Abstract
The investigation on the genetic diversity of grapevine germplasm is crucial for a more efficient use of grapevine genetic resources in light of changing environmental conditions. Here, we used simple sequence repeats (SSRs) coupled with single nucleotide polymorphism (SNP) markers to disclose grapevine genetic diversity of a collection of Apulian minor/neglected genotypes. Their relationships with national or international cultivars were also examined. Genetic diversity was investigated using 10 SSR markers and 1,178 SNPs generated by genotyping by sequencing (GBS). Based on the SSR data, the 128 genotypes were classified into six main genetic clusters. Twenty-four putative cases of synonymy and 2 of misnamings were detected. Ten “unknown” autochthonous genotypes did not show high similarity to Apulian, national, or international varieties. We took advantage of available GBS-derived SNP data points for only forty genotypes to better investigate the genetic distance among them, identify private SNP alleles, and divergent loci putatively under selection. Based on SNP alleles, two interesting gene pools of minor/neglected Apulian samples were identified. Genetic divergence was investigated by FST and allowed the detection of loci capable of differentiating the gene pools. Overall, this work emphasizes the need for recovering the untapped genetic variability that characterizes minor/neglected grapevine Apulian genotypes and the requirement to preserve and use more efficiently grapevine genetic resources in breeding programs.
Keywords:
grapevine; SSR; genotyping by sequencing; SNP; genetic diversity; private alleles; divergent loci 1. Introduction
Grapevine (Vitis vinifera subsp. vinifera, 2n = 2x = 38) domestication occurred in the Near East and Caucasian area during the Neolithic (ca. 8500–4000 BC) from wild forms (Vitis vinifera L. subsp. sylvestris (Gmelin) Hegi) [1]. Then, grapevine was spread by humans in the Middle East and Central Europe, which are considered secondary domestication centers [2].
Three processes had a considerable impact on the development of cultivated grapevines: sexual reproduction, vegetative propagation, and somatic mutations. New combinations obtained by sexual reproduction produce novel genotypes. Then, plants are usually maintained and multiplied by vegetative propagation (asexual). The occurrence of somatic mutations in one clone and not in others might eventually lead to plants of the same cultivar showing a different phenotype. This is referred to as clonal variation [3].
At present, about 24,500 varieties are registered in the Vitis International Variety Catalogue [4], and 1902 are officially authorized for cultivation in the European Union [5]. Nonetheless, most of the production is based on a few cultivars such as Cabernet Sauvignon, Merlot, Chardonnay Blanc, Sauvignon Blanc, and Syrah, while many genotypes, largely cultivated in the past, are confined to very small areas [6].
The use of a restricted panel of grape cultivars could reduce the resilience of plants to pest attacks, fungal contaminations, and virus diseases. A solution to these problems could arise from the use of new resistant hybrids obtained by crosses among different Vitis species. Both Italian and French law, however, does not allow their use in the production of labeled wines. The Italian regulation (law 238 of 12 December 2016) defines “Italian autochthonous grape cultivars” as the key for the setting-up of PDO (Protected Designation of Origin) and PGI (Protected Geographical Indication) wines. Research and investigation on minor/neglected cultivars could enlarge the panel of grapes used for the production of Italian wines strengthening the link with the territory and the possibility to identify new sources of resistant genes.
The particular position of Italy in the middle of the Mediterranean basin has favored the cultivation of a wide array of grapevine varieties with different uses: wine, table, and dessert grapes. As a consequence, Italy is the leading country for grape production, with 7.2 million tons in 2017 from more than 1000 registered varieties [7]. Apulia, in Southern Italy, is a strategic area of the Italian wine industry, with 84,830 hectares cultivated and a wine production of 14 million hectoliters in 2018 [7]. After many years of high yield production of vine grapes suitable for wine blending, Apulia is moving towards a high-quality production. Often it is based on the exploitation of autochthonous varieties, such as Primitivo, Negroamaro, Uva di Troia, Bombino nero, Malvasia nera di Brindisi, which are used to produce PDO and/or PGI wines [8,9].
In the last years, the threats posed by climate change to the wine sector have increased research interest on the effects of such change on grape phenology [10], water management in the vineyards [11,12,13], and diseases development [14,15,16]. These issues gave impetus to projects for collecting, characterizing, and recovering local genotypes and wild relatives as key resources to select and/or develop novel varieties better adapted to changing environmental conditions [17,18,19,20]. The success of future breeding programs will depend upon (i) the continuous recovering and exploitation of genetic resources (i.e., crop wild relatives, landraces, neglected varieties); (ii) the exploitation of knowledge on genome architecture and function, and (iii) the characterization of genetic diversity and the subsequent identification of novel favorable/beneficial alleles through molecular and genomic approaches.
Simple sequence repeats (SSRs/microsatellites) are informative molecular markers used for the genotyping and traceability of many Mediterranean crops [21,22,23,24,25,26,27]. Sometimes, their discriminating power is limited; therefore, they are combined with SNP markers to further improve genotyping [28].
The first draft of the grapevine genome (referred to as 8X genome) was obtained from a highly homozygous accession (Pinot Noir, PN40024) and was published in 2007 [29]. In the same year, Velasco et al. resolved the genome sequence of a highly heterozygous grapevine clone (Pinot Noir, ENTAV115) [30]. An improved version of the PN40024 genome (called the 12X.v0 version) was released into the public domain in 2011 [31], and a further version (referred to as 12X.v2 version) accompanied by a more reliable annotation was published in 2017 [32]. The grapevine genome has an estimated size of ~500 Mb. It is arranged in 19 chromosomes and includes over 33,500 protein-coding genes [33]. The availability of this resource favors the re-sequencing of many individuals to delineate inter- and intra-species genetic variations across the Vitaceae family [34]. It is also facilitating the large-scale genotyping (mainly based on simple sequence repeats (SSRs/microsatellites) and/or SNP markers) of grape collections [35,36,37,38].
In addition, the genetic diversity of the grapevine germplasm from Magna Grecia, the coastal areas of Southern Italy (i.e., Basilicata, Calabria, Campania, Apulia, and Sicily), was investigated using markers spotted onto a 18K SNP array, and provided evidence of gene flow between varieties from Greece and Southern Italy [39]. The identification of divergent loci across germplasm collections may give indications on the genetic differentiation that occurred during the transition from local adapted to modern varieties, as previously investigated in grapevine [40,41,42] and in other Mediterranean crops [43,44].
Herein, we investigated the genetic diversity of a panel of minor/neglected Apulian genotypes collected from marginal areas of the region and previously characterized for ampelographic and morphological traits [45], with the aim to emphasize the need for recovering the untapped genetic variability that often characterize neglected germplasm. We combined SSR and genotyping-by-sequencing-derived SNP markers to (i) characterize the neglected Apulian grapevine genotypes; (ii) disclose relationships between these latter and national or international varieties; (iii) highlight specific molecular patterns that characterize different gene pools, and (iv) detect signatures of genetic divergence.
2. Materials and Methods
2.1. Plant Material
A total of 128 genotypes, conserved at the ex situ collection at Centro di Ricerca, Sperimentazione e Formazione in Agricoltura (CRFSA) “Basile Caramia” located in Locorotondo (40.7559° N, 17.3263° E), Bari, Italy, were subjected to genotyping. The list of genotypes under investigation is detailed in Table S1. Eighty-four were local genotypes (tagged as ‘minor’) collected from small vineyards in different physiographic areas of Apulia during the years 2014–2016 (Figure S1). They were selected based on results from a previous screening for technological, genetic, and morphological traits (data not shown). For 17 samples (tagged as ‘unknown’), only the collection site was known. The collection included also 26 traditional varieties largely cultivated in Apulia and used for renowned wine productions (tagged as ‘Apulian’), and 18 national and international varieties. The latter dataset included two Albanian (Shesh I Bardhe and Vlosh) and one Croatian (Jabixhar) varieties, as Apulia has always had intense commercial exchanges with these two countries.
2.2. SSR Genotyping
Three fresh young leaves of medium size per plant were collected from all the 128 genotypes maintained at the conservation field (CRFSA), and were placed in ice until being stored at −80 °C. DNA was extracted following the procedure described in [46], and its quality and quantity was checked at the Nanodrop 2000C spectrophotometer (Thermo Scientific, Waltham, MA, USA). Samples were analyzed using 10 polymorphic SSR markers. Six of them were suggested for grapevine fingerprinting by the International Organization of Vine and Wine, VvS2, VvMD7, VvMD25, VvMD27, VvMD28, and VvMD32 [47,48,49]; the remaining four, VrZAG21, VrZAG62, VrZAG64, and VrZAG79, were selected based on literature [50] (Table S2). PCR amplifications were performed on a T100™ Thermal Cycler (Bio-Rad, Waltham, MA, USA) using fluorescent-labeled primers (ThermoFisher Scientific, Waltham, MA, USA) using reaction conditions and cycling program as reported in [51]. PCR products were diluted 1:2 in sterile water and 2 µL of these dilutions were mixed with 12 µL Hi-Di™ formamide (Applied Biosystems™, Waltham, MA, USA) and 0.3 µL GeneScan™ 600 LIZ™ dye size standard (Applied Biosystems™, Waltham, MA, USA). DNA fragments were denatured and size-separated using capillary electrophoresis on an ABI PRISM 3130 Avant Genetic Analyzer (Applied Biosystems™, Waltham, MA, USA). Allele size was estimated with the GeneMapper® software version 5.0 (Applied Biosystems™, Waltham, MA, USA), with two independent readings of the chromatograms. Rates of missing data were below 8%. SSR profiles of the minor Apulian genotypes were compared with those of the local, national, or international varieties (the latter recorded in the European Vitis Database; www.eu-vitis.de/index.php).
2.3. Genotyping by Sequencing and SNP Calling
Available genotyping by sequencing (GBS) data for a subset of 40 grapevine genotypes (35 from Apulia and 5 selected among the most widespread Italian and international varieties), were used to corroborate SSR genotyping. DNA was extracted as described before, and GBS was performed by the Elshire group Ltd. (https://www.elshiregroup.co.nz/). GBS tags were aligned onto the PN40024 reference genome and SNP calling was performed by TASSEL-GBS [52]. The list of raw SNP data was filtered for call rate (<20%), minimum depth of coverage (>5), minor allele frequency (≥0.05), and Hardy–Weinberg equilibrium (p ≤ 0.001), using VCFtools [53]. PLINK [54] was run for linkage disequilibrium (LD) pruning (r2 = 0.5).
2.4. Genetic Diversity and Population Structure
The GenAlEx software version 6.5 [55] was used to compute genetic diversity statistics for each SSR locus: number of alleles (Na), effective alleles (Ne), Shannon’s informative index (I), observed (Ho) and expected heterozygosity (He), fixation index (F) [56], number of private alleles (i.e., alleles that are found only in a single population/genotype among a broader collection of populations/genotypes) [57]. Pairwise relatedness was calculated using SSR data to measure the allelic similarity based on the LRM (Lynch and Ritland mean estimator) [58]. The polymorphism information content (PIC) for SSR informativeness was calculated by Cervus v.2.0 [59]. The chromosomal coordinates (start/end position) of each SSR were retrieved from the Vitis vinifera genome browser available at https://urgi.versailles.inra.fr/gb2/gbrowse/vitis_12x_pub/. In addition, for genic SSRs, the putative function of the corresponding genes was described by gene ontology (GO) terms (https://jetgene.bioset.org/genomes/plants/vitis_vinifera/gene_ontology_annotations/GO/30653/go).
Frequency-based genetic distances of the 128 grapevine samples were computed to construct an un-weighted neighbor-joining dendrogram using DARWIN v 6.0.010 (http://darwin.cirad.fr) with 1000 bootstrap replicates and no re-sampling [60].
ADMIXTURE [61] v1.3 was used to infer population structure and assign genotypes to different genetic groups by using the following parameters: 10-fold cross-validation (CV) for sub-populations (K) ranging from K = 1 to 10 and 1000 bootstrap replicates. Cross validation (CV) scores were used to determine the best K value. A qi (coefficient membership) > 0.80 was used to assign individuals to a specific sub-population [35]. AWclust [62] used the gap statistics to select the optimal number of clusters (K) that best fit with the population structure [62]. The fixation index (FST) between pairs of sub-populations as derived from the clustering analysis was estimated using the SNP & Variation Suite (SVS) v8.x (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com). A pairwise identity-by-state (IBS) distance matrix was built using PLINK [56] in order to assess relatedness among the 40 genotypes subjected to GBS and visualized using Morpheus (https://software.broadinstitute.org/morpheus/). Finally, the list of private SNPs was derived using GenAlEx [55].
2.5. Divergent Loci
In order to measure the genetic divergence between the sub-populations identified by AWclust, the FST at individual SNP loci was calculated on the basis of the algorithm in Weir and Cockerham [63] using SVS v8.x (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com). As this parameter ranges from 0 (i.e., no genetic divergence between the populations) to 1 (extreme divergence between populations), we considered a threshold ≥ 0.25 (i.e., strong differentiation) to identify putatively divergent SNP loci [54]; these were mapped on the PN40024 reference genome (http://genomes.cribi.unipd.it/DATA/V2/V2.1/V2.1.gff3) using the vcf-annotate utility with the aim to identify putative candidate genes under selection.
3. Results
3.1. SSR-Based Analysis of Grapevine Genotypes
The ten SSR markers were highly polymorphic and very effective in discriminating the samples. A total of 110 alleles were identified ranging from 7 alleles at VvMD25 and VrZAG64 to 15 alleles at VvMD28 (Table 1). PIC values ranged from 0.721 (VvMD25) to 0.853 (VvZAG79), with an average value of 0.802. SSR analysis revealed high genetic diversity among grapevine samples as confirmed by the Shannon’s information index (1.94 on average) and by both the observed (Ho) and expected (He) heterozygosity. Ho had a value ranging from 0.748 (VvMD25) to 0.914 (VvS2) (mean 0.849); He ranged from 0.758 (VvMD25) to 0.866 (VrZAG79) (mean 0.825). The inbreeding coefficient (F) showed values near zero at all loci.

Table 1.
Genetic diversity indices of the ten simple sequence repeats (SSR) markers used to genotype the grapevine genotypes under study.
All the above-mentioned genetic diversity indices were computed on the dataset that included only the 84 minor/neglected Apulian genotypes (Table S3). Values very similar to those shown in Table 1 were registered. The highest number of private alleles was observed for the SSR marker VvMD28 (four alleles), followed by VVS2 and VVMD7 (three alleles) (Table 2). The highest number of private alleles (no. 22) characterizes the subset of minor/neglected Apulian genotypes. In particular, genotypes with the highest number of private alleles were: Unknown Ibrido Urgesi (four alleles) and Unknown Vite Longo (three alleles) (Table 2). Seven out of ten SSR markers were located in genic regions. The function of these genes was described with gene ontology (GO) annotations (Table S4).

Table 2.
List of genotypes harboring private alleles at different SSR markers.
Estimation of pairwise relatedness revealed 24 putative cases of synonymy (Table 3). The highest LRM value was 0.50 (i.e., strong relationship between two samples; cases of synonymy), whilst the minimum value (−0.153) was recorded for the comparison Bombino nero vs. Jabixhar. The average LRM value was −0.004 (Figure S1 and Table S4). Overall, the frequency distribution of LRM values was skewed toward smaller and negative values (Figure S2).

Table 3.
List of putative cases of synonymy identified based on the Lynch and Ritland mean estimator (LMR = 0.50) using ten SSR markers.
3.2. Genetic Relationships between Grapevine Genotypes Based on SSR Markers
The cluster analysis based on the neighbor-joining method allowed the samples to be grouped into 6 major clusters, each one including groups of identical or very similar genotypes (Figure 1). Overall, it confirmed twenty-three putative cases of synonymy out of 84 minor Apulian grapevines (excluding Pinot), considering only the cases where the genetic distance between individuals was 0 (Figure 1).
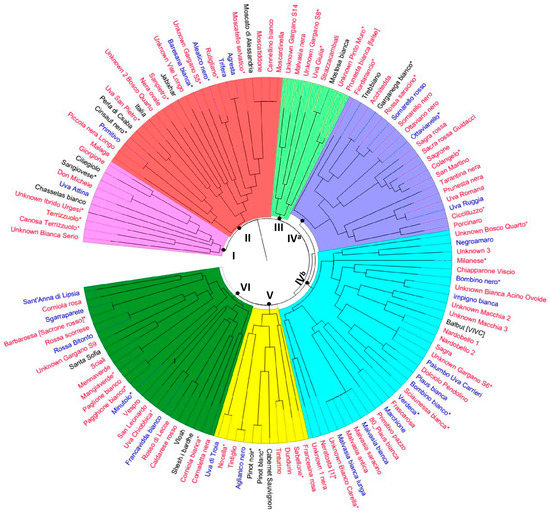
Figure 1.
The dendrogram generated using the neighbor-joining method depicts the genetic relationships among grapevine genotypes based on allele frequencies of 10 SSR markers. Colors distinguish “minor/neglected” genotypes (red), Apulian genotypes (blue), national and international genotypes (black). The asterisk “*” indicates those genotypes for which genotyping-by-sequencing data are available.
In particular, Cluster-I comprised samples closely related to Sangiovese, such as Ciliegiolo and others considered putative cases of synonymy. All Muscat genotypes (Moscardinella, Cannellino bianco, Moscatiddone, Moscato di Alessandria, and Moscatello selvatico) were grouped together in Cluster II. In cluster III, the pairs of samples Strazzacambiali/Mostosa bianca, and Unknown Pinto/Prunesta bianca (false), showed a very similar SSR-based molecular profile. Cluster IV is split into sub-clusters IVa and IVb. As for sub-cluster IVa, Acchitedda and Garganega bianco, a typical variety from the Veneto region (Northern Italy) should be considered as synonymous. Cluster IVb grouped all Malvasia grapes (i.e., bianca, antica, bianca lunga, saracino, and 80-Plaus bianca) and Sagra, unknown Gargano S6, and Palumbo (the latter is a local variety traditionally cultivated in the district of Bari, also known as Uva Carrieri) which should be considered as synonymous. As for misnaming, Primitivo Pazzo and Sanpietro, being completely different from the corresponding references Primitivo and Uva San Pietro, certainly represent different genotypes.
SSR-based genotyping failed to distinguish genotypes with different berry skin color, such as the well-known case of Pinot blanc and Pinot noir (cluster V), and Somarello nero from Somarello rosso (cluster IVa). Interestingly, none of the 17 genotypes tagged as “unknown” were similar to known varieties (Figure 1).
3.3. GBS-Derived SNP Analysis
To confirm and integrate the results obtained by the analysis of SSR markers, we took advantage of available GBS-derived SNP data points for only 40 grapevine samples, including 29 minor/neglected genotypes and 4 well-known Apulian, 4 Italian, and 3 international varieties (Table S1). These 40 samples are part of the entire panel of 128 that underwent SSR genotyping.
Genotyping by sequencing generated about ~900 million reads and 182,710 SNPs were called using PN40024 as the reference genome. After the filtering process, 24,558 high-quality SNPs were obtained. This dataset was then subjected to LD pruning (r2 = 0.5), resulting in 1,178 SNPs, of which 672 SNPs fall within annotated genes and 528 SNPs fall within coding sequences (CDS). Most of the identified SNPs (62.6%) were transitions (A/G or C/T). The most observed substitution type was C/T (31.6%), whereas the least common substitution type was C/G transversion (6.5%). The frequency of transitions was higher than transversions, with a transition/transversion ratio of 1.68.
3.4. Genetic Diversity Assessed by SNPs
ADMIXTURE was run to assess the population structure of the 40 genotypes belonging to the entire panel of 128 that underwent SSR genotyping. The cross validation (CV) indicated K = 2 as the best number of sub-populations in which the 40 genotypes are divided (Figure S3). At K = 2 (Figure 2), the first sub-population grouped four genotypes: Pinot Noir, Pinot Blanc, Uva San Pietro, and Bombino Bianco. The second one grouped 23 genotypes, while 13 were considered admixed at qi > 0.80. AWclust was run using 1,178 GBS-derived SNPs to assess the structure of the population made up of 40 genotypes (Figure 3). The gap statistics indicated K = 5 as the optimal number of clusters in which samples can be grouped (Figure S4). Overall, the use of SNP markers allowed to clearly distinguish all genotypes from one another. Based on a hierarchical plot function, the red line across the tree indicates the level of sub-division at K = 5. Firstly, the samples were separated into two main branches. The first branch (A) included three clusters. Cluster C-I comprised five samples from the Gargano Cape (Northern Apulia, district of Foggia): the table grapes Unknown Gargano S6 and Unknown Gargano S8, cultivated exclusively in this area, the table grape Nocella, typical of the ‘Daunia’, and the varieties Sciaunessa Bianca and Bombino nero, the latter cultivated throughout Apulia for dual purpose (table/wine).
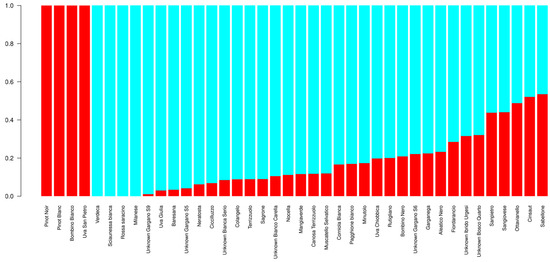
Figure 2.
Bar plot showing the genetic structure of the 40 genotypes at K = 2. The vertical bars represent the membership coefficients (qi) of each individual. K1 = red and K2 = cyan.
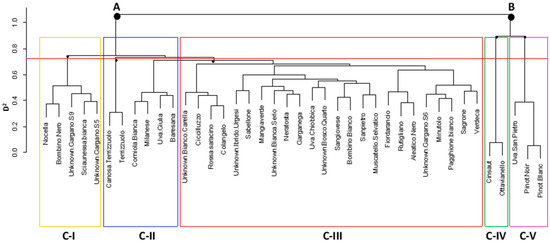
Figure 3.
AWclust-derived dendrogram plot showing how the 40 genotypes cluster into sub-populations. The red line across the tree indicates the level of sub-division at K = 5.
Cluster C-II grouped six minor genotypes, two of which, Canosa Terrizzuolo and Terrizzuolo, are almost identical. Cluster C-III included 24 samples. The genotype Colangelo is very similar to Rossa Saracino, while a sub-cluster included the Muscat genotypes Rutigliano, Aleatico nero, and Fiordarancio. The second branch (B) was split into two clusters. Cluster C-IV grouped Cinsaut Nero and Ottavianello, which are considered synonymous based on ampelographic traits [9]. Finally, Pinot noir and Pinot blanc, strongly related with Uva San Pietro, were grouped in C-V.
FST was calculated to assess differentiation among sub-populations due to genetic structure and to corroborate the results obtained by the clustering analysis. For methodological reasons, we merged clusters IV and V, namely C-IV/V. Pairwise FST values are listed in Table S5: they ranged from 0.029 (C-II vs C-III) to 0.135 (C-I vs C-IV/V). Identity-by-state (IBS: allele sharing distance) estimates for all pairs of the 40 genotypes were obtained to further investigate the genetic distance among samples (Figure S4). IBS values ranged from 0.67 to 0.97, with the majority (82%) of the values in the bin 0.73–0.76 (Figure S4). Pairwise samples with allele sharing distance > 0.80, were listed in Table S6. The analysis confirmed that Pinot noir and Pinot blanc were almost identical (0.97) as well as Cinsaut Nero and Ottavianello (0.96), while Rutigliano and Sabellone have the highest degree of genetic difference (0.67).
3.5. Detection of Private SNP Alleles and Divergent Loci
Private alleles were detected for each of the four clusters investigated in the FST analysis mentioned above. The number of private alleles ranged from a minimum of 12 (C-IV/V) to a maximum of 237 (C-III). For each cluster, we evaluated the distribution of private alleles across the chromosomes (Figure S6). C-II and C-III groups had private alleles on all chromosomes. No private alleles were detected on both chromosomes 9 and 17 in C-I, while C-IV/V had private alleles only on chromosomes 3, 5, 7, 12, 14, 15, 16, 18, and 19. For each genotype, the number of private alleles ranged from 1 (Bombino Nero) to 51 (Sabellone), with an average number of 19.7 (Figure 4).
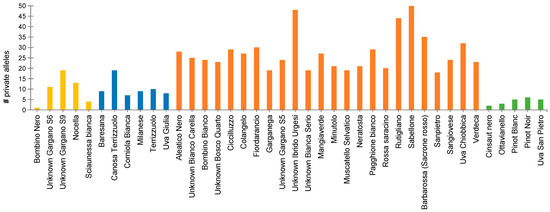
Figure 4.
Distribution of private Single nucleotide polymorphism (SNP) alleles in the forty grapevines subjected to genotyping by sequencing. Different colors indicate samples belonging to different AWclust-based groups: C-I = yellow; C-II = blue; C-III = orange; and C-IV/V (mixed C-IV and C-V) = green.
Genetic differentiation between the four sub-populations aforementioned was further investigated by the analysis of Wright Fixation Index (FST) at individual loci, using FST ≥ 0.25 as threshold (Figure 5 and Table S8). A total of 670 divergent SNPs were detected using pairwise comparisons. Among these, 383 SNPs were non redundant; 66% of those variants were in genic regions, whereas the remaining were intergenic (Table S7). The lowest number (36) of divergent loci was detected in C-II vs C-III, while the highest (218) was detected in C-I vs C-IV. Two SNPs (S9_21838021 and S19_1360860), which mapped on chromosomes 9 and 19, showed strong differentiation FST values (~ 0.90).
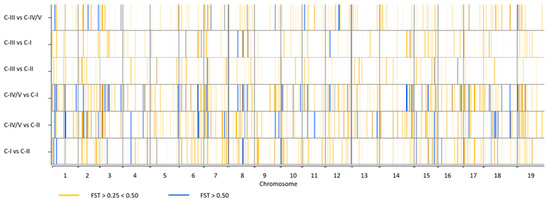
Figure 5.
Graphical representation of pairwise FST values at single SNP loci across the 19 grapevine chromosomes. Vertical lines indicate the divergent loci detected. The white background is for loci with FST < 0.25. The orange lines indicate loci with FST between 0.25 and 0.50 (from the lightest to the darkest). The blue lines (from the lightest to the darkest) indicate loci with FST > 0.50.
4. Discussion
Apulia, stretching into the Mediterranean Sea, has a strategic geographical position, being located on commercial routes of ancient populations such as Phoenicians, Greeks, and Romans. A recent study remarked the central role of Magna Graecia in the spread of grapevine to Western Europe, corroborating the hypothesis that grapevine originated in the Caucasus and then spread to the regions of Southern Italy [39].
Apulia includes five physiographic areas: the Daunia mountainous region, the “Tavoliere delle Puglie” plain, the Gargano Cape (North), the Murgia plateau (Central South), and the Salento peninsula (South). This environmental variability is reflected in the large crop biodiversity that characterizes the Apulian landscape; indeed, the presence of different environments and micro-climates have favored the differentiation of several genetic pools that are a mixture of ancient and more recently introduced varieties [64,65].
In Italy, Apulia is the second wine producer after the Veneto region [66], but only few PDO wines are produced with a restricted number of autochthonous varieties [67]. It is essential to investigate the autochthonous grapevine germplasm in order to identify cultivated varieties able to enlarge the production protocols of PDO wines. This study fits in this context as it deals with the genetic characterization of 84 minor/neglected Apulian grapevine genotypes.
Clustering analysis, identification of private alleles, and detection of divergent loci revealed that the Apulian grapevine germplasm is a source of under-explored genetic diversity.
4.1. Cultivar Identification and Genetic Diversity by Means of SSRs
In previous studies, a limited number of Apulian ancient grapevines was characterized by using SSR markers, thus unveiling putative cases of synonymy and the relationships between Apulian and national/international varieties [8,9,68]. Herein, a larger panel of 84 minor/neglected Apulian grapevines was investigated. Overall, genetic diversity indices estimated by SSR markers revealed the existence of a large genetic variability within Apulian germplasm that could be exploited to broaden the genetic base of grapevine. This result was expected as the high level of polymorphism rate in grapevine has been proven in several works [17,18,40,41,68,69].
The SSR-based clustering disclosed the genetic relationships between Apulian and national and international varieties, revealing six major clusters. Geographical origin did not affect the genetic clustering, although several genotypes are cultivated in specific areas. Therefore, clustering does not reflect geographical origin most likely because those genotypes originated from a few common ancestors. Then, migration, selection, and breeding, combined with epigenetic factors, could have influenced the response of plant to environment-specific adaptation and differentiation [70,71], thus determining the observed variability. Notably, SSR clustering discriminated the majority of minor/neglected Apulian genotypes (75%), suggesting how genetically different the germplasm under study is.
Several putative cases of synonymy were disclosed. The grapevine don Michele was found to be synonymous of Sangiovese, a renowned variety largely cultivated in Central Italy, confirming that, in the past, this variety was popular also in Southern Italy, although renamed locally [8]. In other cases, some Apulian genotypes were found to be synonymous of varieties largely cultivated in the 19th century, such as Uva sacra. Indeed, the dendrogram (Figure 1) brought out the near identity among San Martino, Colangelo, Sagrone, and Sagra rossa with Sacra rossa, which is the local name for Uva sacra, very common in the north of the Bari district [8]. Uva Ruggia/Prunesta nera showed a high degree of genetic similarity with Uva Romana, a table grape largely renowned in the area of Rome since the first half of the 1900s with the name of Pergolese di Tivoli. It is known that both these varieties were largely cultivated in Apulia in the past [8], and our findings show that they still survive in marginal vineyards. Indeed, the region was, until the end of the 1800s, a great wine producer and exporter, and owned a very rich array of varieties [72]. Following the phylloxera vineyards destruction, the monovarietal vineyards become prevalent due to the necessity to use grafted plants. This agronomic practice determined the loss of the finest and most aromatic “autochthonous” grapes, which probably survived in marginal areas of the region [72]. Finally, some genotypes turned out to be synonymous of international varieties, in particular those cultivated in trans-Adriatic countries (i.e., Croatia, Dalmatia, Albania, Greece), such as Balbut, Lianoroi, etc., as a consequence of the intense trading exchanges [73].
4.2. High Throughput SNP Genotyping Reveals Patterns of Genetic Divergence in Apulian Minor/Neglected Varieties
Overall, 35 out of 84 minor/neglected genotypes were identified as known varieties, based on the SSR profiles, while most of the “unknown” genotypes showed a peculiar genetic profile. SSR markers failed also to discriminate Pinot blanc from Pinot noir and Somarello nero from Somarello rosso, confirming that SSR markers are not suitable for differentiating berry color variants [74]. To obtain further insights on the collection under study, a subset of 40 genotypes were further assessed by 1,178 GBS-derived SNP markers.
Population structure inferred by ADMIXTURE highlighted two main sub-populations (Figure S3, Figure 2); however, the Bayesian model did not suggest any robust clustering hypothesis.
We just observed that in the two sub-populations there were varieties (i.e., Pinot blanc, Pinot noir, Bombino bianco, and Uva san pietro in K = 1 as well as Verdeca, Sciaunessa bianca, Rossa saracino, and Milanese in K = 2) that shared the entire proportion of ancestry (qi = 1). A possible explanation could be that these genotypes have been largely used in ancient local cross programs.
In this study, the AWclust clustering method remarked the powerful resolution of SNPs compared with SSRs, in distinguishing all genotypes. In addition, the population structure derived from non-parametric clustering was more informative. Five genetic clusters were identified, thus revealing the presence of different gene pools. Notably, the FST among sub-populations highlighted a moderate genetic differentiation between C-I and C-IV/V clusters (FST = 0.135). Cluster C-I included four table grapes typical of the Gargano Cape (Northern Apulia), an area with peculiar and distinguishing pedoclimatic conditions, caused by the geographic isolation due to the presence of mountains and sea barriers. The history of the Mediterranean settlement, trade, and cultural influence supports this hypothesis. Greeks colonized the southern area of Apulia, and evidence of gene flow between Greece and Apulia has been observed [39]. Conversely, there is minor evidence of gene flow in the Gargano, due to a tardive colonization by the Greeks (from the end of V century B.C.). In addition, Gargano has been marginally touched by the Phylloxera epidemic; therefore, it is possible that some ancient genotypes have been saved from disappearance [72] and then they genetically differentiated because of the exclusive Gargano habitat. The detection of private alleles and divergent loci corroborated the idea that the genotypes retrieved from the Gargano Cape stand out from modern varieties (in C-IV and C-V) and represent an under-explored gene pool that deserves to be studied to search for new and beneficial alleles for facing incoming needs (i.e., fruit-bearing, vegetative and reproductive growth responses, resistance traits, etc.).
The clusters C-II and C-III originated from the same node and grouped samples with probable ancestors of Greek origin. However, only C-II showed a differentiation from C-IV/V (FST = 0.127). C-II included the Apulian cultivar Baresana bianca, which is synonymous of many Greek cultivars [9], and another four minor/neglected Apulian grapes, probably closely related to Baresana Bianca, with which they form a separate genetic pool.
C-III showed the lowest values of FST, revealing that the genetic composition of this sub-population was a result of a miscellaneous structure derived from hybridization and cross-pollination with unknown and renowned materials, most likely from Greece [39]. The highest number of private alleles (19.7 per genotype on average) supported this finding. In particular, Unknown Ibrido Urgesi, Rutigliano, and Sabellone showed a number of private alleles ≥ 40.
The use of Wright fixation index (FST) to detect divergent loci between sub-populations has been previously used in grapevine [42], but no indication of divergent loci has ever been reported in V. vinifera, especially with respect to Apulian cultivars. In this work, we used FST analysis to support the existence of different gene pools in Apulia. The highest number of divergent loci was detected between C-I and C-IV/V, followed by C-II and C-IV/V (Table S7). This finding supports the evidence of at least two separate gene pools in the Apulian germplasm. We found divergent loci associated with genes with a role in plant development, carbohydrate metabolic processes, nitrogen pathway, defense response, stomatal movement, and ripening process (Table S7). Most of these genes belong to the same metabolic pathway identified by Marrano et al. [42], following the comparison between V. sativa and V. sylvestris. The two SNP loci with FST > 0.90 fall within two genes (VIT_209s0054g00970 = mediator subunit 8, and VIT_219s0014g01250 = tubby-like f-box protein 7-like), which are involved in the defense response to fungal infections and in regulation of flower and pollen development [75].
Two SNP loci associated with genes involved in nitrogen metabolism were identified only in C-I vs. C-II (VIT_207s0031g02930 = ammonium transporter amt2,) and C-I vs. C-IV/C-V (VIT_216s0098g00290 = glutamate synthase 1). The importance of nitrate and ammonium for plant growth is well known, and it has been demonstrated that ammonium uptake capacity and response of cytosolic glutamine synthetase to ammonium supply are key factors in the adaptation of ammonium nutrition in Arabidopsis thaliana [76]. Furthermore, several genes involved in nitrogen uptake, assimilation, and remobilization have been found under selection in durum wheat [77], revealing that natural and selective pressures, as well as random mutation and genetic drift, shaped the expression of nitrogen genes. The nitrogen compounds play a pivotal role in photosynthesis as signals in regulating the responses of plants to environmental changes, such as resistance of plants to adverse conditions, including water stress. Thus, our finding suggests the importance to further investigate the influence of nitrogen genes in grape differentiation and adaptation.
Several SNP loci associated with genes involved in carbohydrate metabolic processes were found divergent. This finding was expected, as the effect of artificial selection on sugar metabolism and transporter genes in grape is well known [75]. Overall, the knowledge on genomic regions associated with divergent selection represents a valuable source, which needs to be explored to identify beneficial alleles for plant adaptation and for responding to the new challenges of breeding [78].
5. Conclusions
There is a growing interest in understanding the origin and investigating genetic diversity of grapevine germplasms confined in marginal geographical areas as they represent under-exploited genetic resources. Within this motivating context, the genetic diversity of a large collection of minor/neglected Apulian grapevine genotypes was evaluated by combining microsatellite and SNP markers. The evidence we gathered suggests the existence in Apulia of different grapevine gene pools that are characterized by a large genetic variability, and that could be exploited in future breeding programs as a source of potentially favorable/beneficial alleles. The detection of divergent loci related to the signature of selection may give advantageous indications on the development of varieties that are able to adapt to the new environmental conditions imposed by climate change and to respond to the new challenges of breeding programs.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/10/4/563/s1, Table S1: List of grapevine genotypes analyzed in this study. The 40 samples subjected to both SSR analysis and genotyping by sequencing are marked with asterisk. W = wine, T = table; VIVC = Vitis International Variety Catalogue; n.a. = not available.; Table S2: List of the 10 microsatellite markers (SSR) used for genotyping. For each SSR, the identification code, oligo sequences, the expected fragment size range, annealing temperature, and bibliographic reference are indicated; Table S3. Genetic diversity indices evaluated on the 84 minor/neglected Apulian grapevine genotypes; Table S4: Gene ontology (GO) analysis. Chromosome, position (start/stop), annotation, and gene ontology terms for the 10 SSRs used in the analysis of grapevine germplasm. n.a. = not available; Table S5: Matrix of the pairwise FST genetic distances among the four groups detected by AWclust (C-IV and C-V were merged in a unique cluster renamed C-IV/V); Table S6: List of pairs of genotypes with allele sharing distance (IBS; identity-by-state) ≥ 0.80; Table S7: List of divergent SNP loci derived from pairwise comparisons between clusters identified by AWclust. (C-IV and C-V were merged in a unique cluster renamed C-IV/V). Chromosome, position (bp), FST value, gene ID and annotation were reported; Figure S1: Graphical representation of the five physiographic areas of Apulia region. Yellow = Daunia mountainous region; light blue = Tavoliere delle Puglie plain; green = Gargano cape (North); red = Murgia plateau (Central South); grey = Salento peninsula (South). Red dots represent the geographical coordinates of sampling sites; Figure 2: Frequency distribution of Lynch and Ritland estimator (LRM) values across the Apulian grapevine germplasm; Figure 3: Cross-validation (CV) plot of ADMIXTURE analysis for K values from 1 to 10; Figure 4: Gap statistic plot indicates K=5 as the best value of K in which to divide the population (Figure 3). The number of inferred clusters (K) ranging from 1 to 10 is shown in the graph. A) The blue and red curves are the estimated expectation of log (Wk) and the observed log (Wk), respectively. B) The x-axis indicates different possible K (K = 5 is the best value) and gap value is on the y-axis; Figure S5: Heatmap illustrating the pair-wise allele sharing distances (i.e., identity-by-state values) for the 40 grapevine genotypes subjected to genotyping-by-sequencing; Figure S6: Stacked bar chart describing the distribution of private alleles on the 19 grape chromosomes. Percentage (%) of private alleles for each of the four clusters identified by AWclust (Figure 2) is reported. C-IV and C-V were merged in a unique cluster renamed C-IV/V.
Author Contributions
Conceptualization, C.M., V.N.S. V.F. and P.L.N.; Resources, P.V., V.R. and C.P.; Investigation, M.M.M., V.d.R., C.M. and P.R.; Formal analysis, M.M.M., V.d.R., F.T. and N.D.; Writing – Original Draft, M.M.M., F.T., N.D. and V.d.R.; Writing – Review & Editing, M.M.M., N.D., C.M., and F.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the project Recupero del Germoplasma Viticolo Pugliese (Re.Ge.Vi.P.)—(Bando del PSR Puglia 2007/2013 Misura 214 az. 4- Progetti integrati per la biodiversità). Azione 4 sub azione 4c Caratterizzazione genetica—identificazione varietale di vitigni autoctoni mediante analisi SSR and ‘Progetti integrati per la biodiversità, Recupero del Germoplasma Viticolo Pugliese Re.Ge.Vi.P, prosecuzione delle attività, funded by Region Apulia Rural Development Programme. http://www.fruttiantichipuglia.it/recuperiamo/.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McGovern, P.E.; Glusker, D.L.; Exner, L.J.; Voigt, M.M. Neolithic resinated wine. Nature 1996, 381, 480–481. [Google Scholar] [CrossRef]
- Arroyo-García, R.; Ruíz-García, L.; Bolling, L.; Ocete, R.; López, M.A.; Arnold, C.; Ergul, A.; Söylemezoğlu, G.; Uzun, H.I.; Cabello, F.; et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 2006, 15, 3707–3714. [Google Scholar] [CrossRef] [PubMed]
- This, P.; Lacombe, T.; Thomas, M.R. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006, 22, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Maul, E.; Toepfer, R. Vitis International Variety Catalogue (VIVC): A Cultivar Database Referenced by Genetic Profiles and Morphology. In Proceedings of the BIO Web of Conferences, Mainz, Germany, 5–10 July 2015. [Google Scholar] [CrossRef]
- Registro Nazionale Delle Varietà Di Vite, Ministero Delle Politiche Agricole, Alimentari e Forestali. Available online: http://catalogoviti.politicheagricole.it/catalogo.php (accessed on 14 April 2020).
- Lacombe, T.; Audeguin, L.; Boselli, M.; Bucchetti, B.; Cabello, F.; Chatelet, P.; Crespan, M.; D’Onofrio, C.; Dias, J.E.; Ercisli, S.; et al. Grapevine European catalogue: Towards a comprehensive list. Vitis 2011, 50, 65–68. [Google Scholar]
- Sardaro, R.; Bozzo, F.; Petrillo, F.; Fucilli, V. Measuring the financial sustainability of vine landraces for better conservation programs of Mediterranean agro-biodiversity. Land Use Policy 2017, 68, 160–167. [Google Scholar] [CrossRef]
- La Notte, P.; Civita, F.; Raimondi, S.; Schneider, A. Atlante Dei Vitigni Tradizionali Di Puglia 2018; Caramia, B., Ed.; CRSFA: Locorotondo (BR), Italy, 2018. [Google Scholar]
- Schneider, A.; Raimondi, S.; Pirolo, C.S.; Marinoni, D.; Ruffa, P.; Venerito, P.; La Notte, P. Genetic characterization of grape cultivars from Apulia (Southern Italy) and synonymies in other Mediterranean regions. Am. J. Enol. Vitic. 2014, 65, 244–249. [Google Scholar] [CrossRef]
- Van Leeuwen, C.; Schultz, H.R.; de Cortazar-Atauri, I.G.; Duchene, E.; Ollat, N.; Pieri, P.; Bois, B.; Goutouly, J.P.; Quénol, H.; Touzard, J.M.; et al. Why climate change will not dramatically decrease viticultural suitability in main wine-producing areas by 2050. Proc. Natl. Acad. Sci. USA 2013, 110, E3051–E3052. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Lux, R.; Schultz, H.R. Constructing a framework for risk analyses of climate change effects on the water budget of differently sloped vineyards with a numeric simulation using the Monte Carlo method coupled to a water balance model. Front. Plant Sci. 2014, 5, 645. [Google Scholar] [CrossRef]
- Di Rienzo, V.; Montemurro, C.; Strippoli, G.; Fanelli, V.; de Giovanni, C.; Antonacci, D.; Vivaldi, G.A.; Pellegrini, G.; Blanco, A.; Camposeo, S.; et al. Ecophysiological response to water stress and regulation of gene expression for a 9-cis-epoxycarotenoid dioxygenase in Vitis vinifera L. ‘Italia’. Acta Hortic. 2015, 1082, 277–284. [Google Scholar] [CrossRef]
- De Giovanni, C.; Di Rienzo, V.; Miazzi, M.; Fanelli, V.; Blanco, A.; Montemurro, C. A DNA methylation survey of NCED genes in Vitis vinifera L. under stress conditions. Acta Hortic. 2015, 1082, 277–284. [Google Scholar] [CrossRef]
- Hajjeh, H.; Miazzi, M.; Faretra, F. Overwintering of Erysiphe necator Schw. in southern Italy. J. Plant Pathol. 2009, 90, 323–330. [Google Scholar]
- Miazzi, M.; Hajjeh, H.R. Differential sensitivity to triadimenol of Erysiphe necator isolates belonging to different genetic groups. J. Plant Pathol. 2011, 93, 729–735. [Google Scholar]
- Sanzani, S.M.; Miazzi, M.M.; Di Rienzo, V.; Fanelli, V.; Gambacorta, G.; Taurino, M.R.; Montemurro, C. A rapid assay to detect toxigenic Penicillium spp. contamination in wine and musts. Toxins 2016, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- Marrano, L.; Grzeskowiak, P.; Sanz, M.; Lorenzi, S.; Prazzoli, M.L.; Arzumano, A.; Amanova, M.; Failla, O.; Maghradze, D.; Grando, M.S. Genetic diversity and relationships in the grapevine germplasm collection from Central Asia. Vitis 2015, 54, 233–237. [Google Scholar]
- Gristina, A.S.; De Michele, R.; Garfı, G.; La Mantia, T.; Fontana, I.; Spinelli, P.; Motisi, A.; Carimi, F. Urgent need for preservation of grapevine (Vitis vinifera L. subsp. vinifera) germplasm from small circum-Sicilian islands as revealed by SSR markers and traditional use investigations. Genet. Resour. Crop Evol. 2016. [Google Scholar] [CrossRef]
- Drori, E.; Rahimi, O.; Marrano, A.; Henig, Y.; Brauner, H.; Salmon-Divon, M.; Netzer, Y.; Prazzoli, M.L.; Stanevsky, M.; Failla, O.; et al. Collection and characterization of grapevine genetic resources (Vitis vinifera) in the Holy Land, towards the renewal of ancient winemaking practices. Sci. Rep. 2017, 7, 4463. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, V.; Savoia, M.A.; Gadaleta, S.; Piarulli, L.; Montemurro, C.; La Notte, F.; Miazzi, M.M.; Bruno, M.; Falbo, M.; Petrillo, F.; et al. Molecular characterization of wine grape cultivars from Calabria. Acta Hortic. 2019, 1248. [Google Scholar] [CrossRef]
- Halász, J.; Kodad, O.; Galiba, G.M.; Skola, I.; Ercisli, S.; Ledbetter, C.A. Genetic variability is preserved among strongly differentiated and geographically diverse almond germplasm: An assessment by simple sequence repeat markers. Tree Gen. Genom. 2019, 15, 12. [Google Scholar] [CrossRef]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Costantini, L.; Crespan, M.; Eisenheld, C.; Grando, M.S.; Lacombe, T.; Lacou, V.; et al. Development of a common set of standard varieties and standardized method of scoring microsatellites markers for the analysis of grapevine genetic resources. Theor. Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef]
- Boucheffa, S.; Tamendjari, A.; Sanchez-Gimeno, A.C.; Rovellini, P.; Venturini, S.; di Rienzo, V.; Miazzi, M.M.; Montemurro, C. Diversity assessment of Algerian wild and cultivated olives (Olea europaea L.) by molecular; morphological and chemical traits. Eur. J. Lipid Sci. Technol. 2019, 1800302. [Google Scholar] [CrossRef]
- Pasqualone, A.; Di Rienzo, V.; Nasti, R.; Blanco, A.; Gomes, T.; Montemurro, C. Traceability of Italian Protected Designation of Origin (PDO) table olives by means of microsatellite molecular markers. J. Agric. Food Chem. 2013, 61, 3068–3073. [Google Scholar] [CrossRef] [PubMed]
- Sabetta, W.; Miazzi, M.M.; di Rienzo, V.; Fanelli, V.; Pasqualone, A.; Montemurro, C. Development and application of protocols to certify the authenticity and the traceability of Apulian typical products in olive sector. Rivista Italiana Delle Sostanze Grasse 2017, 94, 37–43. [Google Scholar]
- Pasqualone, A.; Di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Caponio, F.; Montemurro, C. High resolution melting analysis of DNA microsatellites in olive pastes and virgin olive oils obtained by talc addition. Eur. J. Lipid Sci. Technol. 2015, 117, 2000–2010. [Google Scholar] [CrossRef]
- Di Rienzo, V.; Fanelli, V.; Miazzi, M.M.; Sabetta, W.; Montemurro, C. A reliable analytical procedure to discover table grape DNA adulteration in industrial wines and musts. Acta Hortic. 2017, 1188, 365–370. [Google Scholar] [CrossRef]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Grando, M.S.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Troggio, M.; Cartwright, D.A.; Cestaro, A.; Pruss, D.; Pindo, M.; FitzGerald, L.M.; Vezzulli, S.; Reid, J.; et al. A high-quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2007, 2, e1326. [Google Scholar] [CrossRef]
- Adam-Blondon, A.F.; Jaillon, O.; Vezzulli, S.; Zharkikh, A.; Troggio, M.; Velasco, R. Genome sequence initiatives. In Genetics Genomics and Breeding of Grapes; Adam-Blondon, A.-F., Martinez-Zapater, J.M., Chittaranjan, K., Eds.; Science Publishers, CRC Press: Boca Raton, FL, USA, 2011; pp. 211–234. [Google Scholar]
- Canaguier, A.; Grimplet, J.; Di Gaspero, G.; Scalabrin, S.; Duchêne, E.; Choisne, N.; Mohellibi, N.; Guichard, C.; Rombauts, S.; Le Clainche, I.; et al. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genom. Data 2017, 14, 56–62. [Google Scholar] [CrossRef]
- Roach, M.J.; Johnson, D.L.; Bohlmann, J.; van Vuuren, H.J.J.; Jones, S.J.M.; Pretorius, S.I.; Schmidt, S.A.; Borneman, A.R. Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar Chardonnay. PLoS Genet. 2018, 14, e1007807. [Google Scholar] [CrossRef]
- Liang, Z.; Duan, S.; Sheng, J.; Zhu, S.; Ni, X.; Shao, J.; Mao, R.; Zhu, Y.; Deng, W.; Yang, M.; et al. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat. Commun. 2019, 10, 1190. [Google Scholar] [CrossRef]
- Laucou, V.; Launay, A.; Bacilieri, R.; Lacombe, T.; Adam-Blondon, A.F.; Berard, A.; Le Paslier, M.C. Extended diversity analysis of cultivated grapevine Vitis vinifera with 10 K genome-wide SNPs. PLoS ONE 2018, 13, e0192540. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.L.; Zhao, H.L.; Li, Q.; Zhang, G.H.; Jiang, J.F.; Liu, C.H.; Yu, Y.H. Genome-wide association study of berry-related traits in grape [Vitis vinifera L.] based on genotyping-by-sequencing markers. Hortic. Res. 2019, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Lijavetzky, D.; Cabezas, J.A.; Ibáñez, A.; Rodriguez, V.; Martínez-Zapater, J.M. High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genom. 2007, 8, 424. [Google Scholar] [CrossRef] [PubMed]
- Miazzi, M.M.; D’Agostino, N.; Gadaleta, S.; di Rienzo, V.; Fanelli, V.; Sabetta, W.; Montemurro, C.; Taranto, F. GBS-derived SNP catalogue from a wide grape (V. vinifera L.) germplasm collection that includes the most representative Apulian autochthonous varieties. Acta Hortic. 2019, 22, 1248. [Google Scholar] [CrossRef]
- De Lorenzis, G.; Mercati, F.; Bergamini, C.; Cardone, M.F.; Lupini, A.; Mauceri, A.; Caputo, A.F.; Abbate, L.; Barbagallo, M.G.; Antonacci, D.; et al. SNP genotyping elucidates the genetic diversity of Magna Graecia grapevine germplasm and its historical origin and dissemination. BMC Plant Biol. 2019, 19, 5–15. [Google Scholar] [CrossRef]
- Cipriani, G.; Spadotto, A.; Jurman, I.; Di Gaspero, G.; Crespan, M.; Meneghetti, S.; Frare, E.; Vignani, R.; Cresti, M.; Morgante, M.; et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 2010, 121, 1569–1585. [Google Scholar] [CrossRef]
- Lacombe, T.; Boursiquot, J.M.; Laucou, V.; Di Vecchi-Staraz, M.; Péros, J.-P.; This, P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor. Appl. Genet. 2013, 126, 401–414. [Google Scholar] [CrossRef]
- Marrano, A.; Micheletti, D.; Lorenzi, S.; Neale, D.; Grando, M.S. Genomic signatures of different adaptations to environmental stimuli between wild and cultivated Vitis vinifera L. Hortic. Res. 2018, 5, 34. [Google Scholar] [CrossRef]
- Corrado, G.; Piffanelli, P.; Caramante, M.; Coppola, M.; Rao, R. SNP genotyping reveals genetic diversity between cultivated landraces and contemporary varieties of tomato. BMC Genom. 2013, 14, 835. [Google Scholar] [CrossRef]
- Akagi, T.; Hanada, T.; Yaegaki, H.; Gradziel, T.M.; Tao, R. Genome-wide view of genetic diversity reveals paths of selection and cultivar differentiation in peach domestication. DNA Res. 2016, 23, 271–282. [Google Scholar] [CrossRef]
- De Palma, L.; Poli, G.; Lopriore, G.; Tarantino, A.; Tarricone, L.; Soleti, F. Individuazione, Studio e Caratterizzazione Di Vitigni Di Antica Coltivazione Nell’Alto Tavoliere Pugliese; Centro Grafico Francescano: Foggia, Italy, 2008. [Google Scholar]
- Spadoni, A.; Sion, S.; Gadaleta, S.; Savoia, M.; Piarulli, L.; Fanelli, V.; di Rienzo, V.; Taranto, D.; Miazzi, M.; Montemurro, C.; et al. A simple and rapid method for genomic DNA extraction and microsatellite analysis in tree plants. J. Agric. Sci. Technol. 2019, 21, 1215–1226. [Google Scholar]
- OIV Statistical Report on World Vitiviniculture. Available online: http://www.oiv.int/en/ (accessed on 14 April 2020).
- Thomas, M.R.; Scott, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theor. Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Dangl, G.S.; Vignani, R.; Meredith, C.P. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 1996, 39, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Sefc, K.M.; Regner, F.; Turetschek, E.; Glossl, J.; Steinkellner, H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 1999, 42, 367–373. [Google Scholar] [CrossRef]
- Di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Savino, V.; Pollastro, S.; Colucci, F.; Miccolupo, A.; Blanco, A.; Pasqualone, A.; Montemurro, C. An enhanced analytical procedure to discover table grape DNA adulteration in industrial musts. Food Control 2016, 60, 124–130. [Google Scholar] [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Hang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1949, 15, 323–354. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef]
- Gao, X.; Starmer, J. AWclust: Point-and-click software for non-parametric population structure analysis. BMC Bioinform. 2008, 9, 77. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Calò, A.; Scienza, A.; Costacurta, A.; Molina, M. Vitigni d’Italia; Edagricole, Ed.; Agricole Calderini S.R.L.: Bologna, Italy, 2001. [Google Scholar]
- Lentini, A. Archaeobotany brewing and winemaking in Mediterranean Basin and Transcaucasus area. In Cyprus in the Prehistory of Wine; Flourentzos, P., Belgiorno, M.R., Micosia, L.A., Eds.; de Strobel Publ: Nicosia, Cyprus, 2005; pp. 6–11. [Google Scholar]
- Istituto Di Servizi Per Il Mercato Agricolo Alimentare. Available online: www.ismea.it (accessed on 14 April 2020).
- Voluntary Consortia National Confederation for the Protection of the Italian Wines. Available online: www.federdoc.com (accessed on 14 April 2020).
- Zulini, L.; Russo, M.; Peterlunger, E. Genotyping wine and table grape cultivars from Apulia (Southern Italy) using microsatellite markers. Vitis Geilweilerhof 2002, 41, 183–188. [Google Scholar]
- Di Vecchi Staraz, M.; Bandinelli, R.; Boselli, M.; This, P.; Boursiquot, J.M.; Laucou, V.; Lacombe, T.; Varès, D. Genetic structuring and parentage analysis for evolutionary studies in grapevine: Kin group and origin of the cultivar Sangiovese revealed. J. Am. Soc. Hortic. Sci. 2007, 132, 514–524. [Google Scholar] [CrossRef]
- Bacilieri, R.; Lacombe, T.; Le Cunff, L.; Di Vecchi-Staraz, M.; Laucou, V.; Genna, B.; Péros, J.-P.; This, P.; Boursiquot, J.M. Genetic structure in cultivated grapevines is linked to geography and human selection. BMC Plant Biol. 2013, 13, 25. [Google Scholar] [CrossRef]
- Kapazoglou, A.; Ganopoulos, I.; Tani, E.; Tsaftaris, A. Epigenetics, epigenomics and crop improvement. Adv. Bot. Res. 2018, 86, 287–324. [Google Scholar]
- Biscotti, N.; Delviscio, G.; Bonsanto, D.; Casavecchia, S.; Biondi, E. Indagini su popolazioni selvatiche di Vitis vinifera L. rinvenute nel Parco Nazionale del Gargano (Foggia), in Puglia. Inf. Bot. Ital. 2015, 4, 179–186. [Google Scholar]
- Zdunić, G.; Preece, J.E.; Aradhya, M.; Velasco, D.; Koehmstedt, A. Genetic diversity and differentiation within and between cultivated (Vitis vinifera L. ssp. sativa) and wild (Vitis vinifera L. ssp. sylvestris) grapes. Vitis 2013, 52, 29–32. [Google Scholar]
- Migliaro, D.; De Nardi, B.; Vezzulli, S.; Crespan, M. An upgraded core set of 11 SSR markers for grapevine cultivar identification: The case of berry color mutants. Am. J. Enol. Vitic. 2017, 68, 496–498. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, J.; Zhu, W.; Wang, N.; Fang, P.; Han, Y.; Li, S. The effects of artificial selection on sugar metabolism and transporter genes in grape. Tree Gen. Genom. 2013, 9, 1343–1349. [Google Scholar] [CrossRef]
- Yasuda, T.; Konishi, N.; Kojima, S. Ammonium uptake capacity and response of cytosolic glutamine synthetase 1; 2 to ammonium supply are key factors for the adaptation of ammonium nutrition in Arabidopsis thaliana. Soil Sci. Plant Nutr. 2017, 63. [Google Scholar] [CrossRef]
- Taranto, F.; D’Agostino, N.; Rodriguez, M.; Pavan, S.; Minervini, A.P.; Pecchioni, N.; Papa, R.; De Vita, P. Whole genome scan reveals molecular signatures of divergence and selection related to important traits in durum wheat germplasm. Front. Genet. (accepted).
- Taranto, F.; Nicolia, A.; Pavan, S.; De Vita, P.; D’Agostino, N. Biotechnological and digital revolution for climate-smart plant breeding. Agronomy 2018, 8, 277. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).