Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Pathogen Stress Treatment
2.2. Identifying Functional Genes Adjacent to Differentially Expressed Long Noncoding Transcripts (lncRNAs)
2.3. Real-Time Quantitative PCR Analysis
3. Results
3.1. Identification of Transcription Factor Genes Adjacent to Differentially Expressed Long Non-Coding RNA in Wheat Responding to Pathogen Infection
3.2. Co-Expression of Long Non-Coding RNAs with Adjacent Functional Genes
3.3. Co-Expression of Long Non-Coding RNAs and Allele-Specific Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DE | differentially expressed |
| ncRNA | non-coding RNA |
| lincRNAs | long intergenic non-coding RNA |
| linncRNAs | long intron non-coding RNA |
References
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.D.; Sung, S. Long noncoding RNA: Unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012, 17, 16–21. [Google Scholar] [CrossRef]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariel, F.; Romero-Barrios, N.; Jegu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 1–10. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Wang, C.; Xu, Z.; Wang, Y.; Liu, X.; Kang, Z.; Ji, W. Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2013, 40, 6245–6253. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [Green Version]
- Shuai, P.; Liang, D.; Tang, S.; Zhang, Z.; Ye, C.Y.; Su, Y.; Xia, X.; Yin, W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014, 65, 4975–4983. [Google Scholar] [CrossRef]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.C.; Kean, M.J.; Xu, J.; Chua, N.H. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014, 24, 444–453. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Li, J.; Yang, Y.; Tan, C.; Zhu, Y.; Hu, L.; Qi, Y.; Lu, Z.J. Stress-responsive regulation of long non-coding RNA polyadenylation in Oryza sativa. Plant J. 2018, 93, 814–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2010, 331, 76–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Fan, X.; Lin, F.; He, G.; Terzaghi, W.; Zhu, D.; Deng, X.W. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 2014, 111, 10359–10364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hu, W.; Hao, J.; Lv, S.; Wang, C.; Tong, W.; Wang, Y.; Wang, Y.; Liu, X.; Ji, W. Genome-wide identification and functional prediction of novel and fungi-responsive lincRNAs in Triticum aestivum. BMC Genomics 2016, 17, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent, S.G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef]
- Beltran, M.; Puig, I.; Pena, C.; Garcia, J.M.; Alvarez, A.B.; Pena, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [Green Version]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Chen, J.; Lv, Q.; Qin, J.; Huang, Y.; Yu, M.; Zhong, M. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/beta-catenin signaling pathway. Cancer Lett. 2019, 440–441, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat. Commun. 2018, 9, 3516. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Diloknawarit, P.; Park, B.S.; Chua, N.H. ELF18-INDUCED LONG NONCODING RNA 1 evicts fibrillarin from mediator subunit to enhance PATHOGENESIS-RELATED GENE 1 (PR1) expression. New Phytol. 2019, 221, 2067–2079. [Google Scholar] [CrossRef] [PubMed]
- Bourras, S.; McNally, K.E.; Ben-David, R.; Parlange, F.; Roffler, S.; Praz, C.R.; Oberhaensli, S.; Menardo, F.; Stirnweis, D.; Frenkel, Z.; et al. Multiple Avirulence Loci and Allele-Specific Effector Recognition Control the Pm3 Race-Specific Resistance of Wheat to Powdery Mildew. Plant Cell 2015, 27, 2991–3012. [Google Scholar] [CrossRef] [Green Version]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [Green Version]
- Han, D.J.; Wang, Q.L.; Chen, X.M.; Zeng, Q.D.; Wu, J.H.; Xue, W.B.; Zhan, G.M.; Huang, L.L.; Kang, Z.S. Emerging Yr26-Virulent Races of Puccinia striiformis f. tritici Are Threatening Wheat Production in the Sichuan Basin, China. Plant Dis. 2015, 99, 754–760. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Wang, M.; Chen, X.; Kang, Z. Role of Alternate Hosts in Epidemiology and Pathogen Variation of Cereal Rusts. Annu. Rev. Phytopathol. 2016, 54, 207–228. [Google Scholar] [CrossRef]
- Xue, F.; Ji, W.; Wang, C.; Zhang, H.; Yang, B. High-density mapping and marker development for the powdery mildew resistance gene PmAS846 derived from wild emmer wheat (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2012, 124, 1549–1560. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, 661–674. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Shafiq, S.; Li, J.; Sun, Q. Functions of plants long non-coding RNAs. Biochim. Biophys. Acta 2016, 1859, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Sharma, S.; Taneja, M.; Tyagi, S.; Singh, K.; Upadhyay, S.K. Survey of High Throughput RNA-Seq Data Reveals Potential Roles for lncRNAs during Development and Stress Response in Bread Wheat. Front. Plant Sci. 2017, 8, 1019. [Google Scholar] [CrossRef]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, S.S.; Kayani, M.A.; Amjad, M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol. Prog. 2011, 27, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Lv, S.; Wang, C.; Ji, W. The role of transcription factor in wheat defense against pathogen and its prospect in breeding. J. Plant Biol. Crop Res. 2018, 1, 1005. [Google Scholar]
- Ishihama, N.; Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [Green Version]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A Natural Allele of a Transcription Factor in Rice Confers Broad-Spectrum Blast Resistance. Cell 2017, 170, 114–126. [Google Scholar] [CrossRef] [Green Version]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. 2010, 220, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Juan, L.; Wang, G.; Radovich, M.; Schneider, B.P.; Clare, S.E.; Wang, Y.; Liu, Y. Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med. Genomics 2013, 6, S7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
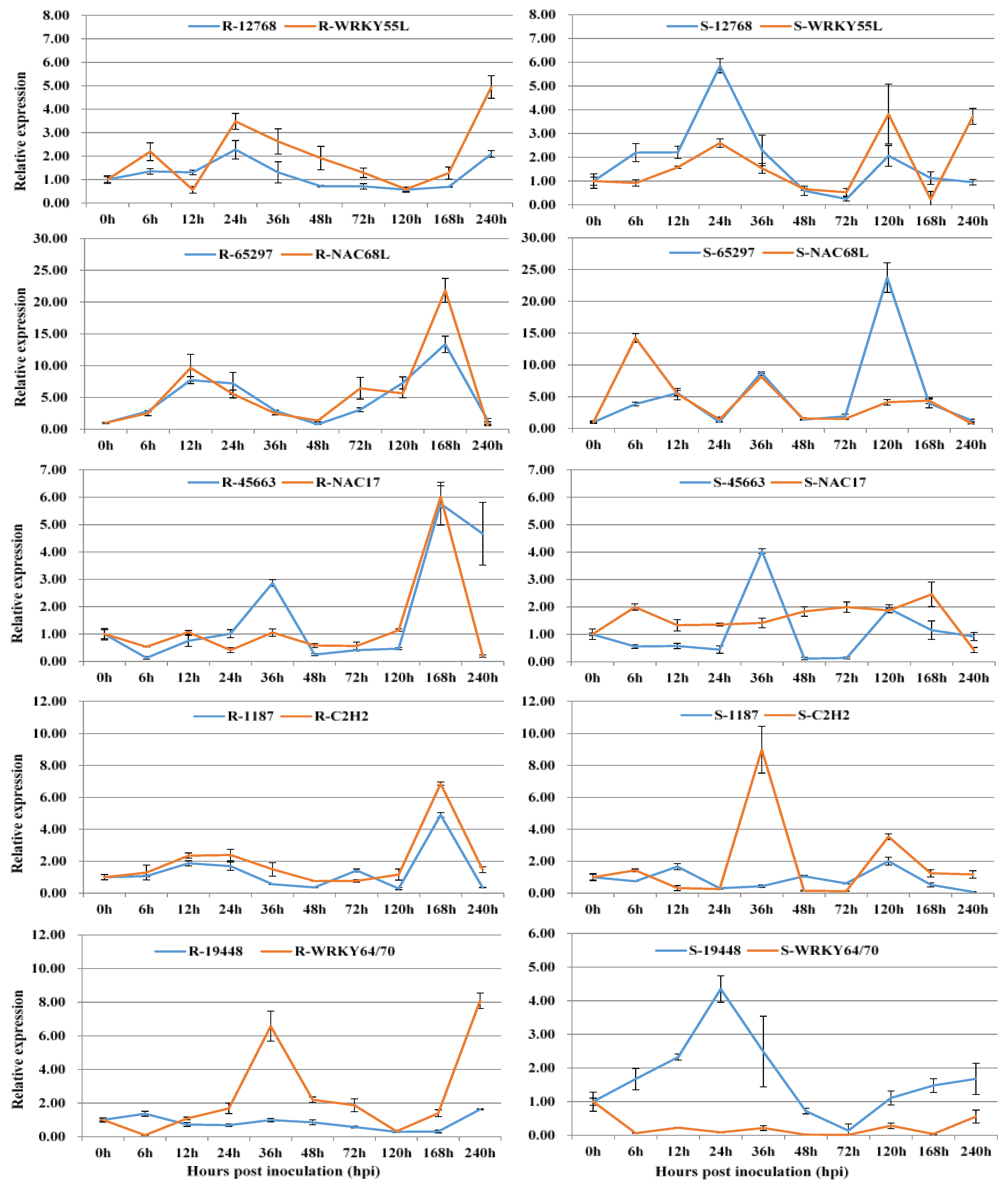

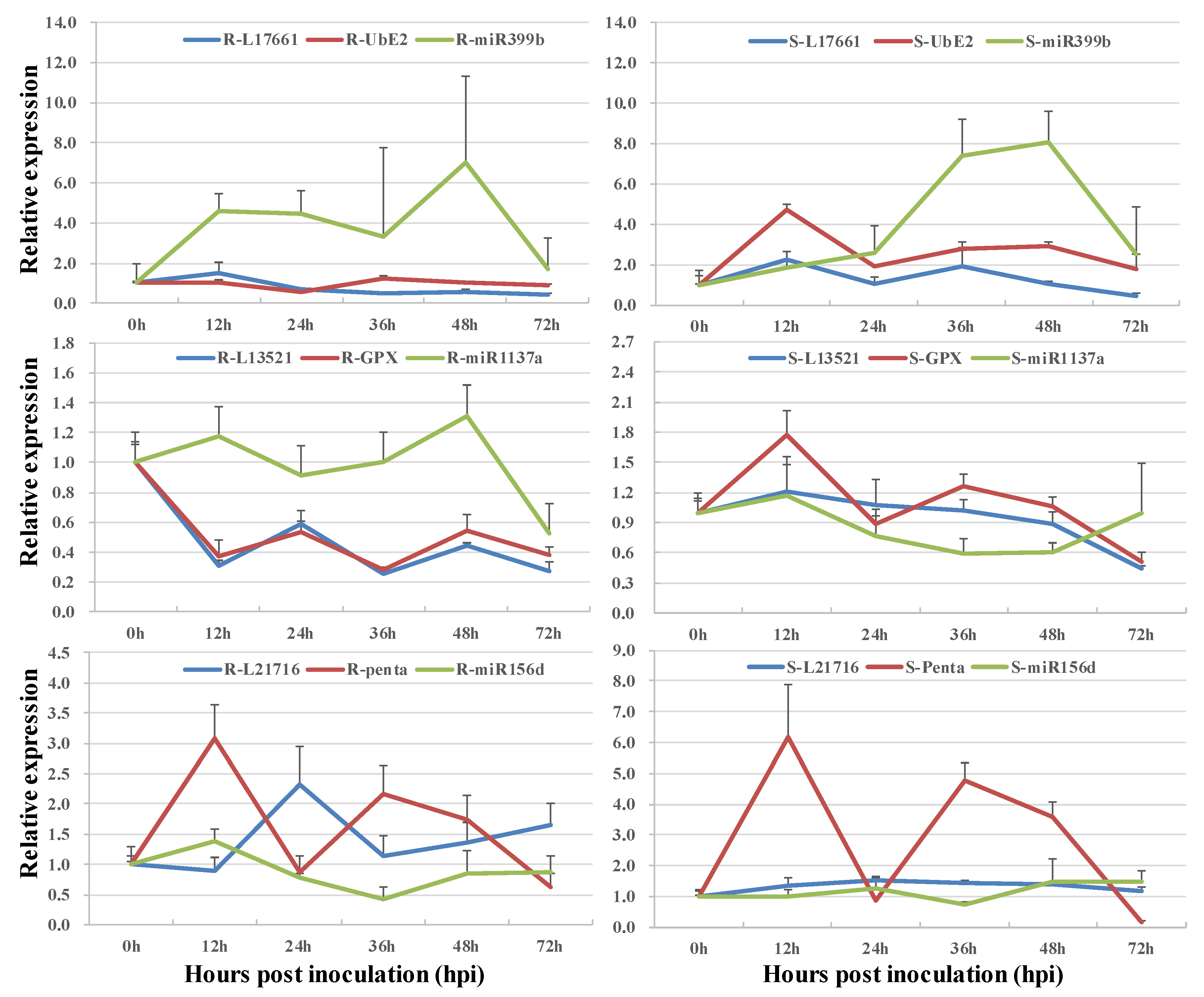
| LncRNA ID | Adjacent Functional Gene | TF Type | LncRNA Type |
|---|---|---|---|
| T4_Unigene_BMK.9130 | Ta_TraesCS1A01G200500.1 | B3 | LincRNA |
| T16_Unigene_BMK.1187 | Ta_TraesCS1B01G146800.1 | C2H2 | LincRNA |
| T10_Unigene_BMK.12768 | Ta_TraesCS1B01G243100.1 | WRKY55L | LincRNA |
| T10_Unigene_BMK.12768 | Ta_TraesCS1B01G243200.1 | AP2/ERF-ERF | LincRNA |
| T4_Unigene_BMK.17456 | Ta_TraesCS1B01G273100.1 * | CSD | LuncRNA |
| T4_Unigene_BMK.17456 | Ta_TraesCS1D01G262500.1 | CSD | LincRNA |
| T19_Unigene_BMK.34110 | Ta_TraesCS2A01G319700.1 | GNAT | LincRNA |
| T13_Unigene_BMK.49502 | Ta_TraesCS3A01G421400.1 * | bHLH | LpncRNA |
| T16_Unigene_BMK.67438 | Ta_TraesCS3A01G432900.1 | MADS-M-type | LinncRNA |
| T4_Unigene_BMK.30836 | Ta_TraesCS3D01G136600.1 | NF-X1 | LincRNA |
| T10_Unigene_BMK.65297 | Ta_TraesCS3D01G333100.1 * | NAC68L/4L | LinncRNA |
| T16_Unigene_BMK.92879 | Ta_TraesCS3D01G365300.1 | B3 | LincRNA |
| T4_Unigene_BMK.9309 | Ta_TraesCS4A01G211100.1 | MYB | LinncRNA |
| T13_Unigene_BMK.19448 | Ta_TraesCS4D01G172200.1 | WRKY64/70 | LincRNA |
| T13_Unigene_BMK.40522 | Ta_TraesCS4D01G265400.1 | GNAT | LincRNA |
| T19_Unigene_BMK.49358 | Ta_TraesCS5A01G312000.1 * | AP2/ERF-ERF | LpncRNA |
| T4_Unigene_BMK.45663 | Ta_TraesCS5D01G279100.2 * | NAC17L | LinncRNA |
| T4_Unigene_BMK.47960 | Ta_TraesCS6A01G085800.1 | BES1 | LincRNA |
| T16_Unigene_BMK.71332 | Ta_TraesCS6B01G219200.1 | mTERF | LincRNA |
| T13_Unigene_BMK.34604 | Ta_TraesCS6B01G237700.1 | AP2/ERF-ERF | LincRNA |
| T19_Unigene_BMK.51118 | Ta_TraesCS6D01G121100.1 | AP2/ERF-ERF | LpncRNA |
| T19_Unigene_BMK.54493 | Ta_TraesCS6D01G217800.1 | AP2/ERF-ERF | LincRNA |
| T16_Unigene_BMK.22544 | Ta_TraesCS7A01G326400.1 | TUB | LinncRNA |
| T16_Unigene_BMK.22544 | Ta_TraesCS7B01G227000.1 * | TUB | LinncRNA |
| T16_Unigene_BMK.22544 | Ta_TraesCS7D01G323100.1 | TUB | LinncRNA |
| T16_Unigene_BMK.23889 | Ta_TraesCS7D01G166500.1 | MYB | LincRNA |
| T13_Unigene_BMK.30347 | Ta_TraesCS7D01G269300.1 | bZIP | LincRNA |
| LncRNA ID | Adjacent Functional Gene | Definition | LncRNA Type |
|---|---|---|---|
| T16.92969 | TraesCS5B01G026400 | uncharacterized protein LOC109734965 | lpncRNA |
| T13.33010 | TraesCS5B01G036800 * | chloroplast stem-loop binding protein of 41 kDa | linncRNA |
| T16.69540 | TraesCS5B01G076100 * | Putative lipid-transfer protein DIR1 | luncRNA |
| T13.22353 | TraesCS5B01G098000 | peptidyl-prolyl cis-trans isomerase G | linncRNA |
| T13.42814 | TraesCS5B01G117000 | uncharacterized protein LOC109768056 isoform X1 | linncRNA |
| T4.63565 | TraesCS5B01G121400 | 2-oxoisovalerate dehydrogenase | linncRNA |
| T19.46503 | TraesCS5B01G134500 | uncharacterized protein LOC109774113 | lincRNA |
| T19.46503 | TraesCS5B01G134600 * | uncharacterized protein LOC109774111 isoform X1 | lincRNA |
| T16.29097 | TraesCS5B01G177300 | mediator complex subunit 25 | linncRNA |
| T16.6266 | TraesCS5B01G208100 * | cysteine endopeptidase EP gamma | linncRNA |
| T7.1464 | TraesCS5B01G232600 * | 1-aminocyclopropane-1-carboxylate oxidase 1-like | lpncRNA |
| T16.68333 | TraesCS5B01G300100 | uncharacterized protein LOC109733149 isoform X2 | lincRNA |
| T16.68333 | TraesCS5B01G300200 * | CDGSH iron-sulfur domain-containing protein NEET | lincRNA |
| T10.61842 | TraesCS5B01G302500 | GDP-mannose 3,5-epimerase 2 | linncRNA |
| T1.39963 | TraesCS5B01G404600 * | subtilisin-like protease SBT1.7 | linncRNA |
| T1.37489 | TraesCS5B01G453800 * | Lr10 disease-resistance locus receptor-like protein kinase 1.5 | linncRNA |
| T10.43083 | TraesCS5B01G478100 * | uncharacterized protein LOC109760008 | linncRNA |
| T13.86345 | TraesCS5B01G488300 | protein synthesis inhibitor II-like | linncRNA |
| T13.38179 | TraesCS5B01G535800 | pirin-like protein isoform X1 | lpncRNA |
| T13.49097 | TraesCS5B01G547000 | cinnamoyl-CoA reductase 1-like | luncRNA |
| T16.26398 | TraesCS5B01G565100 | MAP kinase kinase | linncRNA |
| T16.83333 | TraesCS1B01G069300 | uncharacterized protein LOC109765977 | linncRNA |
| T16.13852 | TraesCS1B01G110200 | Alanyl-tRNA synthetase | lincRNA |
| T16.13852 | TraesCS1B01G110300 | hypothetical protein BRADI_2g36145v3 | lincRNA |
| T16.32365 | TraesCS1B01G113500 | glutathione S-transferase 4-like | luncRNA |
| T16.1187 | TraesCS1B01G146700 | uncharacterized protein LOC109755951 | lincRNA |
| T19.42425 | TraesCS1B01G163000 | uncharacterized protein LOC100846051 isoform | lincRNA |
| T19.42425 | TraesCS1B01G163100 | uncharacterized protein LOC109772407 | lincRNA |
| T16.3724 | TraesCS1B01G174600 * | uncharacterized protein LOC109772407 | linncRNA |
| T16.16515 | TraesCS1B01G200700 | endonuclease MutS2 isoform X1 | lincRNA |
| T16.16515 | TraesCS1B01G200800 | polyprotein/retrotransposon protein, unclassified | LincRNA |
| T16.89858 | TraesCS1B01G276800 * | putative proteinase inhibitor-related protein | luncRNA |
| T7.4304 | TraesCS1B01G276900 * | wali6/Al-inducible genes | luncRNA |
| T13.23369 | TraesCS1B01G289100 | tankyrase-1-like isoform X4 | linncRNA |
| T13.24210 | TraesCS1B01G289600 * | guanylyl cyclase | linncRNA |
| T13.23192 | TraesCS1B01G384100 | chaperone protein dnaJ 10-like | lincRNA |
| T13.23192 | TraesCS1B01G384200 | unnamed protein product | lincRNA |
| T10.79431 | TraesCS1B01G394900 | Lr10 disease-resistance locus receptor-like protein kinase 1.2 | linncRNA |
| T13.51457 | TraesCS1B01G410300 * | tropinone reductase homolog At5g06060-like isoform X3 | linncRNA |
| T16.7005 | TraesCS1B01G410300 * | tropinone reductase homolog At5g06060-like isoform X3 | luncRNA |
| T16.21355 | TraesCS1B01G416400 | Pre-mRNA-processing-splicing factor 8A | lincRNA |
| T16.21355 | TraesCS1B01G416500 | ferredoxin-3, chloroplastic-like isoform X2 | lincRNA |
| T13.26624 | TraesCS1B01G433300 * | Chlorophyll a-b binding protein WCAB precursor | linncRNA |
| T16.538 | TraesCS1B01G433300 * | Chlorophyll a-b binding protein WCAB precursor | linncRNA |
| miRNA | lncRNA Mimic | lncRNA | Functional Gene |
|---|---|---|---|
| tae-miR1137a | T16.13521 | TraesCS6A02G246400.1 | |
| ath-miR414 | T13.49993 | TraesCS7B02G044200.1 | |
| ath-miR5658 | T13.33064 | TraesCS1D02G123200.1 | |
| osa-miR1439 | T19.34869 | 1B: 392147777-392149747 | |
| hvu-miR5049f | T1.48244 | TraesCS1B02G377700.1 | |
| bdi-miR394 | T1.37489 | TraesCS3D02G428200.1 | |
| tae-miR167a | T10.71969 | 7A:267192134-267192333 | |
| ath-miR390a-3p | T10.3513 | TraesCS6D02G306000.1 | |
| ata-miR160a-3p | T19.51118 | TraesCS1B02G080500.1 | |
| ata-miR395c-5p | T13.34604 | 1D:247796039-247796286 | |
| ath-miR399b | T13.17661 | TraesCS1B02G415800.1 | |
| ata-miR156d-3p | T13.21716 | TraesCS2D02G400500.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; Wang, G.; Wang, S.; Nie, X.; Wang, C.; Wang, Y.; Zhang, H.; Ji, W. Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection. Agronomy 2020, 10, 896. https://doi.org/10.3390/agronomy10060896
Hu W, Wang G, Wang S, Nie X, Wang C, Wang Y, Zhang H, Ji W. Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection. Agronomy. 2020; 10(6):896. https://doi.org/10.3390/agronomy10060896
Chicago/Turabian StyleHu, Weiguo, Guanghao Wang, Siwen Wang, Xiaojun Nie, Changyou Wang, Yajuan Wang, Hong Zhang, and Wanquan Ji. 2020. "Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection" Agronomy 10, no. 6: 896. https://doi.org/10.3390/agronomy10060896
APA StyleHu, W., Wang, G., Wang, S., Nie, X., Wang, C., Wang, Y., Zhang, H., & Ji, W. (2020). Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection. Agronomy, 10(6), 896. https://doi.org/10.3390/agronomy10060896






