Soil Microbial Community Profiling and Bacterial Metabolic Activity of Technosols as an Effect of Soil Properties following Land Reclamation: A Case Study from the Abandoned Iron Sulphide and Uranium Mine in Rudki (South-Central Poland)
Abstract
:1. Introduction
2. Materials and Methods
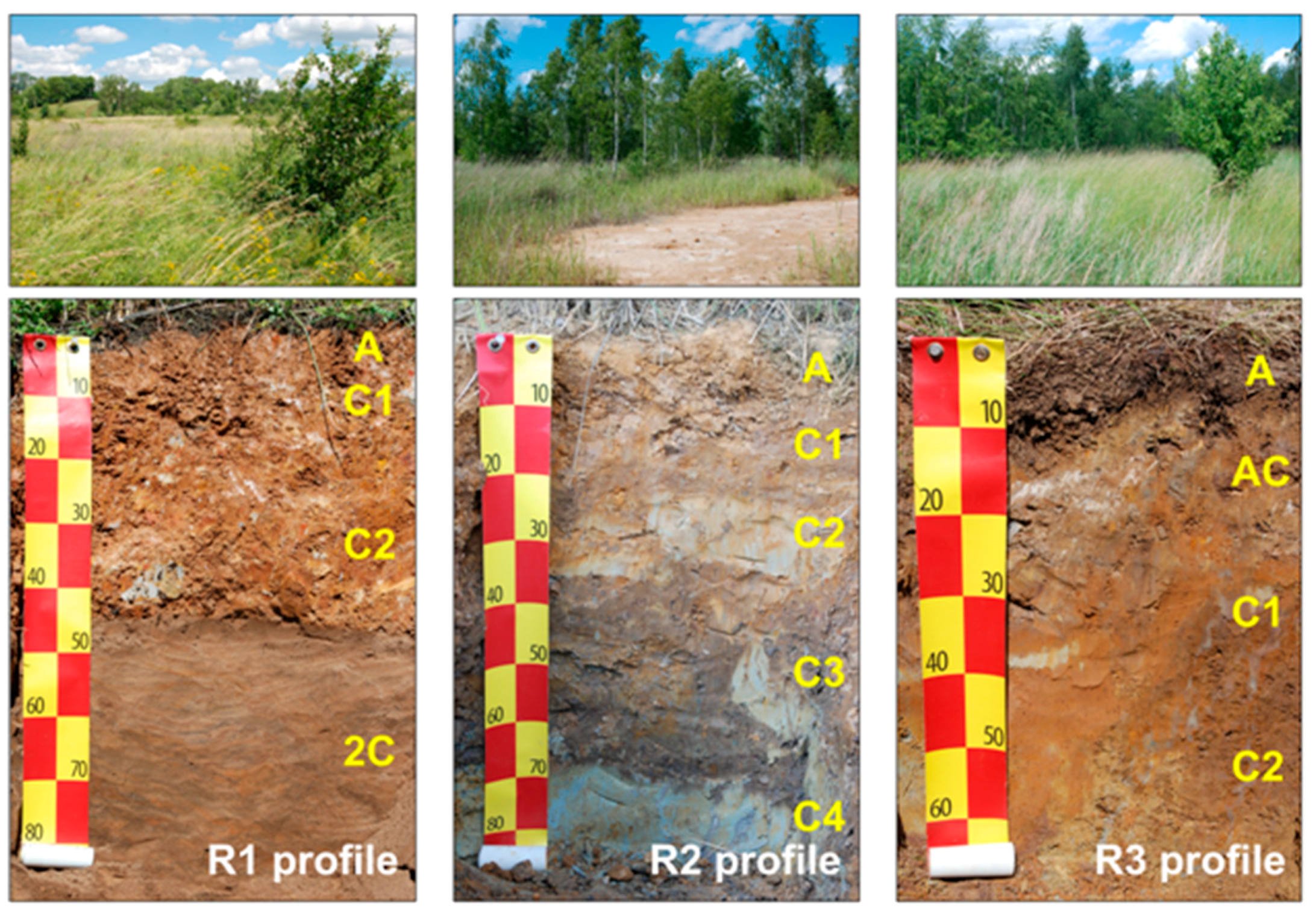
2.1. Study Site Description and Soil Sampling
2.2. DNA Extraction and Next Generation Sequencing (NGS)
2.3. Bioinformatic Analysis
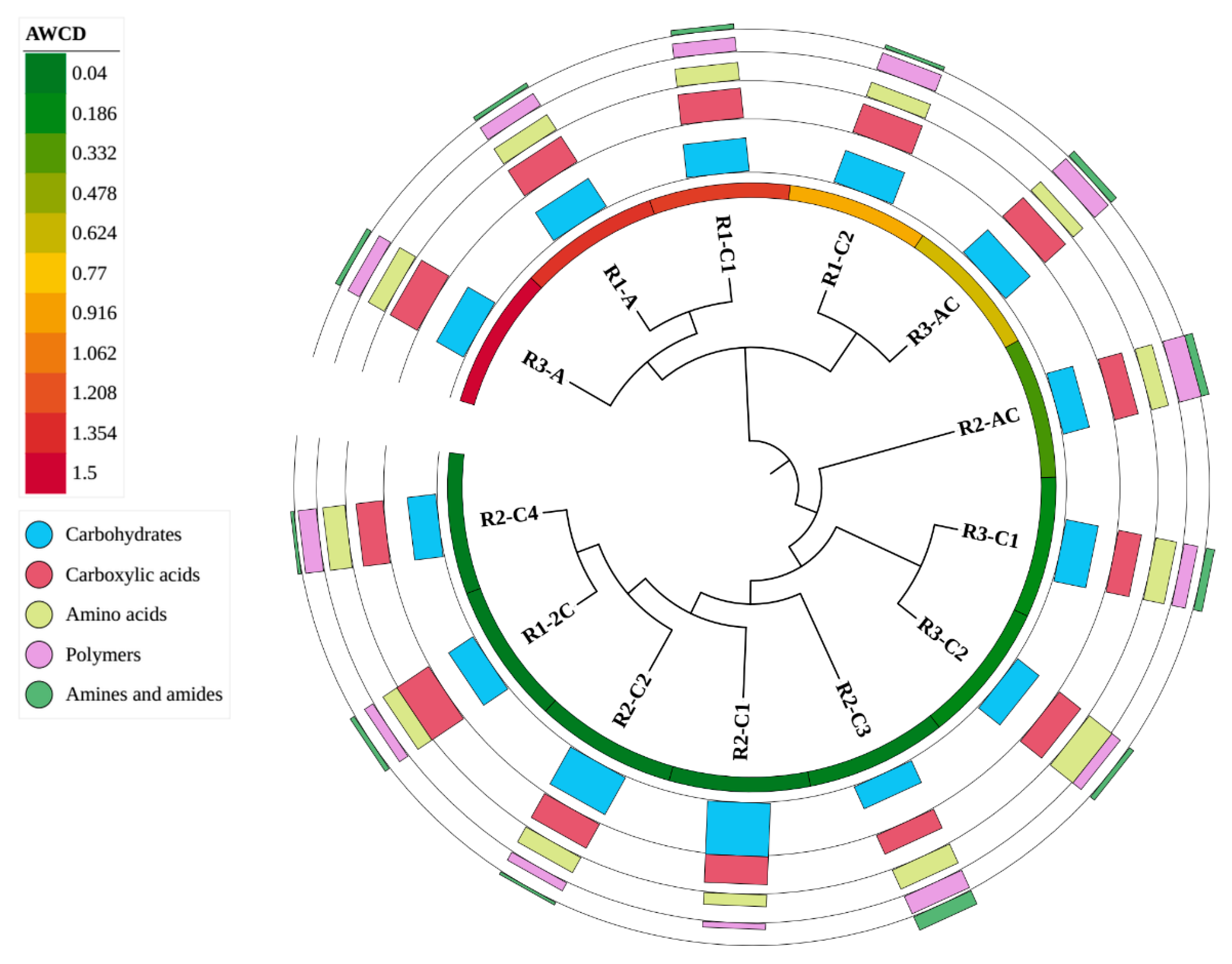
2.4. Bacterial Community-Level Physiological Profiles—Biolog®EcoPlates™
3. Results
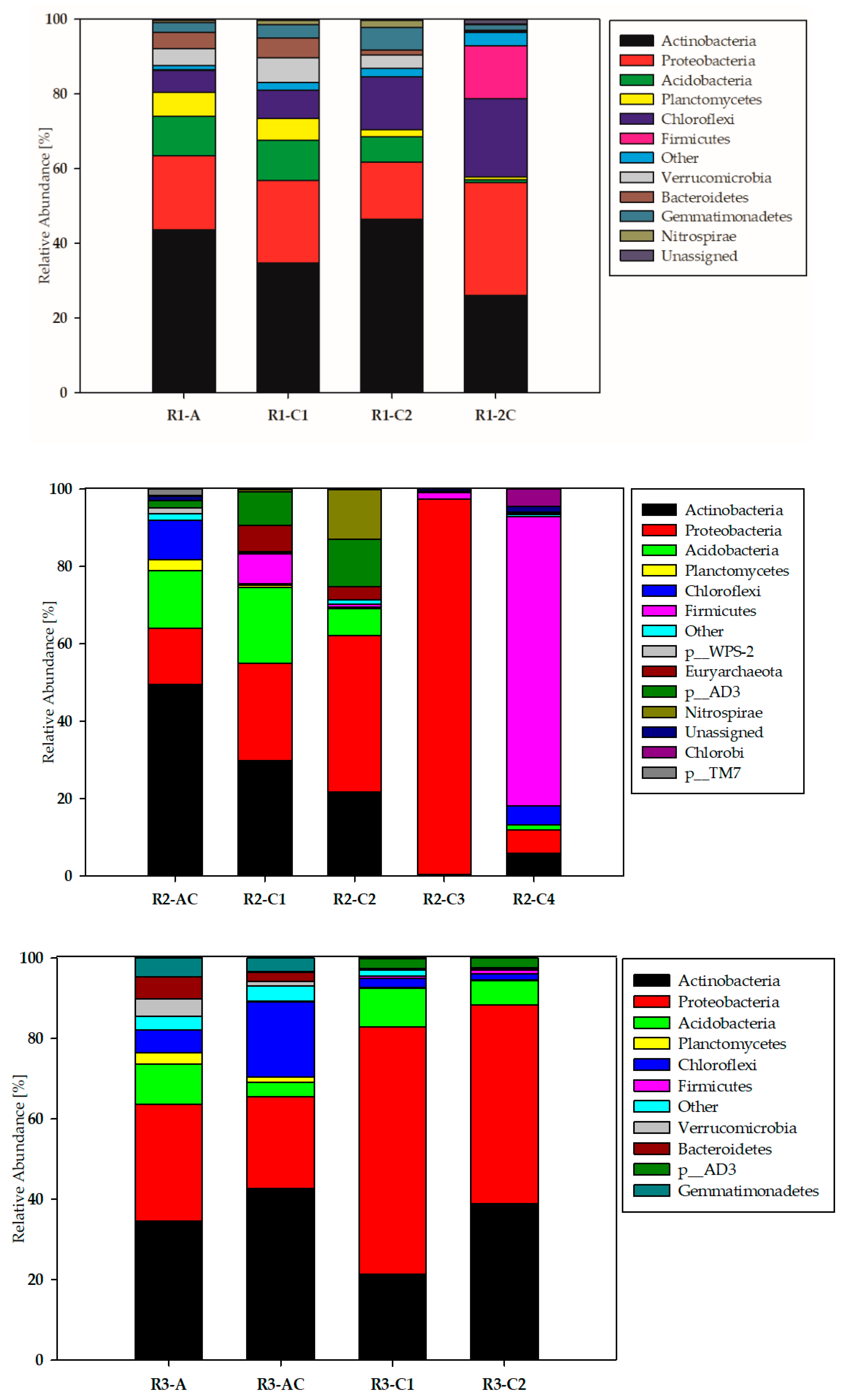
3.1. Biodiversity of the Studied Technosols—Phyla Taxonomic Level
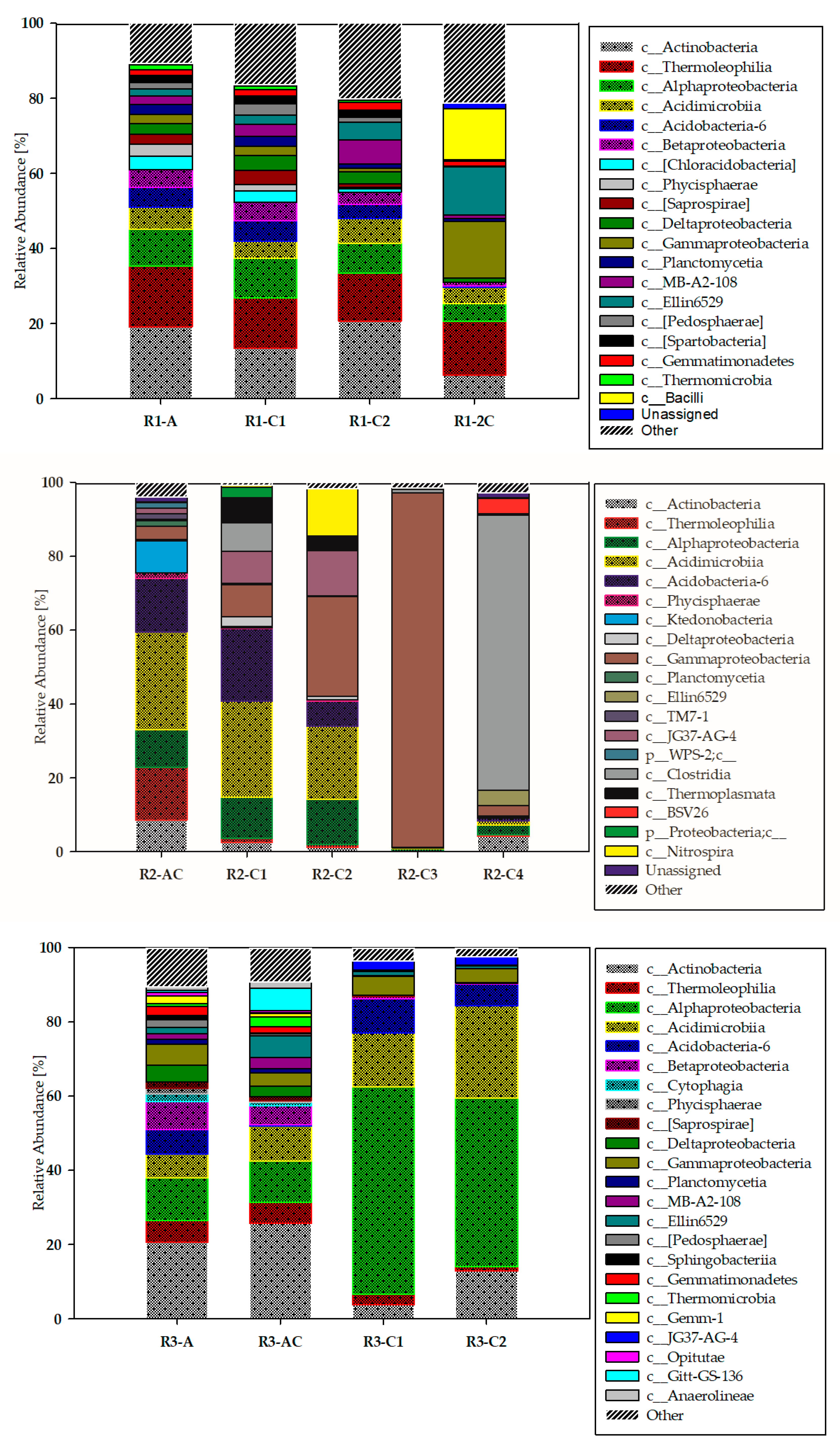
3.2. Biodiversity of the Studied Technosols—Classes Taxonomic Level
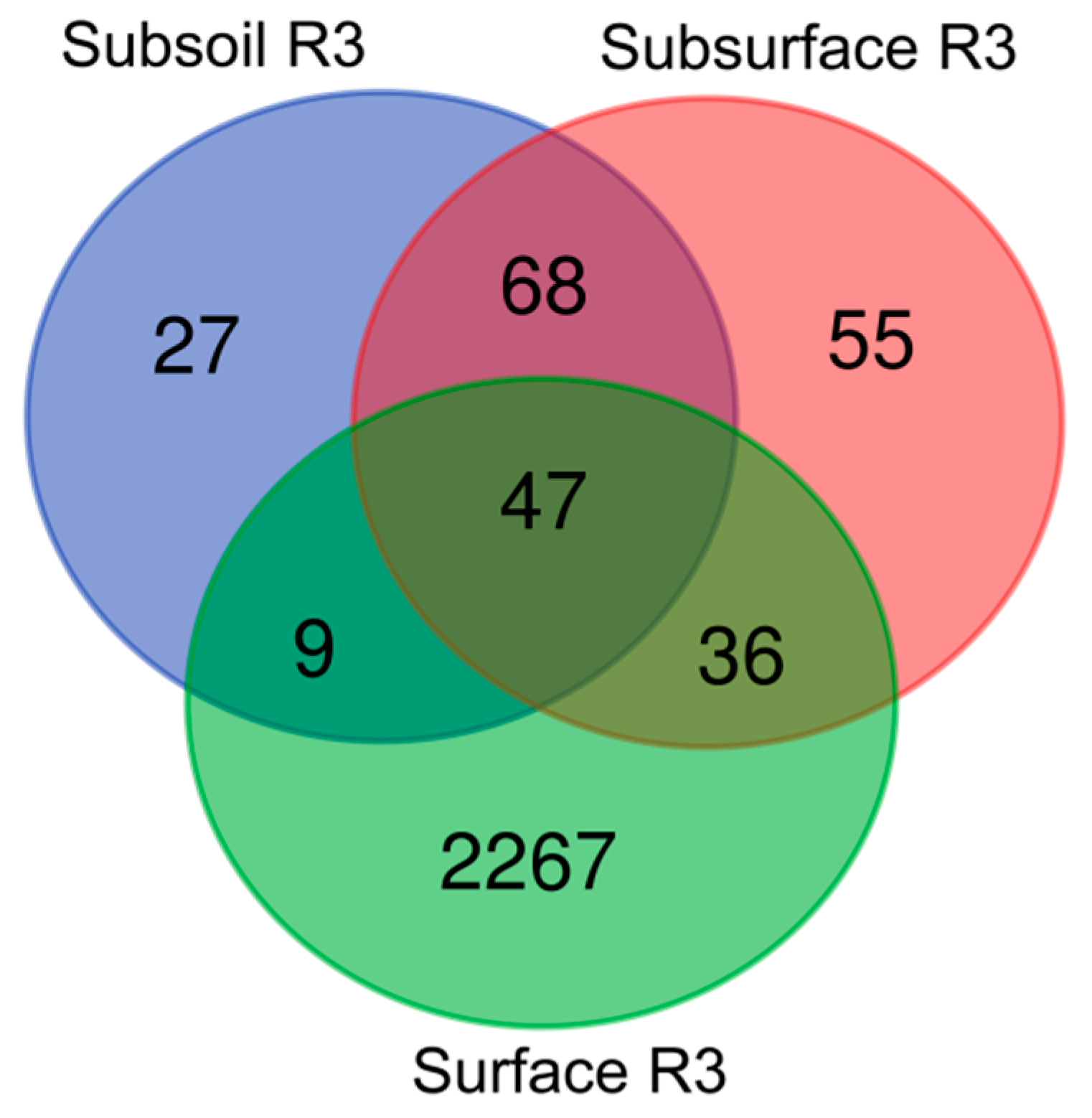
3.3. Beta-Diversity in the Studied Technosols—Genera Taxonomic Level
3.4. Bacterial Community-Level Physiological Profiles—Biolog®EcoPlates™
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- IUSS Working Group, WRB. World Reference Base for Soil Resources 2014, update 2015. In International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; World Soil Rep.: Rome, Italy, 2015. [Google Scholar]
- Uzarowicz, Ł. Technogenic soils developed on mine spoils containing iron sulfides in select abandoned industrial sites: Environmental hazards and reclamation possibilities. Pol. J. Environ. Stud. 2011, 20, 771–782. [Google Scholar]
- Uzarowicz, Ł.; Skiba, S. Technogenic soils developed on mine soils containing iron sulphides: Mineral transformation as indicator of pedogenesis. Geoderma 2011, 163, 95–108. [Google Scholar] [CrossRef]
- Uzarowicz, Ł. Microscopic and microchemical study of iron sulphide weathering in a chronosequence of technogenic and natural soils. Geoderma 2013, 197–198, 137–150. [Google Scholar] [CrossRef]
- Uzarowicz, Ł.; Wolińska, A.; Błońska, E.; Szafranek-Nakonieczna, A.; Kuźniar, A.; Słodczyk, Z.; Kwasowski, W. Technogenic soils (Technosols) developed from mine spoils containing Fe sulphides: Microbiological activity as an indicator of soil development following land reclamation. Appl. Soil Ecol. 2020, 156, 103699. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Izak, D.; Szafranek-Nakonieczna, A.; Banach, A.; Błaszczyk, M. Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by culture independent approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, J.; Zhu, Q.; Zhang, Z.; Lin, X. pH is the primary determinant of the bacterial community structure in agricultural soils impacted by polycyclic aromatic hydrocarbon pollution. Sci. Rep. 2017, 7, 40093. [Google Scholar] [CrossRef] [Green Version]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Błaszczyk, M. Indicators of arable soils fatigue—bacterial families and genera: A metagenomic approach. Ecol. Ind. 2018, 93, 490–500. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Gałązka, A. Biodiversity in the rhizosphere of selected winter wheat (Triticum aestivum L.) cultivars—Genetic and catabolic fingerprinting. Agronomy 2020, 10, 953. [Google Scholar] [CrossRef]
- Poomthongdee, N.; Duangmal, K.; Pathom-aree, W. Acidophylic actinomycetes from rhizosphere soil: Diversity and properties beneficial for plants. J. Antibiot. 2015, 68, 106–114. [Google Scholar] [CrossRef]
- Panyushkina, A.E.; Tsaplina, I.A.; Kondrat’eva, T.F.; Bulaev, A.G. Physiological and morphological characteristics of acidophilic bacteria Leptospirillumferriphilum and Acidithiobacillusthiooxidans, members of a chemolithotrophic microbial consortium. Microbiology 2018, 87, 326–338. [Google Scholar] [CrossRef]
- Wang, R.; Lin, J.Q.; Liu, X.M.; Pang, X.; Zhang, C.J.; Yang, C.L.; Gao, X.Y.; Lin, C.M.; Li, Y.Q.; Li, Y.; et al. Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 2019, 9, 3290. [Google Scholar] [CrossRef]
- Mahdavi, A.; Sajedi, R.H.; Rassa, M. Investigation of acid-neutralizing property of Bacillus cereus GUF8. Biomacromol. J. 2017, 3, 18–25. [Google Scholar]
- Plotnikova, E.G.; Rybkina, D.O.; Anan’ina, L.N.; Yastrebova, O.V.; Demakov, V.A. Characteristics of microorganisms isolated from technogenic soils of the Kama region. Rus. J. Ecol. 2006, 37, 233–240. [Google Scholar] [CrossRef]
- Mangova, K.; Lintnerova, O. Environmental aspects of the low-sulphide post-flotation tailings transformation into anthropogenic soils (Smolnik, Slovakia). Acta Geol. Slov. 2015, 7, 195–212. [Google Scholar]
- Santos, E.S.; Abreu, M.M.; Macías, F.; de Varennes, A. Chemical quality of leachates and enzymatic activities in Technosols with gossan and sulphide wastes from the São Domingos mine. J. Soils Sedim. 2016, 16, 1366–1382. [Google Scholar] [CrossRef]
- Moreno-Barriga, F.; Díaz, V.; Acosta, J.A.; Muñoz, Á.; Faz, Á.; Zornoza, R. Organic matter dynamics, soil aggregation and microbial biomass and activity in Technosols created with metalliferous mine residues, biochar and marble waste. Geoderma 2017, 301, 19–29. [Google Scholar] [CrossRef]
- Ondoño, S.; Bastida, F.; Moreno, J.L. Microbiological and biochemical properties of artificial substrates: A preliminary study of its application as Technosols or as a basis in Green Roof Systems. Ecol. Eng. 2014, 70, 189–199. [Google Scholar] [CrossRef]
- Zornoza, R.; Acosta, J.A.; Faz, A.; Bååth, E. Microbial growth and community structure in acid mine soils after addition of different amendments for soil reclamation. Geoderma 2016, 272, 64–72. [Google Scholar] [CrossRef]
- Šimonovičová, A.; Ferianc, P.; Vojtková, H.; Pangallo, D.; Hanajík, P.; Kraková, L.; Feketeová, Z.; Čerňanský, S.; Okenicová, L.; Žemberyová, M.; et al. AlkalineTechnosol contaminated by former mining activity and its culturable autochtonous microbiota. Chemosphere 2017, 171, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Kumar Awasthi, M.; Ravindran, B.; Sarsaiya, S.; Chen, H.; Wainaina, S.; Singh, E.; Liu, T.; Kumar, S.; Pandey, A.; Singh, L.; et al. Metagenomics for taxonomy profiling: Tools and approaches. Bioengineered 2020, 11, 356–374. [Google Scholar] [CrossRef] [Green Version]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, 245–249. [Google Scholar] [CrossRef] [Green Version]
- Cowan, D.A.; Arslanoglu, A.; Burton, S.G. Metagenomics, gene discovery, and the ideal biocatalyst. Biochem. Soc. Trans. 2004, 32, 298–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, E.E.; Banfield, J.F. Community genomics in microbial ecology and evolution. Nat. Rev. Microbiol. 2005, 3, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Wolińska, A. Metagenomic achievements in microbial diversity determination in croplands: A review. In Microbial Diversity in Genomic Era; Das, S., Dash, H.R., Eds.; Academic Press Elsevier: Cambridge, MA, USA, 2019; Volume 2, pp. 15–35. [Google Scholar]
- Pershina, E.; Valkonen, J.; Kurki, P.; Ivanova, E.; Chirak, E.; Korvigo, I.; Provorov, N.; Andronov, E. Comparative analysis of prokaryotic communities associated with organic conventional farming systems. PLoS ONE 2015, 10, e0145072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazarevic, V.; Whiteson, K.; Gaïa, N.; Gizard, Y.; Hernandez, D.; Farinelli, L.; Østerås, M.; François, P.; Schrenzel, J. Analysis of the salivary microbiome using culture-independent techniques. J. Clin. Bioinf. 2012, 2, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neelkanta, G.; Sultana, H. The use of metagenomic approaches to analyze changes in microbial communities. Microbiol. Ins. 2013, 6, 37–48. [Google Scholar] [CrossRef]
- Garland, J.L.; Millis, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [CrossRef] [Green Version]
- Gałązka, A.; Gawryjołek, K.; Grządziel, J.; Frąc, M.; Księżak, J. Microbial community diversity and their interaction of soil under maize growth in different cultivation techniques. Plant Soil Environ. 2017, 63, 264–270. [Google Scholar]
- Wolińska, A.; Gałązka, A.; Kuźniar, A.; Goraj, W.; Jastrzębska, N.; Grządziel, J.; Stępniewska, Z. Catabolic fingerprinting and diversity of bacteria in MollicGleysol contaminated with petroleum substances. Appl. Sci. 2018, 8, 1970. [Google Scholar] [CrossRef] [Green Version]
- Grządziel, J.; Furtak, K.; Gałązka, A. Microorganisms from different types of soil that are characteristic to Poland—A long-term microplot experiment. Sustainability 2019, 11, 56. [Google Scholar] [CrossRef] [Green Version]
- Kuźniar, A.; Banach, A.; Stępniewska, Z.; Frac, M.; Oszust, K.; Gryta, A.; Kłos, M.; Wolińska, A. Community-level physiological profiles of microorganisms inhabiting soil contaminated with heavy metals. Int. Agrophys. 2018, 32, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Furtak, K.; Gawryjołek, G.; Gajda, A.M.; Gałązka, A. Effects of maize and winter wheat grown under different cultivation techniques on biological activity of soil. Plant Soil Environ. 2017, 63, 449–454. [Google Scholar]
- Chou, Y.M.; Shen, F.T.; Chiang, S.C.; Chang, C.M. Functional diversity and dominant populations of bacteria in banana plantation soils as influenced by long-term organic and conventional farming. Appl. Soil Ecol. 2017, 110, 21–33. [Google Scholar] [CrossRef]
- Lladó, S.; Baldrian, P. Community-level physiological profiling analyses show potential to identify the copiotrophic bacteria present in soil environments. PLoS ONE 2017, 12, e0171638. [Google Scholar] [CrossRef]
- Stefanowicz, A. The Biolog Plate technique as a tool in ecological studies of microbial communities. Pol. J. Environ. Stud. 2006, 15, 669–676. [Google Scholar]
- Stępniewska, H.; Uzarowicz, Ł.; Błońska, E.; Kwasowski, W.; Słodczyk, Z.; Gałka, D.; Hebda, A. Fungal abundance and diversity as influenced by properties of Technosols developed from mine wastes containing iron sulphides: A case study from abandoned iron sulphide and uranium mine in Rudki, south-central Poland. Appl. Soil Ecol. 2020, 145, 103349. [Google Scholar] [CrossRef]
- Skawina, T.; Trafas, M.; Gołda, T. Recultivation of after-mine areas of the pyrite mine “Siarkopol” at Rudki near Kielce. Zesz. Nauk. AGH 1974, 466, 9–21, (in Polish with English abstract). [Google Scholar]
- Gałuszka, A.; Migaszewski, Z.; Duczmal-Czernikiewicz, A.; Dołęgowska, S. Geochemical background of potentially toxic trace elements in reclaimed soils of the abandoned pyrite-uranium mine (south-central Poland). Int. J. Environ. Sci. Technol. 2016, 13, 2649–2662. [Google Scholar] [CrossRef] [Green Version]
- Warda, A. Assessment of reclamation effects in “Staszic” mine in Rudki near Kielce. Geomat. Environ. Engin. 2007, 1, 181–196. [Google Scholar]
- Soliman, T.; Yang, S.Y.; Yamazaki, T.; Jenke-Kodama, H. Profiling soil microbial communities with next-generation sequencing: The influence of DNA kit selection and technician technical expertise. Peer J. 2017, 5, e4178. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Schlatter, D.; Kennedy, P.; Kinkel, L.L.; Kistler, H.C.; Nguyen, N.; Bates, S.T. Effort versus reward: Preparing samples for fungal community characterization in high-throughput sequencing surveys of soils. PLoS ONE 2015, 10, e0127234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thikjs, S.; Op De Beeck, M.; Beckers, B.; Truyens, S.; Stevens, V.; Van Hamme, J.D.; Weyens, N.; Vangronsveld, J. Comparative evaluation of four bacteria-specific primers pairs for 16S rRNA gene surveys. Front. Microbiol. 2017, 8, 494. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Meth. 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Team, R.C.R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R.-project.org/ (accessed on 20 April 2020).
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Woźniak, M.; Furtak, K.; Gałązka, A.; Dziadczyk, E.; Skórzyńska-Polit, E.; Wolińska, A. New insight into the composition of wheat seed microbiota. Int. J. Mol. Sci. 2020, 21, 4634. [Google Scholar] [CrossRef] [PubMed]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Pohland, B.; Owen, B. BiologEcoPlates standard methods. TAS Tech. Biul. Biol. 2009, 1, 1–3. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Frąc, M.; Oszust, K.; Lipiec, J. Community level physiological profiles (CLPP), characterization and microbial activity of soil amended with dairy sewage sludge. Sensors 2012, 12, 3253–3268. [Google Scholar] [CrossRef] [Green Version]
- Hafeez, F.; Spor, A.; Breuil, M.C.; Schwartz, C.; Martin-Laurent, F.; Philipoot, L. Distribution of bacteria and nitrogen-cycling microbial communities along constructed Technosol depth-profiles. J. Hazard. Mater. 2012, 231, 88–97. [Google Scholar] [CrossRef]
- Thouin, H.; Battaglia-Brunet, F.; Norini, M.P.; Joulian, C.; Hellal, J.; Le Forestier, L.; Dupraz, S.; Gautret, P. Microbial community response to environmental changes in a technosol historically contaminated by the burning of chemical ammunitions. Sci. Total Environ. 2019, 697, 134108. [Google Scholar] [CrossRef] [Green Version]
- Hafeez, F.; Martin-Laurent, F.; Beguet, J.; Bru, D.; Cortet, J.; Schwartz, C.; Morel, J.L.; Philippot, L. Taxonomic and functional characterization of microbial communities in Technosols constructed for remediation of a contaminated industrial wasteland. J. Soils Sedim. 2012, 12, 1396–1406. [Google Scholar] [CrossRef]
- Piotrowska-Długosz, A.; Charzyński, P. The impact of the soil sealing degree on microbial biomass, enzymatic activity, and physicochemical properties in the Ekranic Technosols of Toruń (Poland). J. Soils Sedim. 2015, 15, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Celestina, C.; Wood, J.L.; Manson, J.B.; Wang, X.; Sale, P.W.G.; Tang, C.; Franks, A.E. Microbial communities in top-and subsoil of repacked soil columns respond differently to amendments but their diversity is negatively correlated with plant productivity. Sci. Rep. 2019, 9, 8890. [Google Scholar] [CrossRef]
- Polain, K.; Knox, O.; Wilson, B.; Pereg, L. Subsoil microbial diversity and stability in rotational cotton systems. Soil Syst. 2020, 4, 44. [Google Scholar] [CrossRef]
- Ivanova, A.A.; Kulichevskaya, I.S.; Merkel, A.Y.; Toshchakov, S.V.; Dedysh, S.N. High diversity of Plantcomycetes in soils of two lichen-dominated sub-arctic ecosystems of northwestern Siberia. Front. Microbiol. 2016, 7, 2065. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, A.A.; Wegner, C.E.; Kim, Y.; Liesack, W.; Dedysh, S.N. Identification of microbial populations driving biopolymer degradation in acidic peatlands by metatranscriptomic analysis. Mol. Ecol. 2016, 25, 4818–4835. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Ivanova, A.A. Planctomycetes in boreal and subarctic wetlands: Diversity patterns and potential ecological functions. FEMS Microbiol. Ecol. 2019, 95, fiy227. [Google Scholar] [CrossRef] [Green Version]
- Hermans, S.M.; Buckley, H.L.; Case, B.S.; Curran-Cournane, F.; Taylor, M.; Lear, G. Bacteria as emerging indicators of soil condition. Appl. Environ. Microbiol. 2017, 83, e02826-16. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.M.; Jung, J.Y.; Yergeau, E.; Hwang, C.Y.; Hinzman, L.; Nam, S.; Hong, S.G.; Kim, O.S.; Chun, J.; Lee, Y.K. Bacterial community structure and soil properties of a subarctic tundra soil in Council, Alaska. FEMS Microbiol. Ecol. 2014, 89, 465–475. [Google Scholar] [CrossRef]
- Sadeghi, P.M.M.; Pourbabaee, A.A.; Alikhani, H.A.; Haidari, A.; Manafi, Z. The diversity of sulfur-oxidizing bacterial populations at an Iranian copper mine and the surrounding agricultural soils. Appl. Ecol. Environ. Res. 2016, 14, 509–533. [Google Scholar] [CrossRef]
- Wu, X.; Wong, Z.L.; Sten, P.; Engblom, S.; Osterholm, P.; Dopson, M.; Nakatsu, C. Microbial community potentially responsible for acid and metal release from an Ostrobothnian acid sulfate soil. FEMS Microbiol. Ecol. 2013, 84, 555–563. [Google Scholar] [CrossRef]
- Yadav, N.; Yadav, A.N. Actinobacteria for sustainable agriculture. J. Appl. Biotechnol. Bioengin. 2019, 6, 38–41. [Google Scholar]
- Wu, M.; Ye, X.; Chen, K.; Li, W.; Yuan, J.; Jiang, X. Bacterial community shift and hydrocarbon transformation during bioremediation of short-term petroleum-contaminated soil. Environ. Pollut. 2017, 223, 657–664. [Google Scholar] [CrossRef]
- Wu, X.; Gu, Y.; Wu, X.; Zhou, X.; Zhou, H.; Amanze, C.; Shen, L.; Zeng, W. Construction of tetracycline degrading bacterial consortium and its application evaluation in laboratory-scale soil remediation. Microorganisms 2020, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Dutta, A.; Sarkar, J.; Panigrahi, M.K.; Sar, P. Low-abundance members of the Firmicutes facilitate bioremediation of soil impacted by highly acidic mine drainage from the Malanjkhand copper project, India. Front. Microbiol. 2018, 9, 2882. [Google Scholar] [CrossRef]
- Morawe, M.; Hoeke, H.; Wissenbach, D.K.; Lentendu, G.; Wubet, T.; Krober, E.; Kolb, S. Acidotolerant bacteria and fungi as a sink of methanol-derived carbon in a deciduous forest soil. Front. Microbiol. 2017, 8, 1361. [Google Scholar] [CrossRef]
- Konopka, A.; Oliver, L.; Turco, R.F. The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb. Ecol. 1998, 35, 103–115. [Google Scholar] [CrossRef]
- Smalla, K.; Wachtendorf, U.; Heuer, H.; Liu, W.; Forney, L. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl. Environ. Microbiol. 1998, 64, 1220–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preston-Mafham, J.; Boddy, L.; Randerson, P.F. Analysis of microbial community functional diversity using sole-carbon-source utilisation profiles—A critique. FEMS Microbiol. Ecol. 2002, 42, 1–14. [Google Scholar] [PubMed]
- Sofo, A.; Ricciuti, P. A standardized method for estimating the functional diversity of soil bacterial community by BiologEcoPlates assay—The case study of a sustainable olive orchard. Appl. Sci. 2019, 9, 4035. [Google Scholar] [CrossRef] [Green Version]
- Zak, J.C.; Willig, M.R.; Moorhead, D.L.; Wildman, H.G. Functional diversity of microbial communities: A quantitative approach. Soil Biol. Biochem. 1994, 26, 1101–1108. [Google Scholar] [CrossRef]
- Kenarova, A.; Radeva, G.; Tryakov, I.; Boteva, S. Community level physiological profiles of bacterial communities inhabiting uranium mining impacted sites. Ecotoxicol. Environ. Saf. 2014, 100, 226–232. [Google Scholar] [CrossRef]
- Martinez-Toledo, A.; Gonzalez-Mille, D.J.; Garcia-Arreola, M.E.; Cruz-Santiago, O.; Trejo-Acevedo, A.; Ilizaliturri-Hernandez, C.A. Patterns in utilization of carbon siurces in soil microbial communities contaminated with solid astes from San Luis Potosi, Mexico. Ecotoxicol. Environ. Saf. 2021, 208, 111493. [Google Scholar] [CrossRef]
- Kawina, R.; Lebeau, M.; Martineau, S.; Amyot, M. Bioremediation of engine-oil contaminated soil using local residual organic matter. Peer J. 2019, 7, e7389. [Google Scholar]
- Gryta, A.; Frąc, M.; Oszust, K. The application of the Biolog EcoPlate approach in ecotoxicological evaluation of dairy sewage sludge. Appl. Biochem. Biotechnol. 2014, 174, 1434–1443. [Google Scholar] [CrossRef] [Green Version]
- Fazekas, J.; Fazekasova, D.; Adamisin, P.; Hulicova, P.; Benkova, E. Functional diversity of microorganisms in metal- and alkali-contaminated soils of central and north-eastern Slovakia. Soil Water Res. 2019, 14, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Pająk, M.; Błońska, E.; Frąc, M.; Oszust, K. Functional diversity and microbial activity of forest soils that are heavily contaminated by lead and zinc. Water Air Soil Pollut. 2016, 227, 348. [Google Scholar] [CrossRef] [Green Version]
- Epelde, L.; Lanzen, A.; Martin, I.; Virgel, S.; Mijangos, I.; Besga, G.; Garbisu, C. The microbiota of technosols resembles that of a nearby forest soil three years after their establishment. Chemosphere 2018, 220, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Markowicz, A.; Płaza, G.; Piotrowska-Seget, Z. Activity and functional diversity of microbial communities in long-term hydrocarbon and heavy metal contaminated soils. Arch. Environ. Prot. 2016, 42, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Wolińska, A.; Frąc, M.; Oszust, K.; Szafranek-Nakonieczna, A.; Zielenkiewicz, U.; Stępniewska, Z. Microbial biodiversity of meadows under different modes of land use: Catabolic and genetic fingerprinting. World J. Microbiol. Biotechnol. 2017, 33, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Soil Zone | Genus | Soil Profile | ||
|---|---|---|---|---|
| R1 | R2 | R3 | ||
| Surface | Conexibacter | + | ||
| Acidothermus | + | |||
| Granulicella | + | |||
| Gaiella | + | |||
| Nocardioides | + | + | ||
| Streptomyces | + | + | ||
| CandidatusUdaerobacter | + | + | ||
| Acidiphilium | + | |||
| Metallibacterium | + | |||
| Sulfurifustis | + | |||
| Subsurface | Acidiphilium | + | + | |
| Acidithiobacillus | + | |||
| Leptospirillum | + | |||
| Metallibacterium | + | |||
| Gaiella | + | |||
| Acidocella | + | |||
| Acidothermus | + | |||
| Kribbella | + | |||
| Subsoil | Desulfosporosinus | + | ||
| Bacillus | + | |||
| Acidithiobacillus | + | |||
| Acidiphilium | + | |||
| Pseudonocardia | + | |||
| Gaiella | + | |||
| Jatrophihabitans | + | |||
| Sulfurifustis | + | |||
| Paenisporosarcina | + | |||
| Alkanibacter | + | |||
| Acidocella | + | |||
| Cellulomonas | + | |||
| Psychrobacter | + | |||
| Serratia | + | |||
| Ferrithrix | + | |||
| Soil Profile | Soil Sample Label | H′ | D | 1/D |
|---|---|---|---|---|
| R1 | R1-A | 5.930 | 0.0101 | 0.9898 |
| R1-C1 | 6.140 | 0.0075 | 0.9924 | |
| R1-C2 | 2.830 | 0.1866 | 0.8134 | |
| R1-2C | 2.339 | 0.3794 | 0.6206 | |
| R2 | R2-AC | 4.265 | 0.0373 | 0.9626 |
| R2-C1 | 3.500 | 0.0492 | 0.9508 | |
| R2-C2 | 3.091 | 0.0822 | 0.9177 | |
| R2-C3 | 0.335 | 0.9119 | 0.0881 | |
| R2-C4 | 1.610 | 0.5222 | 0.4778 | |
| R3 | R3-A | 6.272 | 0.0073 | 0.9927 |
| R3-AC | 5.827 | 0.0094 | 0.9905 | |
| R3-C1 | 5.807 | 0.0094 | 0.9906 | |
| R3-C2 | 2.880 | 0.1365 | 0.8635 |
| Soil Profile | Soil Sample Label | Shannon-Wiener (H′) | Evenness (E) |
|---|---|---|---|
| R1 | R1-A | 3.286 | 0.983 |
| R1-C1 | 3.219 | 0.986 | |
| R1-C2 | 2.701 | 1.248 | |
| R1-2C | 2.636 | n.d. | |
| R2 | R2-AC | 2.836 | 1.066 |
| R2-C1 | 2.713 | n.d. | |
| R2-C2 | 2.263 | n.d. | |
| R2-C3 | 2.574 | n.d. | |
| R2-C4 | 1.875 | n.d. | |
| R3 | R3-A | 3.288 | 0.980 |
| R3-AC | 3.210 | 1.019 | |
| R3-C1 | 2.804 | 1.018 | |
| R3-C2 | 2.736 | 1.338 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolińska, A.; Włodarczyk, K.; Kuźniar, A.; Marzec-Grządziel, A.; Grządziel, J.; Gałązka, A.; Uzarowicz, Ł. Soil Microbial Community Profiling and Bacterial Metabolic Activity of Technosols as an Effect of Soil Properties following Land Reclamation: A Case Study from the Abandoned Iron Sulphide and Uranium Mine in Rudki (South-Central Poland). Agronomy 2020, 10, 1795. https://doi.org/10.3390/agronomy10111795
Wolińska A, Włodarczyk K, Kuźniar A, Marzec-Grządziel A, Grządziel J, Gałązka A, Uzarowicz Ł. Soil Microbial Community Profiling and Bacterial Metabolic Activity of Technosols as an Effect of Soil Properties following Land Reclamation: A Case Study from the Abandoned Iron Sulphide and Uranium Mine in Rudki (South-Central Poland). Agronomy. 2020; 10(11):1795. https://doi.org/10.3390/agronomy10111795
Chicago/Turabian StyleWolińska, Agnieszka, Kinga Włodarczyk, Agnieszka Kuźniar, Anna Marzec-Grządziel, Jarosław Grządziel, Anna Gałązka, and Łukasz Uzarowicz. 2020. "Soil Microbial Community Profiling and Bacterial Metabolic Activity of Technosols as an Effect of Soil Properties following Land Reclamation: A Case Study from the Abandoned Iron Sulphide and Uranium Mine in Rudki (South-Central Poland)" Agronomy 10, no. 11: 1795. https://doi.org/10.3390/agronomy10111795
APA StyleWolińska, A., Włodarczyk, K., Kuźniar, A., Marzec-Grządziel, A., Grządziel, J., Gałązka, A., & Uzarowicz, Ł. (2020). Soil Microbial Community Profiling and Bacterial Metabolic Activity of Technosols as an Effect of Soil Properties following Land Reclamation: A Case Study from the Abandoned Iron Sulphide and Uranium Mine in Rudki (South-Central Poland). Agronomy, 10(11), 1795. https://doi.org/10.3390/agronomy10111795








