Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase (TPS) Gene Family and Expression Profiling of ScTPS Genes in Sugarcane
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Screening and Classification of TPS Gene Family Members in Sugarcane
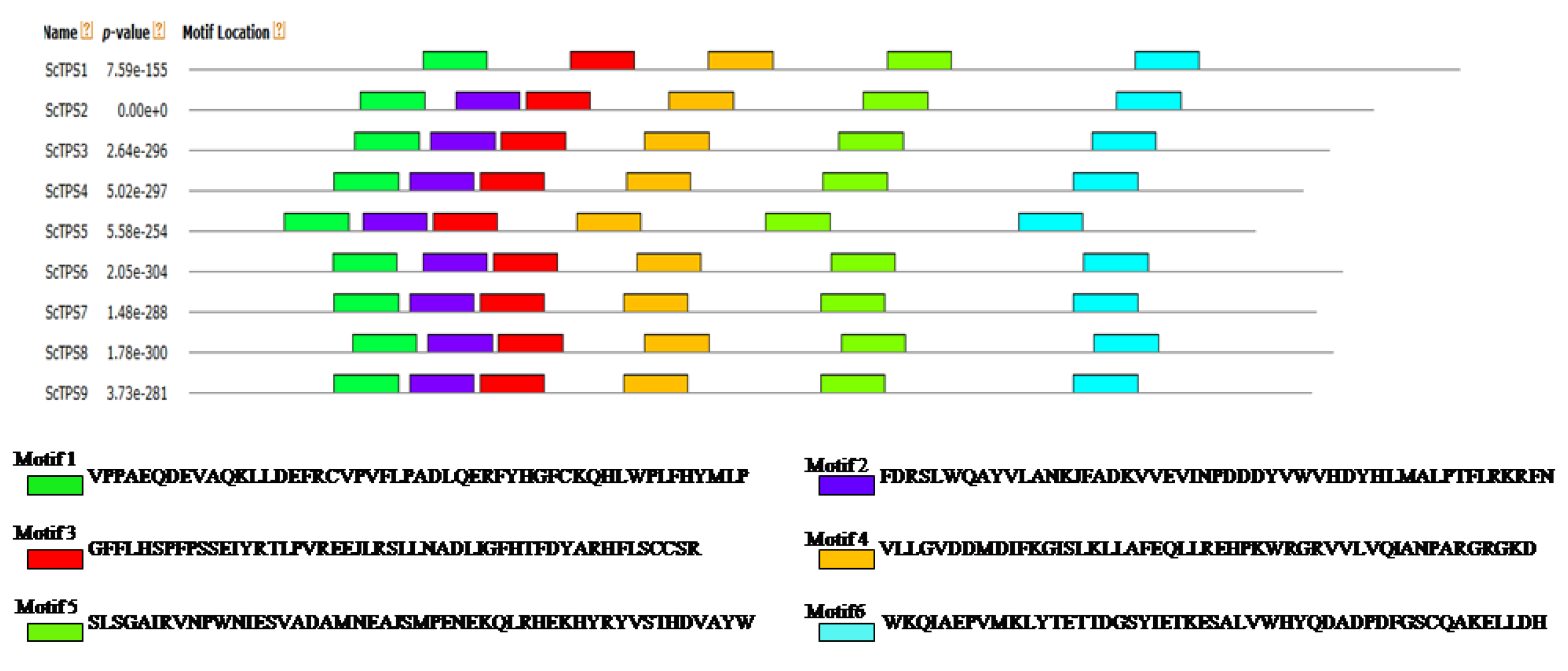
2.3. Motif Analysis of Sugarcane TPS Proteins
2.4. Phylogenetic Analysis of TPS Proteins
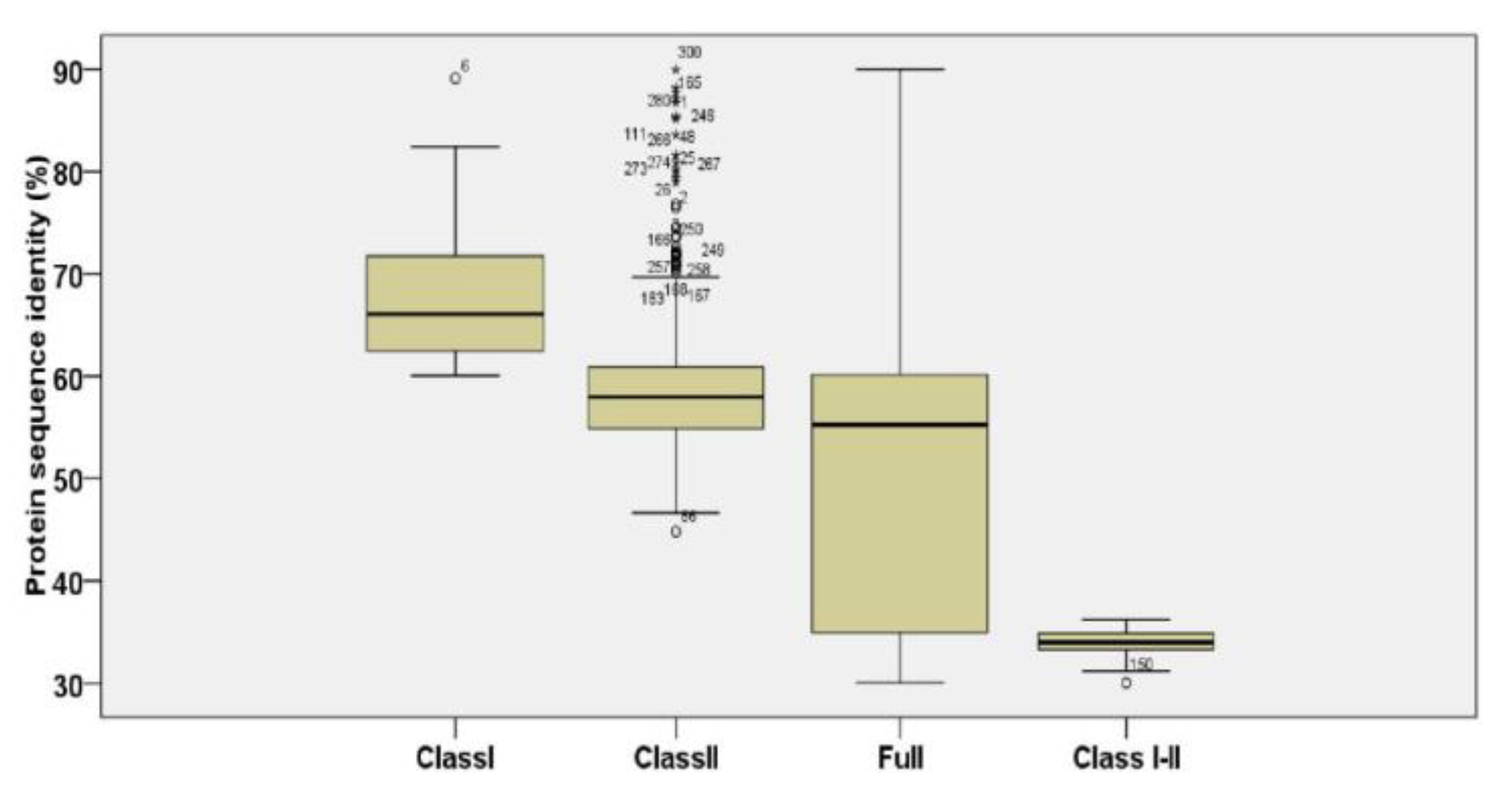
2.5. Selection Assessment and Testing
2.6. Cis-Regulatory Elements Analysis of Sugarcane TPS Gene Families
2.7. Plant Growth and Treatments
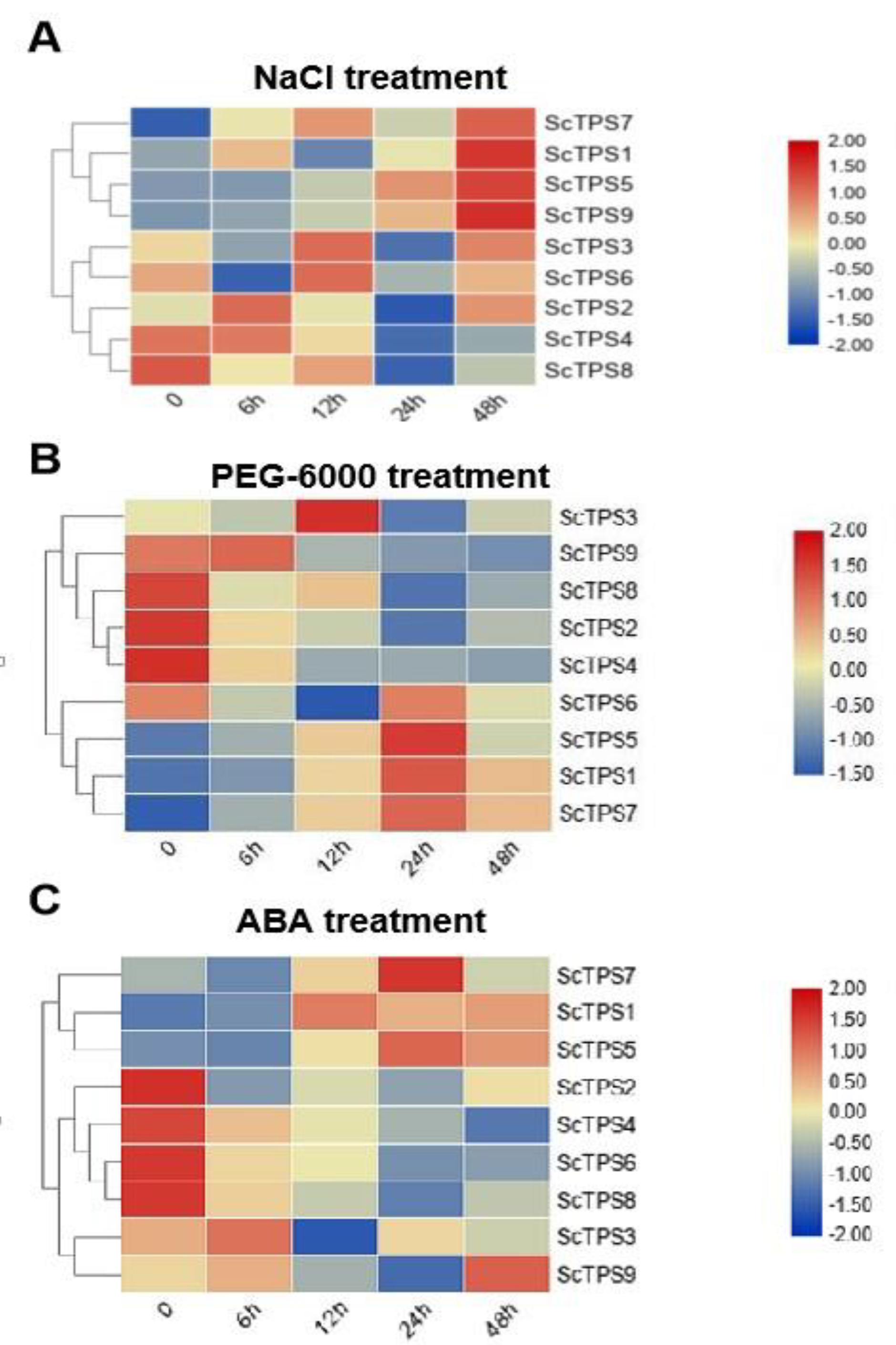
2.8. Expression Analysis of ScTPSs
2.9. Statistical Analysis
3. Results
3.1. Screening TPS Genes for Basic Physic-Chemical Properties of ScTPS Coding Proteins
3.2. Gene Structures and Protein Domains Analysis of ScTPSs
3.3. Evolution Analysis of ScTPS Genes
3.4. Cis-Regulatory Element Analysis of ScTPS Genes
3.5. Analysis of ScTPSs Expression
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pampurova, S.; Van Dijck, P. The desiccation tolerant secrets of Selaginella lepidophylla: What we have learned so far? Plant Physiol. Biochem. 2014, 80, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E. Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol. 2007, 34, 550–563. [Google Scholar] [CrossRef]
- Figueroa, C.M.; Lunn, J.E. A tale of two sugars: Trehalose 6-phosphate and sucrose. Plant Physiol. 2016, 172, 7–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2015, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, M.A.; Santos, E.; Flores, C.-L.; Martínez, M.; Ángeles, L.; Salinas, J.; Gancedo, C. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J. 1998, 13, 685–689. [Google Scholar]
- Zentella, R.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Folch-Mallol, J.; Bonini, B.; Van Vaeck, C.; Gaxiola, R.; Covarrubias, A.A.; Nieto-Sotelo, J.; Thevelein, J.M. A Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tps1 mutant. Plant Physiol. 1999, 119, 1473–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.J.; Hao, Y.J.; Zhang, Z.G.; Chen, T.; Zhang, J.S.; Chen, S.Y. Isolation of trehalose-6-phosphate phosphatase gene from tobacco and its functional analysis in yeast cells. J. Plant Physiol. 2005, 162, 215–223. [Google Scholar] [CrossRef]
- Zang, B.; Li, H.; Li, W.; Deng, X.W.; Wang, X. Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice. Plant Mol. Biol. 2011, 76, 507–522. [Google Scholar] [CrossRef]
- Ling, X.; Wang, Z.X.; Bo, H. Genome-wide identification classification and expression of TPS family genes in soybean. Chin. J. Oil Crop Sci. 2014, 36, 160–167. [Google Scholar]
- Xie, D.W.; Wang, X.N.; Fu, L.S.; Sun, J.; Zheng, W.; Li, Z.F. Identification of the trehalose-6-phosphate synthase gene family in winter wheat and expression analysis under conditions of freezing stress. J. Genet. 2015, 94, 55–65. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Mattson, N.; Yang, L.; Jin, Q. Genome-wide analysis of the Solanum tuberosum (potato) trehalose-6-phosphate synthase (TPS) gene family: Evolution and differential expression during development and stress. BMC Genomics 2017, 18, 926. [Google Scholar] [CrossRef]
- Avonce, N.; Leyman, B.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Thevelein, J.M.; Iturriaga, G. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol. 2004, 136, 3649–3659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.W.; Zang, B.S.; Deng, X.W.; Wang, X.P. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011, 234, 1007–1018. [Google Scholar] [CrossRef]
- Patade, V.Y.; Bhargava, S.; Suprasanna, P. Effects of NaCl and iso-osmotic PEG stress on growth, osmolytes accumulation and antioxidant defense in cultured sugarcane cells. Plant Cell Tissue Organ C (PCTOC) 2012, 108, 279–286. [Google Scholar] [CrossRef]
- Wu, C.W.; Chen, N.W.; Yang, R.Z.; Zhou, Z.L. Effects of water stress and rewatering on growth, development and related physiological indexes in sugarcane. Sugarcane 1998, 5, 6–12. [Google Scholar]
- Deren, C.W.; Snyder, G.H.; Miller, J.D.; Porter, P.S. Screening for and heritability of flood-tolerance in the florida (CP) sugarcane breeding population. Euphytica 1991, 56, 155–160. [Google Scholar] [CrossRef]
- Hemaprabha, G.; Nagarajan, R.; Alarmelu, S.; Natarajan, U.S. Parental potential of sugarcane clones for drought resistance breeding. Sugar Tech. 2006, 8, 59–62. [Google Scholar] [CrossRef]
- Lao, Z.Z.; Lao, F.Y.; Zhou, Y.H.; Li, Q.W.; Deng, H.H.; Huang, H.N.; Fu, C.; Hu, H.X.; Yang, Y.H.; Chen, X.W. Breeding of drought-tolerant sugarcane lines with E. arundinaceus germplasm. Zuo Wu Xue Bao 2002, 28, 841–846. [Google Scholar]
- Wang, L.P.; Jackson, P.A.; Lu, X.; Fan, Y.H.; Foreman, J.W.; Chen, X.K.; Deng, H.H.; Fu, C.; Ma, L.; Aitken, K.S. Evaluation of sugarcane × Saccharum spontaneum progeny for biomass composition and yield components. Crop Sci. 2008, 48, 951–961. [Google Scholar] [CrossRef]
- Pan, Y.-B. Development and integration of an SSR-based molecular identity database into sugarcane breeding program. Agronomy 2016, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Wang, Q.; Xie, J.; Pan, Y.-B.; Zhou, F.; Guo, Y.; Chang, H.; Xu, H.; Zhang, W.; Zhang, C.; et al. SSR marker-assisted management of parental germplasm in sugarcane (Saccharum spp. hybrids) breeding programs. Agronomy 2019, 9, 449. [Google Scholar] [CrossRef] [Green Version]
- Paul, M.J.; Oszvald, M.; Jesus, C.; Rajulu, C.; Griffiths, C.A. Increasing crop yield and resilience with trehalose 6-phosphate: Targeting a feast-famine mechanism in cereals for better source-sink optimization. J. Exp. Bot. 2017, 68, 4455–4462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeekens, S. Drought resistance: Spraying for yield. Nat. Plants 2017, 3, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Z.; Yang, B.P.; Feng, C.L.; Chen, R.K.; Luo, J.P.; Cai, W.W.; Liu, F.H. Expression of the Grifola frondosa trehalose synthase gene and improvement of drought-tolerance in sugarcane (Saccharum officinarum L.). J. Integr. Plant Biol. 2006, 48, 453–459. [Google Scholar] [CrossRef]
- Zhao, P.F.; Liu, J.Y.; Yang, K.; Xia, H.M.; Wu, C.W.; Chen, X.K.; Zhao, J.; Yang, H.C.; Li, J.; Zan, F.G. Registration of ‘YZ05-51‘ sugarcane. J. Plant Regist. 2015, 9, 172–178. [Google Scholar] [CrossRef]
- Yang, H.L.; Liu, Y.J.; Wang, C.L.; Zeng, Q.Y. Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS ONE 2012, 7, e42438. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020, 48, 265–268. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 1987, 13, 1402–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Nielsen, R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 1998, 46, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Fu, L.; Li, R.; Wang, L.N.; Yang, Y.; Liu, N.N.; Zhang, C.M.; Wang, Y.; Liu, P. PAML TBB. 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [PubMed] [Green Version]
- Meijerink, J.; Mandigers, C.; Locht, L.V.D.; Tönissen, E.; Goodsaid, F.; Raemaekers, J. A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J. Mol. Diagn. 2001, 3, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Han, B.; Fu, L.; Zhang, D.; He, X.; Chen, Q.; Peng, M.; Zhang, J. Interspecies and intraspecies analysis of trehalose contents and the biosynthesis pathway gene family reveals roles of trehalose in osmotic-stress tolerance in cassava. Int. J. Mol. Sci. 2016, 17, 1077. [Google Scholar] [CrossRef]
- Henry, C.; Bledsoe, S.W.; Siekman, A.; Kollman, A.; Waters, B.M.; Feil, R.; Stitt, M.; Lagrimini, L.M. The trehalose pathway in maize: Conservation and gene regulation in response to the diurnal cycle and extended darkness. J. Exp. Bot. 2014, 65, 5959–5973. [Google Scholar] [CrossRef] [Green Version]
- Matthew, J.P.; Asier, G.U.; Cara, A.G.; Keywan, H.P. The role of trehalose 6-phosphate in crop yield and resilience. Plant Physiol. 2018, 177, 12–23. [Google Scholar]
- Kumar, A. Bayesian phylogeny analysis of vertebrate serpins illustrates evolutionary conservation of the intron and indels based six groups classification system from lampreys for~500MY. PeerJ 2015, 3, e1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Spitz, M.R.; Amos, C.I.; Lin, J.; Schabath, M.B.; Wu, X. An evolutionary perspective on single-nucleotide polymorphism screening in molecular cancer epidemiology. Cancer Res. 2004, 64, 2251–2257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delorge, I.; Figueroa, C.M.; Feil, R.; Lunn, J.E.; Van Dijck, P. Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem. J. 2015, 466, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Van Dijken, A.J.; Schluepmann, H.; Smeekens, S. Arabidopsis Trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering1. Plant Physiol. 2004, 135, 969–977. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, L.; Paul, M.J.; Wingler, A. How Do Sugars Regulate Plant Growth and Development? New Insight into the Role of Trehalose-6-Phosphate. Mol. Plant 2013, 6, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Yadav, U.P.; Ivakov, A.; Feil, R.; Duan, G.Y.; Walther, D.; Giavalisco, P.; Piques, M.; Carillo, P.; Hubberten, H.-M.; Stitt, M.; et al. The sucrose–trehalose 6-phosphate (Tre6P) nexus: Specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 2014, 65, 1051–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, A.S.N.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, J.M.; Han, P.; Iandolino, A.; Cook, D.R. Characterization and comparison of intron structure and alternative splicing between Medicago truncatula, Populus trichocarpa, Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Pilon-Smits, E.A.H.; Terry, N.; Sears, T.; Kim, H.; Zayed, A.; Hwang, S.; Van Dun, K.; Voogd, E.; Verwoerd, T.C.; Krutwagen, R.W.H.H.; et al. Trehalose-producing transgenic tobacco plants show improved growth performance under drought stress. J. Plant Physiol. 1998, 152, 525–532. [Google Scholar] [CrossRef]
- Schluepmann, H.; Pellny, T.; Van Dijken, A.; Smeekens, S.; Paul, M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6849–6854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junior, N.N.; Nicolau, M.S.; Mantovanini, L.J.; Zingaretti, S.M. Expression analysis of two genes coding for trehalose-6-phosphate synthase (TPS), in sugarcane (Saccharum spp.) under water stress. Am. J. Plant. Sci. 2013, 4, 91–99. [Google Scholar] [CrossRef] [Green Version]



| Protein | Gene Accession Number | Number of Amino Acid | MW KD | TPS Domain Location | TPP Domain Location | pI | GRAVY | Instability Index | Subcellular Location |
|---|---|---|---|---|---|---|---|---|---|
| ScTPS1 | MN365026 | 976 | 108.4 | 129–594 | 653–852 | 6.23 | −0.34 | 50.64 | Cytoplasm |
| ScTPS2 | MT406177 | 909 | 101.8 | 77–577 | 626–860 | 5.19 | −0.233 | 54.21 | Cytomembrane |
| ScTPS3 | MT406178 | 875 | 98.6 | 73–558 | 607–842 | 5.68 | −0.325 | 49.24 | Cytoplasm |
| ScTPS4 | MT406179 | 855 | 96.9 | 56–546 | 595–828 | 5.87 | −0.228 | 46.14 | Cytoplasm |
| ScTPS5 | MT406180 | 818 | 91.1 | 18–502 | 551–787 | 6.63 | −0.165 | 54.84 | Cytoplasm |
| ScTPS6 | MT406181 | 885 | 99.4 | 55–552 | 601–835 | 5.45 | −0.261 | 49.28 | Cytoplasm |
| ScTPS7 | MT406182 | 865 | 98.0 | 58–544 | 593–829 | 6.09 | −0.241 | 41.63 | Nucleus |
| ScTPS8 | MT406183 | 878 | 99.0 | 66–560 | 609–843 | 5.87 | −0.2 | 45.82 | Cytoplasm |
| ScTPS9 | MT406184 | 861 | 97.5 | 57–544 | 593–827 | 6.22 | −0.191 | 36.98 | Cytoplasm |
| Model | p a | Estimates of Parameters | lnL | df | 2|ΔlnL| | p |
|---|---|---|---|---|---|---|
| M0 (one ratio model) | 1 | ω0 = 0.10310 | −37899.458 | - | - | - |
| Branch-specific model (Model2: two ratios) | ||||||
| Estimate ω for A | 2 | ω0 = 0.10296, ω1 = 0.15313 | −37899.349 | 1 | 0.218 | 0.640 |
| Estimate ω for B | 2 | ω0 = 0.10527, ω1 = 0.01488 | −37890.616 | 1 | 17.684 | <0.001 |
| Estimate ω for C | 2 | ω0 = 0.10406, ω1 = 0.05586 | −37897.187 | 1 | 4.542 | 0.033 |
| Estimate ω for D | 2 | ω0 = 0.10741, ω1 = 0.00102 | −37865.102 | 1 | 68.712 | <0.001 |
| Estimate ω for E | 2 | ω0 = 0.10346, ω1 = 0.00759 | −37895.919 | 1 | 7.078 | 0.008 |
| Model | p a | Estimates of Parameters | lnL | df | 2|ΔlnL| | p | Positively Selected Sites |
|---|---|---|---|---|---|---|---|
| Site Class 0 1 2a 2b | |||||||
| Branch-sites test for positive selection of Clade A | |||||||
| null | 3 | proportion 0.90708 0.09292 0.00000 0.00000 background ω 0.09643 1.00000 0.09643 1.00000 foreground ω 0.09643 1.00000 1.00000 1.00000 | −37564.649 | - | - | - | - |
| alternative | 3 | proportion 0.88351 0.08743 0.02644 0.00262 background ω 0.09624 1.00000 0.09624 1.00000 foreground ω 0.09624 1.00000 19.25885 19.25885 | −37557.107 | 1 | 15.084 | 0.0001 | 3(NEB:p > 0.95) 1(BEB:p > 0.95) |
| Branch-sites test for positive selection of Clade B | |||||||
| null | 3 | proportion 0.90708 0.09292 0.00000 0.00000 background ω 0.09643 1.00000 0.09643 1.00000 foreground ω 0.09643 1.00000 1.00000 1.00000 | −37564.649 | - | - | - | - |
| alternative | 3 | proportion 0.90708 0.09292 0.00000 0.00000 background ω 0.09643 1.00000 0.09643 1.00000 foreground ω 0.09643 1.00000 1.00000 1.00000 | −37564.649 | 1 | -2E-06 | 0.9989 | none |
| Branch-sites test for positive selection of Clade C | |||||||
| null | 3 | proportion 0.81228 0.08171 0.09631 0.00969 background ω 0.09485 1.00000 0.09485 1.00000 foreground ω 0.09485 1.00000 1.00000 1.00000 | −37551.627 | - | - | - | - |
| alternative | 3 | proportion 0.84537 0.08458 0.06368 0.00637 background ω 0.09554 1.00000 0.09554 1.00000 foreground ω 0.09554 1.00000 70.65026 70.65026 | −37545.377 | 1 | 12.499 | 0.0004 | 3(NEB:p > 0.95) |
| Branch-sites test for positive selection of Clade D | |||||||
| null | 3 | proportion 0.88381 0.09033 0.02346 0.00240 background ω 0.09606 1.00000 0.09606 1.00000 foreground ω 0.09606 1.00000 1.00000 1.00000 | −37563.772 | - | - | - | - |
| alternative | 3 | proportion 0.86913 0.08823 0.03871 0.00393 background ω 0.09603 1.00000 0.09603 1.00000 foreground ω 0.09603 1.00000 99.32701 99.32701 | −37557.905 | 1 | 11.733 | 0.0006 | none |
| Branch-sites test for positive selection of Clade E | |||||||
| null | 3 | proportion 0.58766 0.06196 0.31697 0.03342 background ω 0.09601 1.00000 0.09601 1.00000 foreground ω 0.09601 1.00000 1.00000 1.00000 | −37534.060 | - | - | - | - |
| alternative | 3 | proportion 0.61213 0.06467 0.29231 0.03088 background ω 0.09664 1.00000 0.09664 1.00000 foreground ω 0.09664 1.00000 34.71583 34.71583 | −37531.563 | 1 | 4.994 | 0.0254 | none |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Wu, Z.-D.; Luo, Z.-Y.; Burner, D.M.; Pan, Y.-B.; Wu, C.-W. Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase (TPS) Gene Family and Expression Profiling of ScTPS Genes in Sugarcane. Agronomy 2020, 10, 969. https://doi.org/10.3390/agronomy10070969
Hu X, Wu Z-D, Luo Z-Y, Burner DM, Pan Y-B, Wu C-W. Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase (TPS) Gene Family and Expression Profiling of ScTPS Genes in Sugarcane. Agronomy. 2020; 10(7):969. https://doi.org/10.3390/agronomy10070969
Chicago/Turabian StyleHu, Xin, Zhuan-Di Wu, Zheng-Ying Luo, David M. Burner, Yong-Bao Pan, and Cai-Wen Wu. 2020. "Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase (TPS) Gene Family and Expression Profiling of ScTPS Genes in Sugarcane" Agronomy 10, no. 7: 969. https://doi.org/10.3390/agronomy10070969
APA StyleHu, X., Wu, Z.-D., Luo, Z.-Y., Burner, D. M., Pan, Y.-B., & Wu, C.-W. (2020). Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase (TPS) Gene Family and Expression Profiling of ScTPS Genes in Sugarcane. Agronomy, 10(7), 969. https://doi.org/10.3390/agronomy10070969






