expression Profiling of Nine Gene Families Associated with Calcium Transport in Soybean. Agronomy, 10(10), 1539. https://doi.org/10.3390/agronomy10101539




