Propagation Fidelity and Kinship of Tomato Varieties ‘UC 82’ and ‘M82’ Revealed by Analysis of Sequence Variation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. SNP Genotyping Data from the Illumina “Solanaceae Coordinated Agricultural Project” (SolCAP) Array
2.3. Whole Genome Resequencing Data
2.4. Sequence Alignment and SNP Calling
2.5. SNP Density Comparison from Resequencing Data
3. Results
3.1. SNP Genotyping Data Analysis
3.2. Sequence Alignment and SNP Calling
3.3. SNP Density Comparison from Sequence Data
3.4. SNP Allele Comparison across the Entire Genome
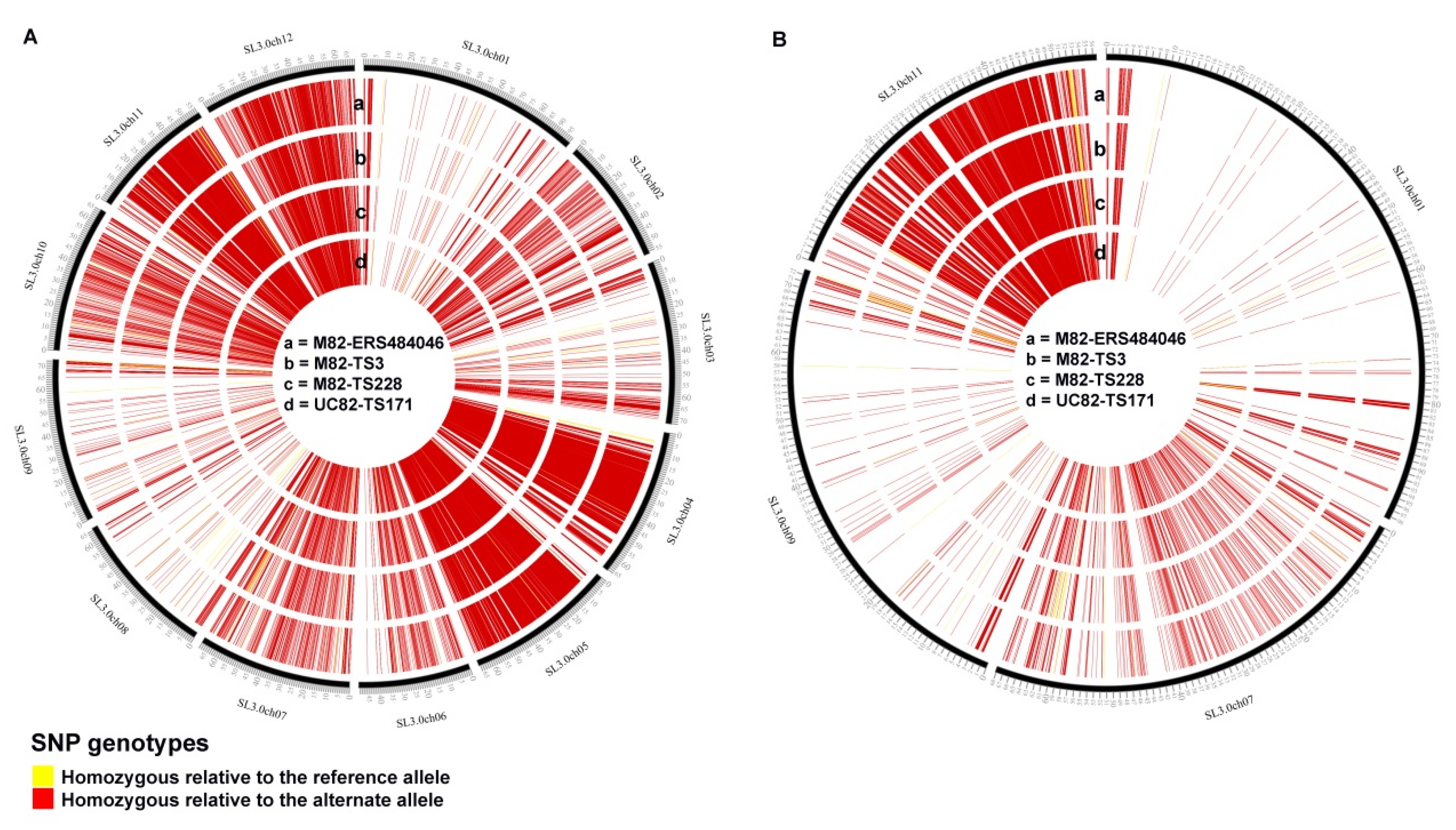
3.5. Comparison of Polymorphic Genomic Regions
3.6. Haplotype Identification Based on SNP Genotyping and Whole Genome Resequence Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Haun, W.J.; Hyten, D.L.; Xu, W.W.; Gerhardt, D.J.; Albert, T.J.; Richmond, T.; Jeddeloh, J.A.; Jia, G.; Springer, N.M.; Vance, C.P.; et al. The Composition and Origins of Genomic Variation among Individuals of the Soybean Reference Cultivar Williams 82. Plant Physiol. 2011, 155, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Schnable, J.C. RNA-seq based analysis of population structure within the maize inbred B73. PLoS ONE 2016, 11, e0157942. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.-R.; Shedge, V.; Kundariya, H.; Lehle, F.R.; Mackenzie, S.A. Ws-2 introgression in a proportion of Arabidopsis thaliana Col-0 stock seed produces specific phenotypes and highlights the importance of routine genetic verification. Plant Cell 2016, 28, 603–605. [Google Scholar] [CrossRef] [PubMed]
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.-C.; Durstewitz, G.; Plieske, J.; Wieseke, R.; Ganal, M.W.; Van Deynze, A.; Hamilton, J.P.; Buell, C.R.; Causse, M.; Wijeratne, S.; et al. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS ONE 2012, 7, e40563. [Google Scholar] [CrossRef]
- Sim, S.-C.; Van Deynze, A.; Stoffel, K.; Douches, D.S.; Zarka, D.; Ganal, M.W.; Chetelat, R.T.; Hutton, S.F.; Scott, J.W.; Gardner, R.G.; et al. High-density SNP genotyping of tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PLoS ONE 2012, 7, e45520. [Google Scholar] [CrossRef]
- Hamilton, J.P.; Sim, S.-C.; Stoffel, K.; Van Deynze, A.; Buell, C.R.; Francis, D.M. Single Nucleotide Polymorphism Discovery in Cultivated Tomato via Sequencing by Synthesis. Plant Genome J. 2012, 5, 17–29. [Google Scholar] [CrossRef]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef]
- Aflitos, S.; Schijlen, E.; de Jong, H.; de Ridder, D.; Smit, S.; Finkers, R.; Wang, J.; Zhang, G.; Li, N.; Mao, L.; et al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 2014, 80, 136–148. [Google Scholar]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sørensen, I.; Lichtenstein, G.; et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef]
- Gao, L.; Gonda, I.; Sun, H.; Bao, K.; Tieman, D.M.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; Theodore, W.; Foolad, M.R.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Tranchida-Lombardo, V.; Aiese Cigliano, R.; Anzar, I.; Landi, S.; Palombieri, S.; Colantuono, C.; Bostan, H.; Termolino, P.; Aversano, R.; Batelli, G.; et al. Whole-genome re-sequencing of two Italian tomato landraces reveals sequence variations in genes associated with stress tolerance, fruit quality and long shelf-life traits. DNA Res. 2018, 25, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Cambiaso, V.; Pratta, G.R.; Pereira da Costa, J.H.; Zorzoli, R.; Francis, D.M.; Rodríguez, G.R. Whole genome re-sequencing analysis of two tomato genotypes for polymorphism insight in cloned genes and a genetic map construction. Sci. Hortic. (Amsterdam) 2019, 247, 58–66. [Google Scholar] [CrossRef]
- Blanca, J.; Montero-Pau, J.; Sauvage, C.; Bauchet, G.; Illa, E.; Díez, M.J.; Francis, D.; Causse, M.; van der Knaap, E.; Cañizares, J. Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genom. 2015, 16, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Razifard, H.; Ramos, A.; Della Valle, A.L.; Bodary, C.; Goetz, E.; Manser, E.J.; Li, X.; Zhang, L.; Visa, S.; Tieman, D.; et al. Genomic Evidence for Complex Domestication History of the Cultivated Tomato in Latin America. Mol. Biol. Evol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Eshed, Y.; Abu-Abied, M.; Saranga, Y.; Zamir, D. Lycopersicon esculentum lines containing small overlapping introgressions from L. pennellii. Theor. Appl. Genet. 1992, 83, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Eshed, Y.; Zamir, D. A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica 1994, 79, 175–179. [Google Scholar] [CrossRef]
- Eshed, Y.; Zamir, D. Introgressions from Lycopersicon pennellii can improve the soluble-solids yield of tomato hybrids. Theor. Appl. Genet. 1994, 88, 891–897. [Google Scholar] [CrossRef]
- Eshed, Y.; Zamir, D. An Introgression Line Population of Lycopersicum pennellii in the Cultivated Tomato Enables the Identification and Fine Mapping of Yield-Associated QTL. Genetics 1995, 141, 1147–1162. [Google Scholar]
- Eshed, Y.; Zamir, D. Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 1996, 143, 1807–1817. [Google Scholar]
- Lippman, Z.B.; Semel, Y.; Zamir, D. An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr. Opin. Genet. Dev. 2007, 17, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Fridman, E.; Pleban, T.; Zamir, D. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. USA 2000, 97, 4718–4723. [Google Scholar] [CrossRef] [PubMed]
- Fridman, E.; Carrari, F.; Liu, Y.; Fernie, A.R.; Zamir, D. Zooming in on a quantitative interspecific introgressions. Science 2004, 305, 1786–1790. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Rodriguez, G.R.; Clevenger, J.P.; Illa-Berenguer, E.; Lin, J.; Blakeslee, J.J.; Liu, W.; Fei, Z.; Wijeratne, A.; Meulia, T.; et al. Candidate gene selection and detailed morphological evaluations of fs8.1, a quantitative trait locus controlling tomato fruit shape. J. Exp. Bot. 2015, 66, 6471–6482. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E.; et al. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 2017, 169, 1–14. [Google Scholar] [CrossRef]
- Stevens, M.A.; Dickinson, G.L.; Aguirre, M.S. UC82—A High Yielding Processing Tomato; UC-Davis Bull. Veg. Crop. Ser. 183; Vegetable Crops Department, University of California: Davis, CA, USA, 1976. [Google Scholar]
- Stevens, M.A.; Dickinson, G.L.; Aguirre, M.S. UC-82 Pedigree. Rep. Tomato Genet. Coop. 1977, 27, 65. [Google Scholar]
- Lawson, D.M.; Lunde, C.F.; Mutschler, M.A. Marker-assisted transfer of acylsugar-mediated pest resistance from the wild tomato, Lycopersicon pennellii, to the cultivated tomato, Lycopersicon esculentum. Mol. Breed. 1997, 3, 307–317. [Google Scholar] [CrossRef]
- Carter, J.D.; Pereira, A.; Dickerman, A.W.; Veilleux, R.E. An Active Ac/Ds Transposon System for Activation Tagging in Tomato Cultivar M82 Using Clonal Propagation. Plant Physiol. 2013, 162, 145–156. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef]
- Sol Genomics Network International Tomato Genome Sequencing Project Release Version 3.0. Available online: https://solgenomics.net/organism/Solanum_lycopersicum/genome (accessed on 8 April 2020).
- Sequence Read Archive Submissions Staff Using the SRA Toolkit to Convert .Sra Files into other Formats. Available online: https://www.ncbi.nlm.nih.gov/books/NBK158900/ (accessed on 8 April 2020).
- Andrews, S. FastQC a Quality Control Tool for High Throughput Sequence Ddata. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 April 2020).
- Babraham Bionformatics Group Trim Galore! Babraham Institute (UK). Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 8 April 2020).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Broad Institute Picard Tools. Available online: http://broadinstitute.github.io/picard/ (accessed on 8 April 2020).
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A mapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Li, H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 2014, 30, 2843–2851. [Google Scholar] [CrossRef]
- Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Ruden, D.M.; Lu, X. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front. Genet. 2012, 3, 1–9. [Google Scholar] [CrossRef]
- Drori, E.; Levy, D.; Rahimi, O. Genome analysis CircosVCF: Circos visualization of whole-genome sequence variations stored in VCF files. Bioinformatics 2017, 33, 1392–1393. [Google Scholar] [CrossRef]
- Raffaele, S.; Farrer, R.A.; Cano, L.M.; Studholme, D.J.; MacLean, D.; Thines, M.; Jiang, R.H.Y.; Zody, M.C.; Kunjeti, S.G.; Donofrio, N.M.; et al. Genome evolution following host jumps in the irish potato famine pathogen lineage. Science 2010, 330, 1540–1543. [Google Scholar] [CrossRef]
- Kosugi, S.; Natsume, S.; Yoshida, K.; MacLean, D.; Cano, L.; Kamoun, S.; Terauchi, R. Coval: Improving Alignment Quality and Variant Calling Accuracy for Next-Generation Sequencing Data. PLoS ONE 2013, 8, e75402. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 11, 1–33. [Google Scholar]
- Rudich, Y.; Harel, S.; Geisenberg, H. M-82-1-8 (VF) A new variety of tomatoes for canning. Hassadeh 1977, 57, 846–850. [Google Scholar]


| Polymorphic Regionsz | Polymorphic SNPs between | ||||||
|---|---|---|---|---|---|---|---|
| Chromosome | Start Position | End Position | Region Size (bp) | UC 82 Accessions | UC 82-COMAV and M82-OSU | UC 82-OSU and M82-OSU | UC 82 and M82y |
| 1 | 80,898,673 | 81,065,071 | 166,398 | 3 | - | 3 | 3 |
| 6 | 37,721 | 37,722 | 1x | 1 | 1 | - | 1 |
| 7 | 60,221,252 | 60,222,929 | 1677 | 2 | 2 | - | 2 |
| 9 | 71,304,279 | 71,953,325 | 649,046 | - | 8 | 8 | 8 |
| 10 | 59,730,745 | 59,731,037 | 292 | - | 2 | 2 | 2 |
| 11 | 7,180,897 | 7,728,808 | 547,911 | 25 | 25 | - | 25 |
| 52,038,184 | 52,836,721 | 798,537 | 48 | - | 48 | 48 | |
| Total | 2,163,862 | 79 | 38 | 61 | 89 | ||
| Percentage different relative to the total genome sequence (%)w | 0.18 | 0.14 | 0.19 | 0.26 | |||
| Sample | Number of Mapped Reads | Genome Coverage (%)z | Mean Depth of Coverage (×) | Raw | Filteredy | Hetetozygous SNPsx | |||
|---|---|---|---|---|---|---|---|---|---|
| Indels | SNPs | Indels | SNPs | Number | % | ||||
| M82-ERS484046 | 511,813,113 | 89.1 | 50.1 | 169,490 | 1,257,174 | 162,203 | 385,236 | 14,082 | 3.8 |
| M82-TS3 | 62,544,931 | 55.3 | 4.6 | 33,423 | 627,865 | 33,396 | 261,324 | 4593 | 5.1 |
| M82-TS228 | 142,963,559 | 70.4 | 16.5 | 97,189 | 1,165,307 | 91,969 | 299,680 | 24,266 | 13.9 |
| UC82-TS171 | 68,783,839 | 61.4 | 8.0 | 93,344 | 1,074,041 | 90,628 | 362,639 | 30,228 | 11.0 |
| Differential Regionsz | Polymorphic SNPs within M82 Samples | Polymorphic SNPs between M82 and UC 82 Samples | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chy | Start Position | End Position | Region Size (bp) | M82-ERS484046 vs. M82-TS228 | M82-ERS484046 vs. M82-TS3 | M82-TS228 vs. M82-TS3 | M82-ERS484046 vs. UC82-TS171 | M82-TS228 vs. UC82-TS171 | M82-TS3 vs. UC82-TS171 | UC 82 vs. M82x |
| 1 | 80,660,651 | 81,107,229 | 446,578 | - | - | - | 71 | 66 | 66 | 71 |
| 3 | 23,871,750 | 23,871,769 | 19 | 5 | - | 5 | - | 5 | - | 5 |
| 4 | 4,575,994 | 5,372,152 | 796,158 | - | - | - | 5 | 5 | 5 | 5 |
| 5 | 27,883,416 | 27,884,031 | 615 | 3 | - | 3 | - | 3 | - | 3 |
| 45,459,503 | 45,459,508 | 5 | 3 | - | 3 | - | 3 | - | 3 | |
| 7 | 60,125,464 | 62,435,965 | 2,310,501 | - | 29 | 29 | - | - | 28 | 29 |
| 9 | 71,180,818 | 71,538,753 | 357,935 | - | 14 | 14 | 14 | 14 | - | 14 |
| 72,124,815 | 72,418,523 | 293,708 | - | 19 | 19 | 19 | 19 | 19 | ||
| 10 | 42,637,890 | 42,637,909 | 19 | 4 | - | 4 | - | 4 | - | 4 |
| 59,465,991 | 59,518,850 | 52,859 | - | 5 | 5 | 5 | 5 | - | 5 | |
| 11 | 7,181,778 | 7,700,078 | 518,300 | - | 16 | 16 | - | - | 16 | 16 |
| 52,035,195 | 52,843,385 | 808,190 | - | - | - | 78 | 78 | 78 | 78 | |
| Total | 5,584,887 | 15 | 83 | 98 | 192 | 202 | 193 | 202 | ||
| Percentage different relative to the total genome sequence (%)w | 0.0001 | 0.43 | 0.43 | 0.33 | 0.33 | 0.59 | 0.67 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cambiaso, V.; Rodríguez, G.R.; Francis, D.M. Propagation Fidelity and Kinship of Tomato Varieties ‘UC 82’ and ‘M82’ Revealed by Analysis of Sequence Variation. Agronomy 2020, 10, 538. https://doi.org/10.3390/agronomy10040538
Cambiaso V, Rodríguez GR, Francis DM. Propagation Fidelity and Kinship of Tomato Varieties ‘UC 82’ and ‘M82’ Revealed by Analysis of Sequence Variation. Agronomy. 2020; 10(4):538. https://doi.org/10.3390/agronomy10040538
Chicago/Turabian StyleCambiaso, Vladimir, Gustavo Rubén Rodríguez, and David Merrill Francis. 2020. "Propagation Fidelity and Kinship of Tomato Varieties ‘UC 82’ and ‘M82’ Revealed by Analysis of Sequence Variation" Agronomy 10, no. 4: 538. https://doi.org/10.3390/agronomy10040538
APA StyleCambiaso, V., Rodríguez, G. R., & Francis, D. M. (2020). Propagation Fidelity and Kinship of Tomato Varieties ‘UC 82’ and ‘M82’ Revealed by Analysis of Sequence Variation. Agronomy, 10(4), 538. https://doi.org/10.3390/agronomy10040538







