Fruit Breeding in Regard to Color and Seed Hardness: A Genomic View from Pomegranate
Abstract
:1. Introduction
2. Assembly and Annotation of the Pomegranate Genomes
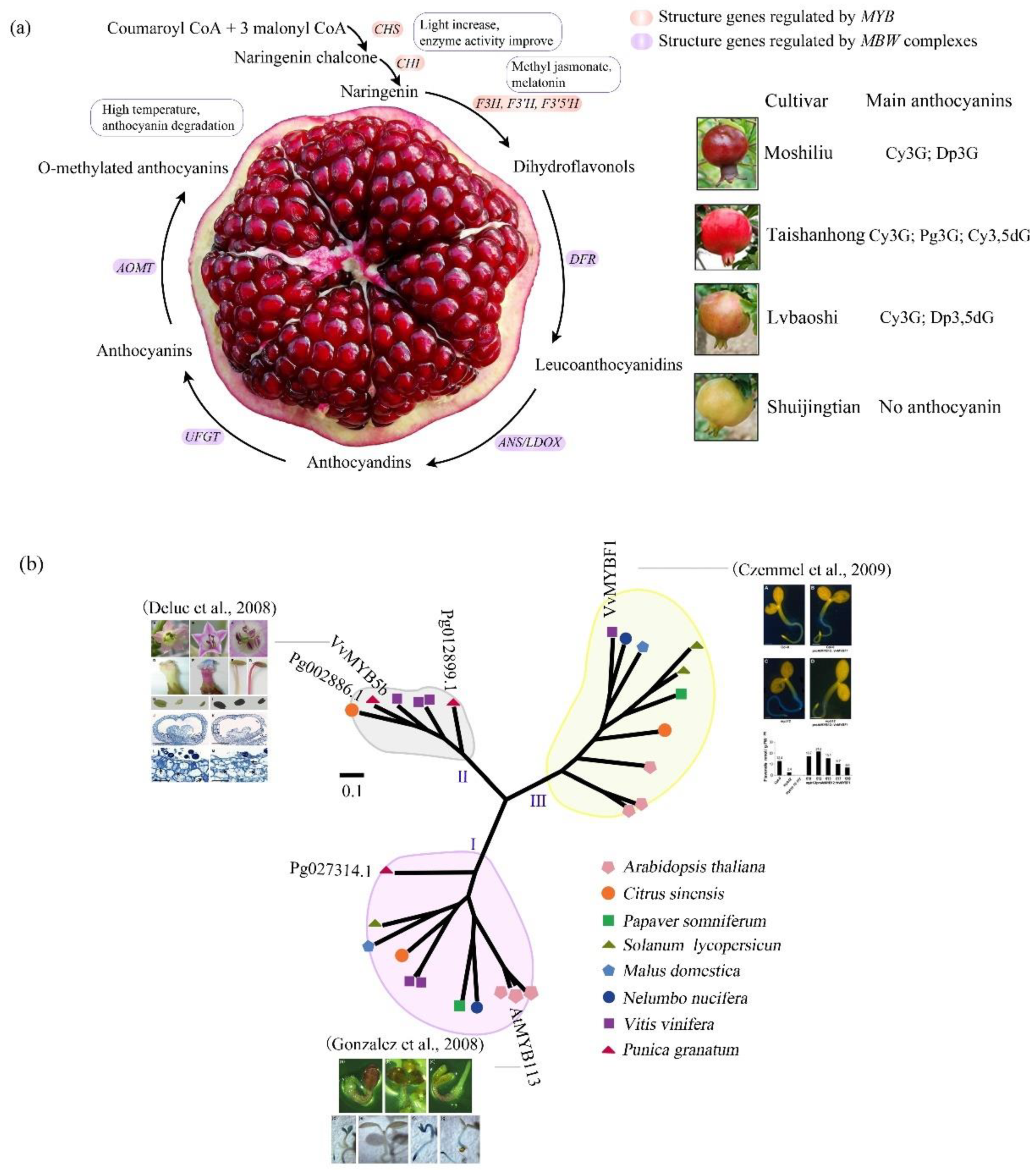
3. The Evolution-Development (Evo-devo) of Anthocyanin Biosynthesis in Pomegranate
4. Pomegranate Population Genetics for Soft Seed Breeding
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Varshney, R.K.; Bansal, K.C.; Aggarwal, P.K.; Datta, S.K.; Craufurd, P.Q. Agricultural biotechnology for crop improvement in a variable climate: Hope or hype? Trends Plant Sci. 2011, 16, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Terauchi, R.; McCouch, S.R. Harvesting the Promising Fruits of Genomics: Applying Genome Sequencing Technologies to Crop Breeding. PLoS Biol. 2014, 12, e1001883. [Google Scholar] [CrossRef] [Green Version]
- Rugini, E.; Cristofori, V.; Silvestri, C. Genetic improvement of olive (Olea europaea L.) by conventional and in vitro biotechnology methods. Biotechnol. Adv. 2016, 34, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Minio, A.; Massonnet, M.; Solares, E.; Lv, Y.; Beridze, T.; Cantu, D.; Gaut, B.S. The population genetics of structural variants in grapevine domestication. Nat. Plants 2019, 5, 965–979. [Google Scholar] [CrossRef]
- Duan, N.; Bai, Y.; Sun, H.; Wang, N.; Ma, Y.; Li, M.; Wang, X.; Jiao, C.; Legall, N.; Mao, L.; et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017, 8, 249. [Google Scholar] [CrossRef] [PubMed]
- Cornille, A.; Gladieux, P.; Smulders, M.J.M.; Roldán-Ruiz, I.; Laurens, F.; Le Cam, B.; Nersesyan, A.; Clavel, J.; Olonova, M.; Feugey, L.; et al. New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties. PLoS Genet. 2012, 8, e1002703. [Google Scholar] [CrossRef] [Green Version]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Zheng, Z.; Wang, L.; Liu, X.; Zhu, G.; Fang, W.; Cheng, S.; Zeng, P.; Chen, C.; Wang, X.; et al. Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol. 2014, 15, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef] [Green Version]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.L.; Chen, J.; Biggers, E.; et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Ding, J.; Deng, D.; Tang, W.; Sun, H.; Liu, D.; Zhang, L.; Niu, X.; Zhang, X.; Meng, M.; et al. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 2013, 4, 2640. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ma, T.; Kang, M.; Ai, F.; Zhang, J.; Dong, G.; Liu, J. A high-quality Actinidia chinensis (kiwifruit) genome. Hortic. Res. 2019, 6, 117. [Google Scholar] [CrossRef] [Green Version]
- Colle, M.; Leisner, C.P.; Wai, C.M.; Ou, S.; Bird, K.A.; Wang, J.; Wisecaver, J.H.; Yocca, A.E.; Alger, E.I.; Tang, H.; et al. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. GigaScience 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Teh, B.T.; Lim, K.; Yong, C.H.; Ng, C.C.Y.; Rao, S.R.; Rajasegaran, V.; Lim, W.K.; Ong, C.K.; Chan, K.; Cheng, V.K.Y.; et al. The draft genome of tropical fruit durian (Durio zibethinus). Nat. Genet. 2017, 49, 1633–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.-M.; Jia, H.-J.; Cai, Q.-L.; Wang, Y.; Zhao, H.-B.; Yang, W.-F.; Wang, G.-Y.; Li, Y.-H.; Zhan, D.-L.; Shen, Y.-T.; et al. The red bayberry genome and genetic basis of sex determination. Plant Biotechnol. J. 2019, 17, 397–409. [Google Scholar] [CrossRef] [Green Version]
- Zhu, T.; Wang, L.; You, F.M.; Rodriguez, J.C.; Deal, K.R.; Chen, L.; Li, J.; Chakraborty, S.; Balan, B.; Jiang, C.-Z.; et al. Sequencing a Juglans regia × J. microcarpa hybrid yields high-quality genome assemblies of parental species. Hortic. Res. 2019, 6, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.-W.; Xu, L.-L.; Li, N.; Yan, P.-C.; Jiang, X.-H.; Woeste, K.E.; Lin, K.; Renner, S.S.; Zhang, D.-Y.; Bai, W.-N. Phylogenomics Reveals an Ancient Hybrid Origin of the Persian Walnut. Mol. Biol. Evol. 2019, 36, 2451–2461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, H.; Sun, L.; Fan, G.; Ye, M.; Jiang, L.; Liu, X.; Ma, K.; Shi, C.; Bao, F.; et al. The genetic architecture of floral traits in the woody plant Prunus mume. Nat. Commun. 2018, 9, 1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.; Liu, Y.; Zhang, Q.; Nie, X.; Sun, Y.; Zhang, Z.; Li, H.; Fang, K.; Wang, G.; Huang, H.; et al. Hybrid de novo genome assembly of Chinese chestnut (Castanea mollissima). GigaScience 2019, 8, giz112. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhang, J.; Wang, S.; Yang, L.; Luo, Y.; Gao, S.; Zhang, M.; Wu, S.; Hu, S.; Sun, H.; et al. The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and beta-carotenoid synthesis. Hortic. Res. 2019, 6, 128. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Tu, X.L.; Dai, H.; Han, F.M.; Lu, B.S.; Wang, M.S.; Nanaei, H.A.; Tajabadipour, A.; Mansouri, M.; Li, X.L.; et al. Whole genomes and transcriptomes reveal adaptation and domestication of pistachio. Genome Biol. 2019, 20, 79. [Google Scholar] [CrossRef]
- Baek, S.; Choi, K.; Kim, G.-B.; Yu, H.-J.; Cho, A.; Jang, H.; Kim, C.; Kim, H.-J.; Chang, K.S.; Kim, J.-H.; et al. Draft genome sequence of wild Prunus yedoensis reveals massive inter-specific hybridization between sympatric flowering cherries. Genome Biol. 2018, 19, 127. [Google Scholar] [CrossRef]
- Lantican, D.V.; Strickler, S.R.; Canama, A.O.; Gardoce, R.R.; Mueller, L.A.; Galvez, H.F. De Novo Genome Sequence Assembly of Dwarf Coconut (Cocos nucifera L.’Catigan Green Dwarf’) Provides Insights into Genomic Variation Between Coconut Types and Related Palm Species. G3 Genes Genomes Genet. 2019, 9, 2377–2393. [Google Scholar]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.; Li, K.; Poudel, K.; Zhao, D. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft-and hard-seeded cultivars. Plant Biotechnol. J. 2019, 18, 955–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Hu, J.; Han, X.; Li, J.; Gao, Y.; Richards, C.M.; Zhang, C.; Tian, Y.; Liu, G.; Gul, H. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Itkin, M.; Yeselson, Y.; Tzuri, G.; Portnoy, V.; Harel-Baja, R.; Lev, S.; Sa’ar, U.; Davidovitz-Rikanati, R.; Baranes, N.; et al. The PH gene determines fruit acidity and contributes to the evolution of sweet melons. Nat. Commun. 2014, 5, 4026. [Google Scholar] [CrossRef] [Green Version]
- Chandra, R.; Babu, D.; Jadhav, V.T.; Jaime, A.; Silva, T. Origin, history and domestication of pomegranate. Fruit Veg. Cereal Sci. Biotechnol. 2010, 2, 1–6. [Google Scholar]
- Graham, S.A.; Graham, A. Ovary, fruit, and seed morphology of the Lythraceae. Int. J. Plant Sci. 2014, 175, 202–240. [Google Scholar] [CrossRef]
- Berger, B.A.; Kriebel, R.; Spalink, D.; Sytsma, K.J. Divergence times, historical biogeography, and shifts in speciation rates of Myrtales. Mol. Phylogenet. Evol. 2016, 95, 116–136. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Mohanty, A.; Lal, A. Pomegranate genetic resources and germplasm conservation: A review. Fruit Veg. Cereal Sci. Biotechnol. 2010, 4, 120–125. [Google Scholar]
- Jalikop, S. Pomegranate breeding. Fruit Veg. Cereal Sci. Biothecnol. 2010, 4, 26–34. [Google Scholar]
- Teixeira da Silva, J.A.; Rana, T.S.; Narzary, D.; Verma, N.; Meshram, D.T.; Ranade, S.A. Pomegranate biology and biotechnology: A review. Sci. Hortic. 2013, 160, 85–107. [Google Scholar] [CrossRef]
- Tzulker, R.; Glazer, I.; Bar-Ilan, I.; Holland, D.; Aviram, M.; Amir, R. Antioxidant Activity, Polyphenol Content, and Related Compounds in Different Fruit Juices and Homogenates Prepared from 29 Different Pomegranate Accessions. J. Agric. Food Chem. 2007, 55, 9559–9570. [Google Scholar] [CrossRef]
- Kazemi, M.; Karim, R.; Mirhosseini, H.; Hamid, A.A. Optimization of pulsed ultrasound-assisted technique for extraction of phenolics from pomegranate peel of Malas variety: Punicalagin and hydroxybenzoic acids. Food Chem. 2016, 206, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Halvorsen, B.L.; Holte, K.; Myhrstad, M.C.W.; Barikmo, I.; Hvattum, E.; Remberg, S.F.; Wold, A.-B.; Haffner, K.; Baugerød, H.; Andersen, L.F.; et al. A systematic screening of total antioxidants in dietary plants. J. Nutr. 2002, 132, 461–471. [Google Scholar] [CrossRef] [Green Version]
- Medjakovic, S.; Jungbauer, A. Pomegranate: A fruit that ameliorates metabolic syndrome. Food Funct. 2013, 4, 19–39. [Google Scholar] [CrossRef]
- Tehranifar, A.; Zarei, M.; Nemati, Z.; Esfandiyari, B.; Vazifeshenas, M.R. Investigation of physico-chemical properties and antioxidant activity of twenty Iranian pomegranate (Punica granatum L.) cultivars. Sci. Hortic. 2010, 126, 180–185. [Google Scholar] [CrossRef]
- Miguel, M.G.; Neves, M.A.; Antunes, M.D. Pomegranate (Punica granatum L.): A medicinal plant with myriad biological properties-A short review. J. Med. Plants Res. 2010, 4, 2836–2847. [Google Scholar]
- Sharma, P.; McClees, S.F.; Afaq, F. Pomegranate for prevention and treatment of cancer: An update. Molecules 2017, 22, 177. [Google Scholar] [CrossRef] [Green Version]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Kovach, A.; Wegrzyn, J.L.; Parra, G.; Holt, C.; Bruening, G.E.; Loopstra, C.A.; Hartigan, J.; Yandell, M.; Langley, C.H.; Korf, I.; et al. The Pinus taeda genome is characterized by diverse and highly diverged repetitive sequences. BMC Genom. 2010, 11, 420. [Google Scholar] [CrossRef] [Green Version]
- Guan, R.; Zhao, Y.; Zhang, H.; Fan, G.; Liu, X.; Zhou, W.; Shi, C.; Wang, J.; Liu, W.; Liang, X.; et al. Draft genome of the living fossil Ginkgo biloba. GigaScience 2016, 5, 49. [Google Scholar] [CrossRef] [Green Version]
- Wan, T.; Liu, Z.-M.; Li, L.-F.; Leitch, A.R.; Leitch, I.J.; Lohaus, R.; Liu, Z.-J.; Xin, H.-P.; Gong, Y.-B.; Liu, Y.; et al. A genome for gnetophytes and early evolution of seed plants. Nat. Plants 2018, 4, 82–89. [Google Scholar] [CrossRef]
- Ouyang, S.; Buell, C.R. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004, 32, D360–D363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Tingey, S.V.; Morgante, M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001, 11, 1660–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, K.; Goertzen, L.R. Spliceosomal intron size expansion in domesticated grapevine (Vitis vinifera). BMC Res. Notes 2011, 4, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ungerer, M.C.; Strakosh, S.C.; Zhen, Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 2006, 16, R872–R873. [Google Scholar] [CrossRef] [Green Version]
- Vitte, C.; Panaud, O. LTR retrotransposons and flowering plant genome size: Emergence of the increase/decrease model. Cytogenet. Genome Res. 2005, 110, 91–107. [Google Scholar] [CrossRef]
- Xia, E.-H.; Zhang, H.-B.; Sheng, J.; Li, K.; Zhang, Q.-J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Zhang, T.; Yuan, Z. Evolution and classification of pomegranate. In Proceedings of the IV International Symposium on Pomegranate and Minor Mediterranean Fruits 1254, Elche, Spain, 18–22 September 2017; pp. 41–48. [Google Scholar]
- Initiative, O.T.P.T. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679. [Google Scholar]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousuf, B.; Gul, K.; Wani, A.A.; Singh, P. Health Benefits of Anthocyanins and Their Encapsulation for Potential Use in Food Systems: A Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, C.; Huang, X.; Zhang, H.; Yuan, Z. Land-plant Phylogenomic and Pomegranate Transcriptomic Analyses Reveal an Evolutionary Scenario of CYP75 Genes Subsequent to Whole Genome Duplications. J. Plant Biol. 2019, 62, 48–60. [Google Scholar] [CrossRef]
- Casedas, G.; Les, F.; Gomez-Serranillos, M.P.; Smith, C.; Lopez, V. Anthocyanin profile, antioxidant activity and enzyme inhibiting properties of blueberry and cranberry juices: A comparative study. Food Funct. 2017, 8, 4187–4193. [Google Scholar] [CrossRef]
- Fang, J. Classification of fruits based on anthocyanin types and relevance to their health effects. Nutrition 2015, 31, 1301–1306. [Google Scholar] [CrossRef]
- Bar-Ya’akov, I.; Tian, L.; Amir, R.; Holland, D. Primary Metabolites, Anthocyanins, and Hydrolyzable Tannins in the Pomegranate Fruit. Front. Plant Sci. 2019, 10, 620. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yuan, Z.; Fang, Y.; Yin, Y.; Feng, L. Characterization and evaluation of major anthocyanins in pomegranate (Punica granatum L.) peel of different cultivars and their development phases. Eur. Food Res. Technol. 2013, 236, 109–117. [Google Scholar] [CrossRef]
- Feng, Z.; Yuan, Z.; Zhao, X.; Yin, Y.; Feng, L. COMPOSITION AND CONTENTS OF ANTHOCYANINS IN DIFFERENT POMEGRANATE CULTIVARS. Acta Hortic. 2015, 10890, 35–41. [Google Scholar] [CrossRef]
- Ben-Simhon, Z.; Judeinstein, S.; Trainin, T.; Harel-Beja, R.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A "White" Anthocyanin-less Pomegranate (Punica granatum L.) Caused by an Insertion in the Coding Region of the Leucoanthocyanidin Dioxygenase (LDOX.; ANS) Gene. PLoS ONE 2015, 10, e0142777. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- García-Pastor, M.E.; Guillén, F.; Zapata, P.J.; Agulló, V.; Valero, D. Melatonin: A new tool to increase yield and quality at harvest and to extend postharvest shelf-life of pomegranate. Acta Hortic. 2019, 289–294. [Google Scholar] [CrossRef]
- Pastor, M.E.; Serrano, M.; Guillén, F.; Giménez, M.; Martínez-Romero, D.; Valero, D.; Zapata, P. Preharvest application of methyl jasmonate increases crop yield, fruit quality and bioactive compounds in pomegranate ‘Mollar de Elche’ at harvest and during postharvest storage. J. Sci. Food Agric. 2019, 100, 145–153. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2394. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Li, N.; Wu, H.; Ding, Q.; Li, H.; Li, Z.; Ding, J.; Li, Y. The heterologous expression of Arabidopsis PAP2 induces anthocyanin accumulation and inhibits plant growth in tomato. Funct. Integr. Genom. 2018, 18, 341–353. [Google Scholar] [CrossRef]
- Carrasco, D.; De Lorenzis, G.; Maghradze, D.; Revilla, E.; Bellido, A.; Failla, O.; Arroyo-García, R. Allelic variation in the VvMYBA1 and VvMYBA2 domestication genes in natural grapevine populations (Vitis vinifera subsp. sylvestris). Plant Syst. Evol. 2015, 301, 1613–1624. [Google Scholar] [CrossRef]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Khaksar, G.; Tabatabaei, B.E.S.; Arzani, A.; Ghobadi, C.; Ebrahimie, E. Functional Analysis of a Pomegranate (Punica granatum L.) MYB Transcription Factor Involved in the Regulation of Anthocyanin Biosynthesis. Iran. J. Biotechnol. 2015, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-Induced Expression of a MYBGene Regulates Anthocyanin Biosynthesis in Red Apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, D.-G.; Sun, C.-H.; Ma, Q.-J.; You, C.-X.; Cheng, L.; Hao, Y.-J. MdMYB1 Regulates Anthocyanin and Malate Accumulation by Directly Facilitating Their Transport into Vacuoles in Apples. Plant Physiol. 2016, 170, 1315–1330. [Google Scholar] [CrossRef] [Green Version]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Brendolise, C.; Carlisle, C.M.; Kumar, S.; De Silva, N.; et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef] [Green Version]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and Functional Analysis of a MYB Transcription Factor Gene that is a Key Regulator for the Development of Red Coloration in Apple Skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Deluc, L.; Barrieu, F.; Marchive, C.; Lauvergeat, V.; Decendit, A.; Richard, T.; Carde, J.-P.; Mérillon, J.-M.; Hamdi, S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006, 140, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.-M.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Zhao, K.; Liu, L.; Zhang, K.; Yuan, H.; Liao, X.; Wang, Q.; Guo, X.; Li, F.; Li, T. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 2014, 55, 862–880. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [Green Version]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahim, M.A.; Busatto, N.; Trainotti, L. Regulation of anthocyanin biosynthesis in peach fruits. Planta 2014, 240, 913–929. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; LI, S.; Zhang, R.; Zhao, J.; Chen, Y.; Zhao, Q.; Yao, Y.I.; You, C.I.; Zhang, X.H.; Hao, Y.I. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; McGhie, T.K.; Wang, M.; Liu, Y.; Warren, B.; Storey, R.; Espley, R.V.; Allan, A.C. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 2014, 5, 651. [Google Scholar] [CrossRef] [Green Version]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005, 10, 621–630. [Google Scholar] [CrossRef] [Green Version]
- Ono, N.N.; Britton, M.T.; Fass, J.N.; Nicolet, C.M.; Lin, D.; Tian, L. Exploring the Transcriptome Landscape of Pomegranate Fruit Peel for Natural Product Biosynthetic Gene and SSR Marker Discovery F. J. Integr. Plant Biol. 2011, 53, 800–813. [Google Scholar] [CrossRef]
- Rouholamin, S.; Zahedi, B.; Nazarian-Firouzabadi, F.; Saei, A. Expression analysis of anthocyanin biosynthesis key regulatory genes involved in pomegranate (Punica granatum L.). Sci. Hortic. 2015, 186, 84–88. [Google Scholar] [CrossRef]
- Arlotta, C.; Puglia, G.D.; Genovese, C.; Toscano, V.; Karlova, R.; Beekwilder, J.; De Vos, R.C.H.; Raccuia, S.A. MYB5-like and bHLH influence flavonoid composition in pomegranate. Plant Sci. 2020, 298, 110563. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, Z.; Feng, L.; Fang, Y. Cloning and expression of anthocyanin biosynthetic genes in red and white pomegranate. J. Plant Res. 2015, 128, 687–696. [Google Scholar] [CrossRef]
- Luo, X.; Cao, D.; Li, H.; Zhao, D.; Xue, H.; Niu, J.; Chen, L.; Zhang, F.; Cao, S. Complementary iTRAQ-based proteomic and RNA sequencing-based transcriptomic analyses reveal a complex network regulating pomegranate (Punica granatum L.) fruit peel colour. Sci. Rep. 2018, 8, 12362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Yuan, Z. Quick convergent evolution of MBW complex for pomegranate fruit coloration. In Proceedings of the IV International Symposium on Pomegranate and Minor Mediterranean Fruits 1254, Elche, Spain, 18–22 September 2017; pp. 135–142. [Google Scholar]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czemmel, S.; Stracke, R.; Weisshaar, B.; Cordon, N.; Harris, N.N.; Walker, A.R.; Robinson, S.P.; Bogs, J. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 2009, 151, 1513–1530. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.-S.; Gugger, P.F.; Wang, Q.-F.; Chen, J.-M. Identification of a R2R3-MYB gene regulating anthocyanin biosynthesis and relationships between its variation and flower color difference in lotus (Nelumbo Adans.). PeerJ 2016, 4, e2369. [Google Scholar] [CrossRef] [Green Version]
- Shaked-Sachray, L.; Weiss, D.; Reuveni, M.; Nissim-Levi, A.; Oren-Shamir, M. Increased anthocyanin accumulation in aster flowers at elevated temperatures due to magnesium treatment. Physiol. Plant 2002, 114, 559–565. [Google Scholar] [CrossRef]
- Mori, K.; Goto-Yamamoto, N.; Kitayama, M.; Hashizume, K. Loss of anthocyanins in red-wine grape under high temperature. J. Exp. Bot. 2007, 58, 1935–1945. [Google Scholar] [CrossRef]
- Poudel, P.R.; Mochioka, R.; Beppu, K.; Kataoka, I. Influence of Temperature on Berry Composition of Interspecific Hybrid Wine Grape; Kadainou R-1; (Vitis ficifoliavar. ganebu; V. vinifera;Muscat of Alexandria;). J. Jpn. Soc. Hortic. Sci. 2009, 78, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Shvarts, M.; Borochov, A.; Weiss, D. Low temperature enhances petunia flower pigmentation and induces chalcone synthase gene expression. Physiol. Plant 1997, 99, 67–72. [Google Scholar] [CrossRef]
- Huh, E.J.; Shin, H.K.; Choi, S.Y.; Kwon, O.G.; Lee, Y.R. Thermosusceptible Developmental Stage in Anthocyanin Accumulation and Color Response to High Temperature in Red Chrysanthemum Cultivars. Korean J. Hortic. Sci. Technol. 2008, 26, 357–361. [Google Scholar]
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef] [Green Version]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamrick, J. Plant population genetics and evolution. Am. J. Bot. 1982, 69, 1685–1693. [Google Scholar] [CrossRef]
- Levin, G.M. Pomegranate Roads: A Soviet Botanist’s Exile from Eden; Floreant Press: Forestville, CA, USA, 2006.
- Levin, G.M. Pomegranate (Punica granatum) plant genetic resources in Turkmenistan. Bull. Des Ressour. Phytogenet. (Ipgri/Fao) Not. de Recur. Fitogeneticos (Ipgri/Fao) 1994, 97, 31–36. [Google Scholar]
- Yuan, Z.; Yin, Y.; Qu, J.; Zhu, L.; Li, Y. Population Genetic Diversity in Chinese Pomegranate (Punica granatum L.) Cultivars Revealed by Fluorescent-AFLP Markers. J. Genet. Genom. 2007, 34, 1061–1071. [Google Scholar] [CrossRef]
- Narzary, D.; Rana, T.S.; Ranade, S.A. Genetic diversity in inter-simple sequence repeat profiles across natural populations of Indian pomegranate (Punica granatum L.). Plant Biol. 2010, 12, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Hasnaoui, N.; Buonamici, A.; Sebastiani, F.; Mars, M.; Zhang, D.; Vendramin, G.G. Molecular genetic diversity of Punica granatum L. (pomegranate) as revealed by microsatellite DNA markers (SSR). Gene 2012, 493, 105–112. [Google Scholar] [CrossRef]
- Sarkhosh, A.; Zamani, Z.; Fatahi, R.; Ranjbar, H. Evaluation of genetic diversity among Iranian soft-seed pomegranate accessions by fruit characteristics and RAPD markers. Sci. Hortic. 2009, 121, 313–319. [Google Scholar] [CrossRef]
- Glozer, K.; Ferguson, L. Pomegranate production in Afghanistan. Ucdavis Coll. Agric. Environ. Sci. 2008, 1–32. [Google Scholar]
- Dardick, C.; Callahan, A.M. Evolution of the fruit endocarp: Molecular mechanisms underlying adaptations in seed protection and dispersal strategies. Front. Plant Sci. 2014, 5, 284. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [Green Version]
- Zarei, A.; Zamani, Z.; Fatahi, R.; Mousavi, A.; Salami, S.A.; Avila, C.; Cánovas, F.M. Differential expression of cell wall related genes in the seeds of soft- and hard-seeded pomegranate genotypes. Sci. Hortic. 2016, 205, 7–16. [Google Scholar] [CrossRef]
- Niu, J.; Cao, D.; Li, H.; Xue, H.; Chen, L.; Liu, B.; Cao, S. Quantitative proteomics of pomegranate varieties with contrasting seed hardness during seed development stages. Tree Genet. Genomes 2018, 14, 14. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Cao, D.; Zhang, J.; Chen, L.; Xia, X.; Li, H.; Zhao, D.; Zhang, F.; Xue, H.; Chen, L.; et al. Integrated microRNA and mRNA expression profiling reveals a complex network regulating pomegranate (Punica granatum L.) seed hardness. Sci. Rep. 2018, 8, 9292. [Google Scholar] [CrossRef] [Green Version]
- Dai, H.; Han, G.; Yan, Y.; Zhang, F.; Liu, Z.; Li, X.; Li, W.; Ma, Y.; Li, H.; Liu, Y.; et al. Transcript Assembly and Quantification by RNA-Seq Reveals Differentially Expressed Genes between Soft-Endocarp and Hard-Endocarp Hawthorns. PLoS ONE 2013, 8, e72910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, H.; Cao, S.; Li, H.; Zhang, J.; Niu, J.; Chen, L.; Zhang, F.; Zhao, D. De novo transcriptome assembly and quantification reveal differentially expressed genes between soft-seed and hard-seed pomegranate (Punica granatum L.). PLoS ONE 2017, 12, e0178809. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Li, H.; Cao, D.; Luo, X.; Yang, X.; Chen, L.; Liu, B.; Wang, Q.; Jing, D.; Cao, S. Characterization of a NAC transcription factor involved in the regulation of pomegranate seed hardness (Punica granatum L.). Plant Physiol. Biochem. 2019, 139, 379–388. [Google Scholar] [CrossRef]
- Hermisson, J.; Pennings, P.S. Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics 2005, 169, 2335–2352. [Google Scholar] [CrossRef] [Green Version]
- Messer, P.W.; Ellner, S.P.; Hairston, N.G. Can Population Genetics Adapt to Rapid Evolution? Trends Genet. 2016, 32, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Xie, H.; Lv, S.; Zheng, Y.; Yu, M.; Shen, L.; Sheng, J. LeMAPK4 participated in cold-induced ethylene production in tomato fruit. J. Sci. Food Agric. 2013, 93, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Polesello, C.; Le Bourg, E. A mild cold stress that increases resistance to heat lowers FOXO translocation in Drosophila melanogaster. Biogerontology 2017, 18, 791–801. [Google Scholar] [CrossRef] [PubMed]
| Dabenzi | Taishanhong | Tunisia | |
|---|---|---|---|
| Sequencing platform | Illumina HiSeq 2000 | Illumina Hiseq 2500 | Pacific Biosciences (PacBio) Sequel platform SMART |
| K-mer | 356.98 | 336.00 | |
| Assembled genome size (Mb) by flow cytometry | 328.13 | 322.70 ± 9.80 | 313.18 |
| Assembly length (Mb) | 328.38 | 274.00 | 320.31 |
| Number of chromosomes (2n) | 18 | 18 | 16 |
| Number of scaffolds | 1111 (≥2 kb) | 2117 (≥1 kb) | 473 |
| Scaffold N50 length (Mb) | 1.89 | 1.70 | 39.96 |
| Longest scaffold (Mb) | - | 7.60 | 55.56 |
| Total size of assembled contigs (Mb) | - | 269.00 | - |
| Number of contigs | - | 7088 (≥1 kb) | 661 |
| Contig N50 length | 66.97 kb | 97.00 kb | 4.49 Mb |
| Longest contig | - | 528.60 kb | 14.77 Mb |
| GC content (%) | 39.40 | 39.20 | 40.38 |
| Percentage of assembly (%) | 94.32 | 94.30 | 97.76 |
| Predicated number of gene models | 29,229 | 30,903 | 33,594 |
| Average gene length (bp) | 2574.61 | 2332.8 | 2229 |
| Average CDS length (bp) | 1077.85 | 1110.40 | 1048.00 |
| Average exon number per gene | 4.31 | 4.52 | - |
| Average exon length (bp) | - | 245.90 | 263.00 |
| Average intron length (bp) | - | 347.60 | - |
| Percentage of contigs anchored on chromosome (%) | 70.32 | - | - |
| Percentage of genes anchored on chromosome (%) | 84.62 | - | 97.76 |
| Pertence of repetitive sequence (%) | 46.10 | 51.20 | 50.93 |
| Percentage of TE to repetitive sequence (%) | 92.62 | 82.10 | 51.80 |
| Percentage of retrotransposons (%) | 40.50 | 35.32 | 24.05 |
| Percentage of DNA transposons (%) | - | 6.35 | 2.33 |
| LTR rate (%) | 17.06 | 17.40 | 24.59 |
| Reference | [30] | [29] | [31] |
| Gene | Species | Function | References |
|---|---|---|---|
| MYB | Pomegranate | PgMYB regulated the accumulation of anthocyanin during reproductive stages. | [82] |
| MYB1 | Apple | MdMYB1 regulated genes coordinately in the anthocyanin pathway response to light in apple skin. | [83,84] |
| MYB3 | Apple | MdMYB3 regulated the anthocyanin accumulation in apple skin. | [80] |
| MYB10 | Apple | MdMYB10 was the key gene that synthesized anthocyanin in red apple fruit. | [81] |
| MYB110a | Apple | MdMYB110a could up-regulate anthocyanin biosynthesis in apple. | [85] |
| MYBA | Apple | MdMYBA was a crucial regulator gene in anthocyanin accumulation in red-peel apple induced by low temperature or UV-B irradiation. | [86] |
| MYB10 | Pear | PyMYB10 promoted anthocyanin accumulation in fruit fresh and foliage induced by light. | [87] |
| MYB5a | Grape | VvMYB5a regulated structural genes expression controlling the phenylpropanoid synthesis, such as anthocyanins, flavonols, tannins. | [88] |
| MYB5b | Grape | VvMYB5b controlled anthocyanin and proanthocyanidin biosynthesis during grape berry development. | [89] |
| MYBA1 | Grape | VvMYBA1 could induce red pigmentation when introduced into white-peel grapes. | [79] |
| MYBA | Sweet cherry | PacMYBA one R2R3-MYB transcription factor from red-colored sweet cherry, played an important role in ABA-regulated anthocyanin biosynthesis. | [90] |
| MYB10.1 | Sweet cherry | PavMYB10.1 played a key role in regulating anthocyanin biosynthesis and determined skin color of sweet cherry. | [91] |
| Ruby | Blood orange | One MYB transcription factor CsRuby contributed to producing anthocyanin induced by cold. | [92] |
| bHLH3 | Peach | Overexpression of MYB10.1/bHLH3 and MYsB10.3/bHLH3 activated anthocyanin production. | [93] |
| bHLH3 | Apple | MdbHLH3 bound to MdMYB1 to regulate low temperature-induced accumulation. | [94] |
| bHLH33 | Strawberry | FvbHLH33, co-expressed with FvbHLH33, strongly activated structural genes in the anthocyanin pathway. | [95] |
| WD40 | Pomegranate | PgWD40 with PgAn1 (bHLH) and PgAn2 (MYB) co-regulated the downstream structural gene expression involved in the anthocyanin synthesis. | [96] |
| Gene | Function | Reference |
|---|---|---|
| PgSND1-like | The overexpression of the NAC transcription factor PgSND1-like enhanced lignin concentration in transgenic plants compared with wild-type Arabidopsis. | [131] |
| SUC6 | SUC6, one sucrose transport protein, which was more highly expressed at 60 days after flowering than 120 days after flowering in ‘Tunisia’ and ‘Sanbai’. | [126] |
| SUC8-like | SUC8-like was important for controlling seed development and was down-regulated significantly in soft-seeded pomegranate ‘Tunisia’ compared to hard-seeded pomegranate ‘Sanbai’. | [31,126] |
| PgL0044640 | These two genes were enriched in the FoxO signaling pathway indicated in the hard-seeded population by KEGG analysis. | [31,134,135] |
| PgL0314990 | [31,134,135] | |
| PgL0044700 | This gene was enriched in the MAPK signaling pathway in the hard-seeded population by KEGG analysis. | [31,134,135] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhao, Y.; Ren, Y.; Wang, Y.; Yuan, Z. Fruit Breeding in Regard to Color and Seed Hardness: A Genomic View from Pomegranate. Agronomy 2020, 10, 991. https://doi.org/10.3390/agronomy10070991
Zhang X, Zhao Y, Ren Y, Wang Y, Yuan Z. Fruit Breeding in Regard to Color and Seed Hardness: A Genomic View from Pomegranate. Agronomy. 2020; 10(7):991. https://doi.org/10.3390/agronomy10070991
Chicago/Turabian StyleZhang, Xinhui, Yujie Zhao, Yuan Ren, Yuying Wang, and Zhaohe Yuan. 2020. "Fruit Breeding in Regard to Color and Seed Hardness: A Genomic View from Pomegranate" Agronomy 10, no. 7: 991. https://doi.org/10.3390/agronomy10070991
APA StyleZhang, X., Zhao, Y., Ren, Y., Wang, Y., & Yuan, Z. (2020). Fruit Breeding in Regard to Color and Seed Hardness: A Genomic View from Pomegranate. Agronomy, 10(7), 991. https://doi.org/10.3390/agronomy10070991






