DiaMOS Plant: A Dataset for Diagnosis and Monitoring Plant Disease
Abstract
:1. Introduction
2. DiaMOS Plant Dataset
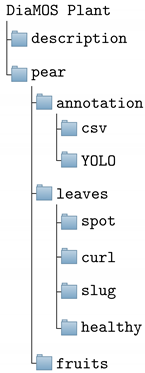
- Description contains the data description;
- Pear contains the data related to the pear tree;
- Annotation contains the annotation files;
- Leaves contains the leaf images;
- Fruits contains the fruit images.
3. Open Dataset for Plant Disease Classification and Detection
3.1. RoCoLe Dataset
3.2. BRACOL Dataset
3.3. Rice Leaf Disease Dataset
3.4. Plant Pathology Dataset
3.5. Citrus Dataset
3.6. APDA Dataset
3.7. PlantVillage Dataset
4. Comparative Analysis
4.1. Dataset Acquisistion
4.2. Symptoms and Diseases
4.3. Technical Dataset Settings
5. Discussion
- Dataset size: The greatest limitation of the current datasets is the small number of disease classes and the sample sizes. Even our proposed dataset, DiaMOS Plant, contains few samples for the “healthy” class. Inevitably, a strong imbalance of classes leads to the model not generalizing well in practical applications. This confirms and demonstrates, in agreement with Lu and Young [9], that although the need for larger datasets is recognized, this task is challenging due to the manual effort and cost required, which in some cases is further exacerbated as very few occurrences in the field can be found for some classes. This technical problem can be mitigated by data augmentation, transfer learning, and fine tuning techniques.
- Representativeness: The most widely adopted acquisition protocol is based on data collection under controlled, laboratory conditions. The representativeness of the dataset is limited by two factors: the place of acquisition and mode of acquisition. Controlled conditions are not able to reflect the spectrum of variability detectable in the field. Algorithms tend to achieve near-perfect accuracy when trained on laboratory datasets, but performance degrades significantly when trained on field datasets [5]. In addition, few datasets took into account the evolution of symptoms during an entire growing season. More efforts should focus on capturing symptoms at an early stage of emergency. In fact, at these stages, digital aids are essential to take timely action to stop the disease proliferation.
- Completeness: In Strong et al. [17], completeness is defined as “the level of breadth, depth, and appropriateness of a datum according to its purpose”. Although some datasets are well constructed, in some cases, we found a lack of completeness in providing ground truth labels. The annotation of multiple symptoms present in the leaf maximizes and completes the informative capacity of the data. Similarly, the presence of bounding-boxes and segmentation masks would extend usability.
- Performance baseline: The availability of a performance baseline can help in the development and validation of new methods that can be applied.
- Disease recognition mobile applications;
- Robotic applications that recognize and identify a disease and spray chemical or natural inputs based on the extent of the damage.
- Defer the angle, focus and position of the leaf in individual frames;
- Portray the disease for an entire growing season, identifying different levels of severity;
- Collect the samples at different times of the day—that is, with different climatic conditions (sunny, cloudy, direct light).
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ML | Machine Learning |
| DL | Deep Learning |
| DA | Digital Agriculture |
| ICT | Information and Communication Technologies |
References
- Fenu, G.; Malloci, F.M. Forecasting plant and crop disease: An explorative study on current algorithms. Big Data Cogn. Comput. 2021, 5, 2. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. The State of the World’s Land and Water Resources for Food and Agriculture: Managing Systems at Risk; Rome and Earthscan: London, UK, 2011. [Google Scholar]
- Fenu, G.; Malloci, F.M. An application of machine learning technique in forecasting crop disease. In Proceedings of the 2019 3rd International Conference on Big Data Research, Cergy-Pontoise, France, 20–22 November 2019; pp. 76–82. [Google Scholar] [CrossRef]
- Fenu, G.; Malloci, F.M. Artificial intelligence technique in crop disease forecasting: A case study on potato late blight prediction. In Proceedings of the International Conference on Intelligent Decision Technologies, Split, Croatia, 17–19 June 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 79–89. [Google Scholar] [CrossRef]
- Fenu, G.; Malloci, F.M. Using Multioutput Learning to Diagnose Plant Disease and Stress Severity. Complexity 2021, 2021, 6663442. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Barbedo, J.G.A. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Lu, Y.; Young, S. A survey of public datasets for computer vision tasks in precision agriculture. Comput. Electron. Agric. 2020, 178, 105760. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–29 June 2009; IEEE: Piscataway, NJ, USA, 2009. [Google Scholar] [CrossRef] [Green Version]
- Parraga-Alava, J.; Cusme, K.; Loor, A.; Santander, E. RoCoLe: A robusta coffee leaf images dataset for evaluation of machine learning based methods in plant diseases recognition. Data Brief 2019, 25, 104414. [Google Scholar] [CrossRef]
- Krohling, R.; Esgario, J.; Ventura, J.A. BRACOL—A Brazilian Arabica Coffee Leaf images dataset to identification and quantification of coffee diseases and pests. Mendeley Data 2019, V1. [Google Scholar] [CrossRef]
- Prajapati, H.B.; Shah, J.P.; Dabhi, V.K. Detection and classification of rice plant diseases. Intell. Decis. Technol. 2017, 11, 357–373. [Google Scholar] [CrossRef]
- Thapa, R.; Zhang, K.; Snavely, N.; Belongie, S.; Khan, A. The Plant Pathology Challenge 2020 data set to classify foliar disease of apples. Appl. Plant Sci. 2020, 8, e11390. [Google Scholar] [CrossRef] [PubMed]
- Rauf, H.T.; Saleem, B.A.; Lali, M.I.U.; Khan, M.A.; Sharif, M.; Bukhari, S.A.C. A citrus fruits and leaves dataset for detection and classification of citrus diseases through machine learning. Data Brief 2019, 26, 104340. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, A.; Khanum, A.; Khan, S.A.; Shaukat, A. Automated plant disease analysis (APDA): Performance comparison of machine learning techniques. In Proceedings of the 2013 11th International Conference on Frontiers of Information Technology, Islamabad, Pakistan, 16–18 December 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 60–65. [Google Scholar]
- Strong, D.M.; Lee, Y.W.; Wang, R.Y. Data quality in context. Commun. ACM 1997, 40, 103–110. [Google Scholar] [CrossRef]

| DiaMOS Plant Dataset | |
|---|---|
| Plant | Pear |
| Cultivar | Septoria Piricola |
| Data Source Location | Sardegna, Italy |
| Type of data | RGB Images |
| Annotation | csv, YOLO |
| ROI (Region of Interest) captured | leaf, fruit |
| Total size | 3505 images (3006 leaves images + 499 fruit images) |
| Data Accessibility | https://doi.org/10.5281/zenodo.5557313 accessed on 17 October 2021 |
| Application | The images are suitable for different machine and deep learning tasks such as images detection and classification. |
| Leaves Images | Leaf Symptoms | Size |
|---|---|---|
| Healthy | 43 | |
| Spot | 884 | |
| Curl | 54 | |
| Slug | 2025 | |
| Severity Levels | Size | |
| 0 | 43 | |
| 1 | 682 | |
| 2 | 1139 | |
| 3 | 699 | |
| 4 | 389 |
| Smartphone Camera | DSRL Camera | |
|---|---|---|
| Image size | 2976 × 3968 | 3456 × 5184 |
| Model device | Honor 6× | Canon EOS 60D |
| Focal length | 3.83 mm | 50 mm |
| Focal ratio | f/2.2 | f/4.5 |
| Color space | RGB | RGB |
| RMSprop (a) | |||
| CNN | Train Acc (%) | Validation Acc (%) | Test Acc (%) |
| EfficientNetB0 | 81.13 | 82.82 | 83.38 |
| InceptionV3 | 81.96 | 79.66 | 82.72 |
| MobileNetV | 85.38 | 81.12 | 83.06 |
| ResNet50 | 68.49 | 67.16 | 68.44 |
| VGG19 | 72.42 | 71.68 | 73.75 |
| Adam (b) | |||
| CNN | Train Acc(%) | Validation Acc (%) | Test Acc (%) |
| EfficientNetB0 | 89.02 | 86.33 | 86.05 |
| InceptionV3 | 84.44 | 80.29 | 83.39 |
| MobileNetV2 | 87.70 | 83.83 | 84.05 |
| ResNet50 | 68.38 | 68.47 | 69.10 |
| VGG19 | 76.66 | 76.53 | 75.75 |
| RMSprop (a) | |||
| CNN | Precision (%) | Recall (%) | F1-Score (%) |
| EfficientNetB0 | 81.14 | 83.38 | 82.23 |
| InceptionV3 | 80.21 | 82.72 | 81.45 |
| MobileNetV2 | 81.35 | 83.05 | 82.07 |
| ResNet50 | 68.27 | 68.43 | 56.67 |
| VGG19 | 70.47 | 73.75 | 71.76 |
| Adam (b) | |||
| CNN | Precision (%) | Recall (%) | F1-Score (%) |
| EfficientNetB0 | 84.42 | 86.04 | 85.03 |
| InceptionV3 | 81.14 | 83.38 | 82.23 |
| MobileNetV2 | 82.37 | 84.05 | 83.06 |
| ResNet50 | 66.38 | 69.10 | 59.51 |
| VGG19 | 72.71 | 75.74 | 74.05 |
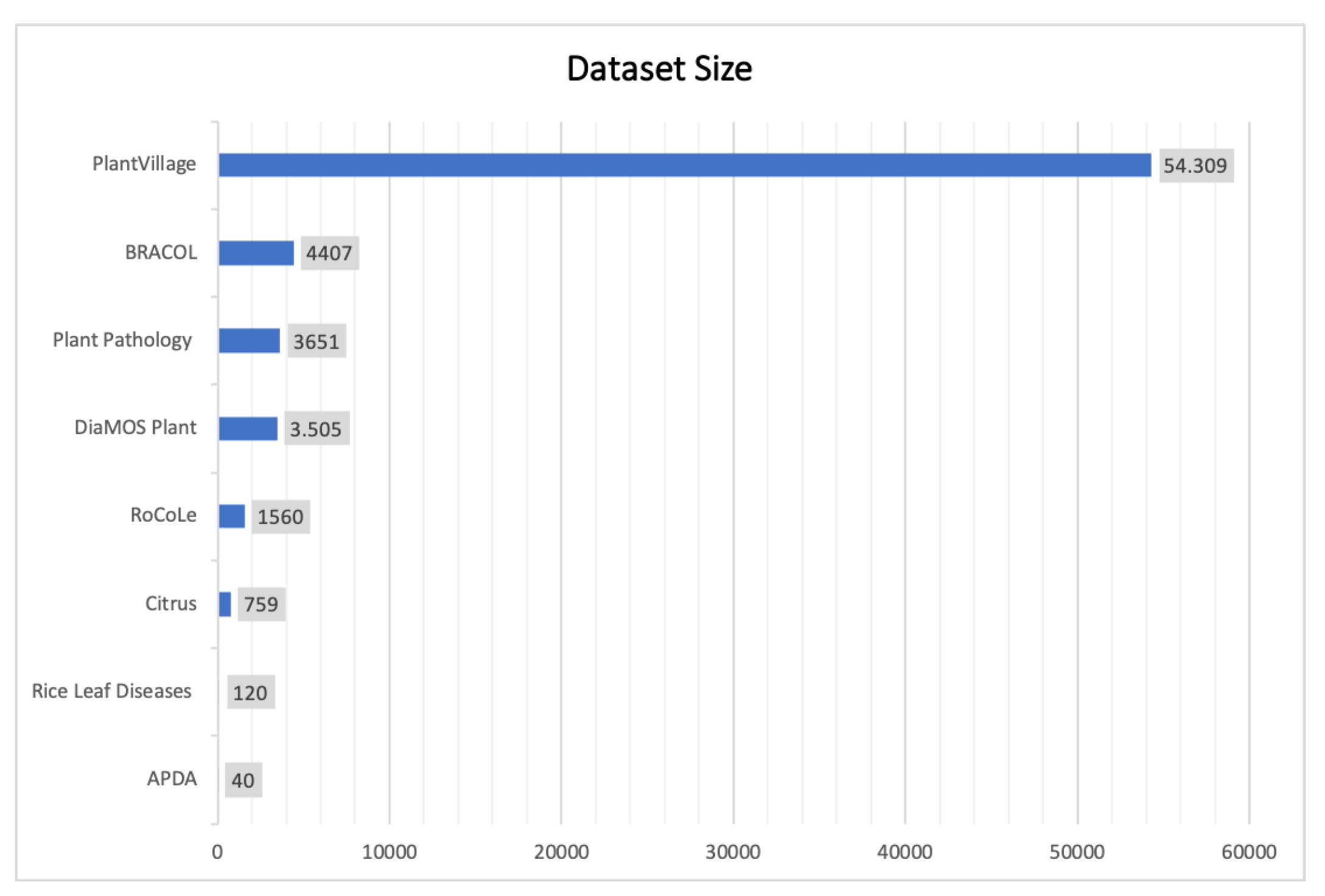
| Dataset | DiaMOSPlant [5] | BRACOL [12] | RoCoLe [11] | Plant Pathology [14] | Rice Leaf Diseases [13] | Citrus [15] | APDA [16] | PlantVillage [7] |
|---|---|---|---|---|---|---|---|---|
| Plant/Crop | Pear | Coffee | Coffee | Apple | Rice | Citrus | Rose | Multiple |
| Dataset size | 3505 (3006 leaf images + 499 fruit images) | 4407 | 1560 | 3651 | 120 | 759 (609 leaf images + 150 fruit images) | 40 | 54.309 |
| No. of symptoms | 4 | 4 | 2 | 3 | 3 | 5 | 2 | 26 |
| Acquisition device | Smarthphone e DSRL | Smartphone | Smartphone | DSLR Camera, Smartphone | DSLR camera | DSLR camera | Smartphone | Smartphone |
| Color | RGB | RGB | RGB | RGB | RGB | RGB | RGB | RGB |
| Image resolution | Multiple | 2048 × 1024 | Multiple | 2048 × 1365 | 2848 × 4288 | 256 × 256 | N.d. | Multiple |
| Annotation | Polygon, Label | Polygon, Label | Polygon, Label | Label | Label | Label | Label | Label |
| Annotation format | csv, YOLO | csv | csv, COCO, JSON, Pascal VOC | csv | Folder structure | Folder structure | N.d. | Folder structure |
| Data sharing platform | Zenodo | GitHub | Mendeley Data | Kaggle | UCI Machine Learning Repository | Mendeley Data | MathWorks | Github |
| Acquisition place | Field | Laboratory | Field | Field | Laboratory | Laboratory | Laboratory | Laboratory |
| Side of the leaf | Adaxial | Abaxial | Adaxial, Abaxial | Adaxial | Adaxial | Adaxial | Adaxial | Adaxial |
| Object of interest | Fruit, leaf | Leaf | Leaf | Leaf | Leaf | Fruit, leaf | Leaf | Leaf |
| Dataset | Online Repository |
|---|---|
| DiaMOSPlant | https://doi.org/10.5281/zenodo.5557313 |
| BRACOL [12] | https://data.mendeley.com/datasets/yy2k5y8mxg/1 |
| RoCoLe [11] | https://data.mendeley.com/datasets/c5yvn32dzg/2 |
| Plant Pathology [14] | https://www.kaggle.com/c/plant-pathology-2020-fgvc7 |
| Rice Leaf Diseases [13] | https://archive.ics.uci.edu/ml/datasets/Rice+Leaf+Diseases |
| Citrus [15] | https://data.mendeley.com/datasets/3f83gxmv57/2 |
| APDA [16] | https://it.mathworks.com/matlabcentral/fileexchange/55098 |
| PlantVillage [7] | https://github.com/spMohanty/PlantVillage-Dataset |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fenu, G.; Malloci, F.M. DiaMOS Plant: A Dataset for Diagnosis and Monitoring Plant Disease. Agronomy 2021, 11, 2107. https://doi.org/10.3390/agronomy11112107
Fenu G, Malloci FM. DiaMOS Plant: A Dataset for Diagnosis and Monitoring Plant Disease. Agronomy. 2021; 11(11):2107. https://doi.org/10.3390/agronomy11112107
Chicago/Turabian StyleFenu, Gianni, and Francesca Maridina Malloci. 2021. "DiaMOS Plant: A Dataset for Diagnosis and Monitoring Plant Disease" Agronomy 11, no. 11: 2107. https://doi.org/10.3390/agronomy11112107
APA StyleFenu, G., & Malloci, F. M. (2021). DiaMOS Plant: A Dataset for Diagnosis and Monitoring Plant Disease. Agronomy, 11(11), 2107. https://doi.org/10.3390/agronomy11112107







