The FBA Motif-Containing Protein NpFBA1 Causes Leaf Curling and Reduces Resistance to Black Shank Disease in Tobacco
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Culture of Pathogens and Production of Spore Suspension
2.3. RNA Extraction and cDNA Synthesis
2.4. NpFBA1 Cloning and Sequence Analysis
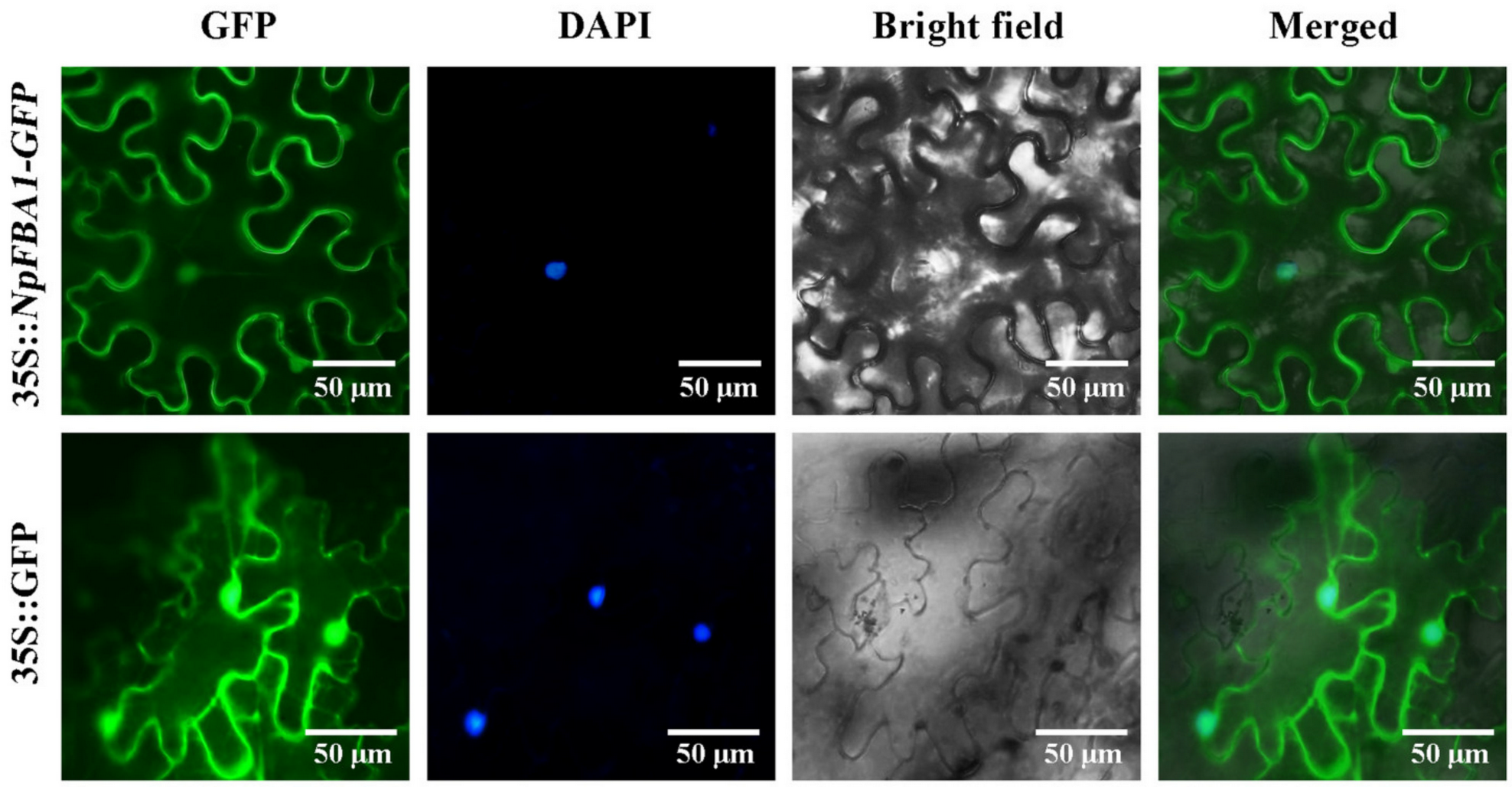
2.5. Subcellular Localization of NpFBA1 Proteins
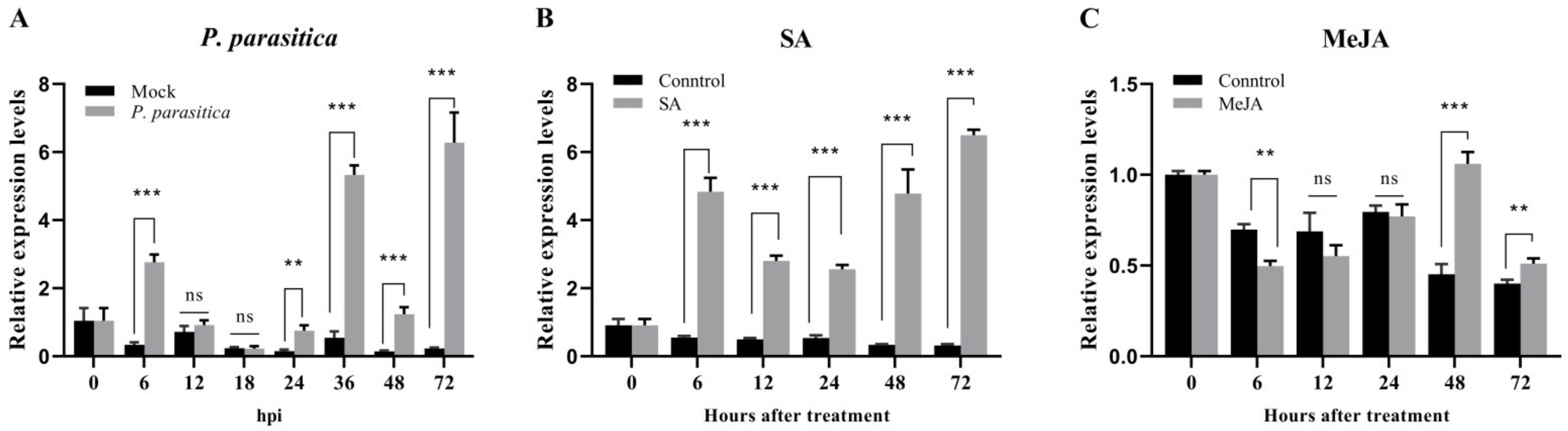
2.6. Hormone Treatment and Pathogen Infection
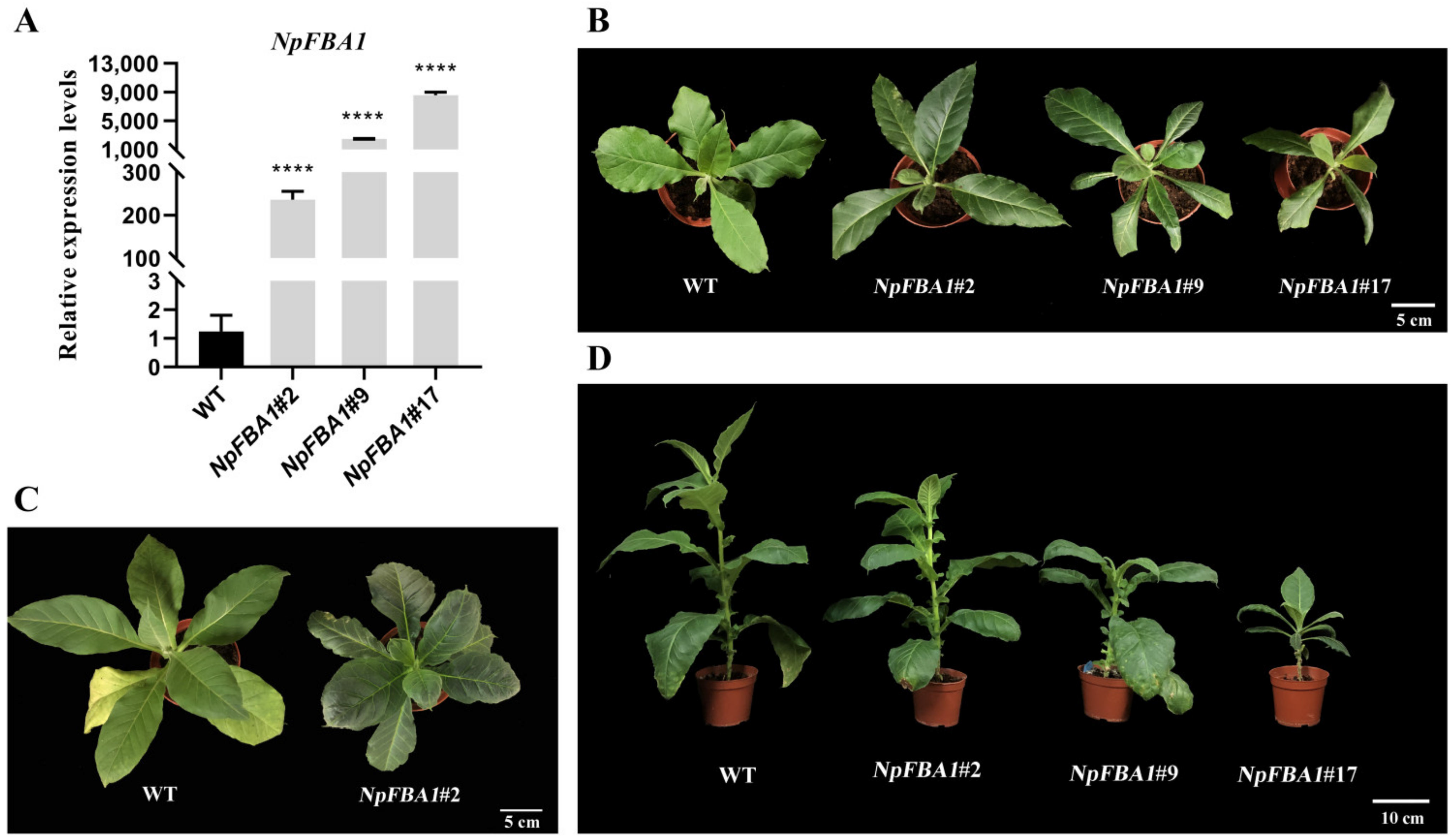
2.7. NpFBA1 Gene Expression Vector Constructs and the Acquisition of Transgenic Tobacco
2.8. DNA Extraction from Transgenic and Wild Tobacco
2.9. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
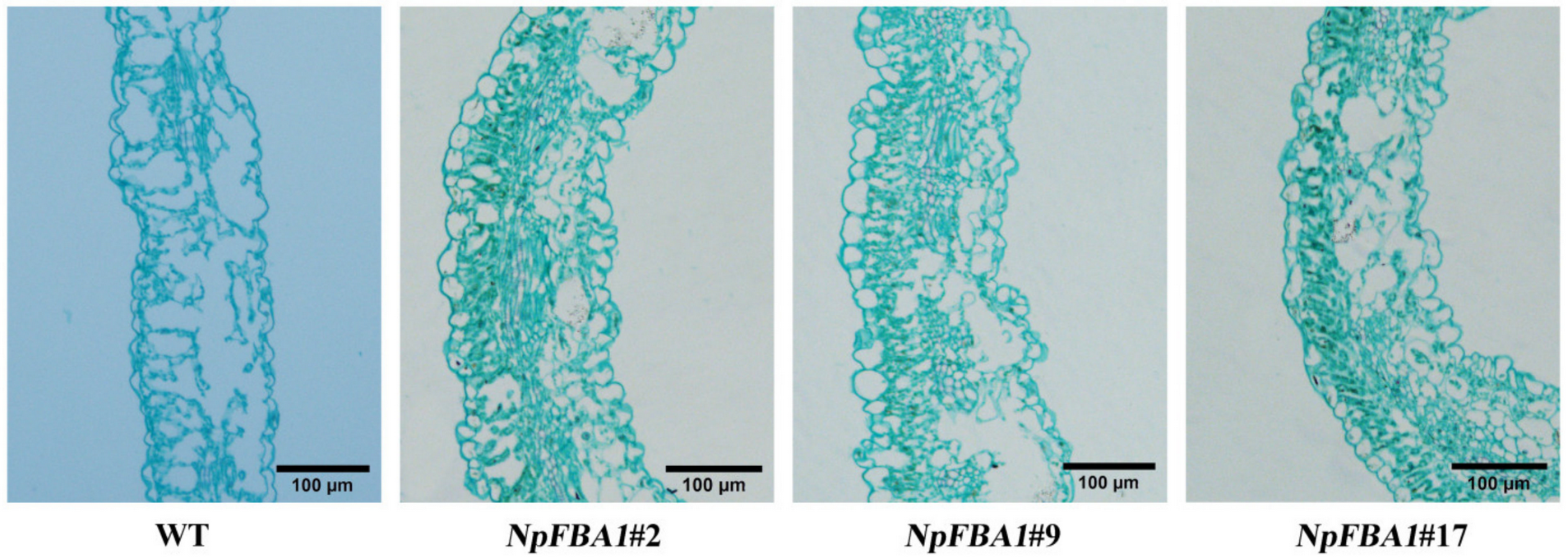
2.10. Leaf Anatomy Observation
2.11. Calculation of Leaf Damage Area after P. parasitica Infection
3. Results
3.1. Identification of the NpFBA1 Gene from Nicotiana plumbaginifolia
3.2. Subcellular Localization of NpFBA1
3.3. NpFBA1 Responds to Pathogen Infection and Exogenous Hormones
3.4. Functional Identification of the NpFBA1 Gene
3.4.1. Effects of the NpFBA1 Gene on Tobacco Growth and Development
3.4.2. Observation of the Leaf Anatomical Structure of WT and Overexpressed Plants
3.4.3. Pathogen Infection Status
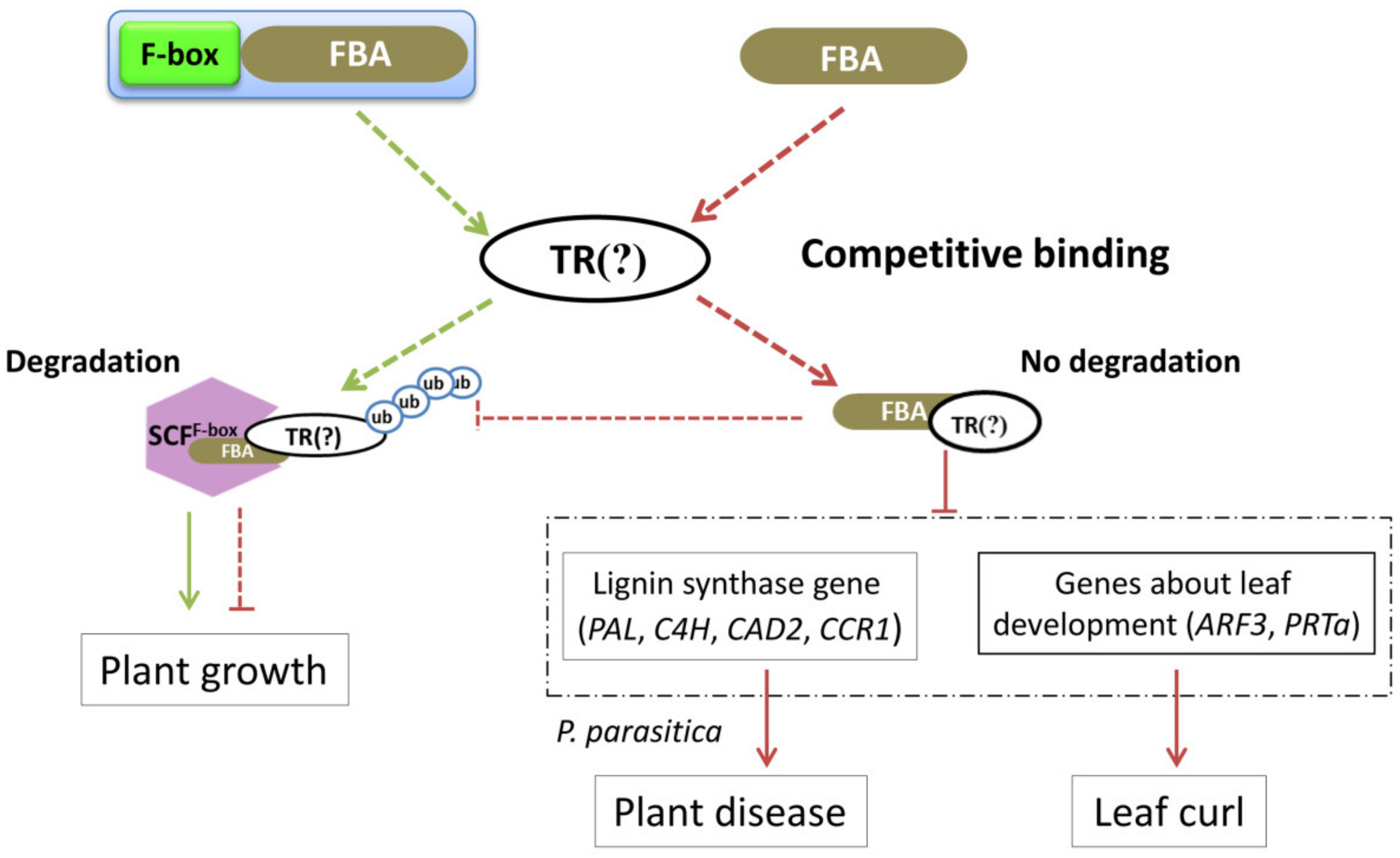
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Sen, P.; Hofmann, K.; Ma, L.; Goebl, M.; Harper, J.W.; Elledge, S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996, 86, 263–274. [Google Scholar] [CrossRef]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef]
- Kuroda, H.; Takahashi, N.; Shimada, H.; Seki, M.; Shinozaki, K.; Matsui, M. Classification and expression analysis of Arabidopsis F-Box-Containing protein genes. Plant Cell Physiol. 2002, 43, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Kipreos, E.T.; Pagano, M. The F-box protein family. Genome Biol. 2000, 1, REVIEWS3002. [Google Scholar] [CrossRef] [PubMed]
- Schumann, N.; Navarro-Quezada, A.; Ullrich, K.; Kuhl, C.; Quint, M. Molecular evolution and selection patterns of plant F-box proteins with C-terminal kelch repeats. Plant Physiol. 2011, 155, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Hauvermale, A.L.; Nelson, S.K.; Hanada, A.; Yamaguchi, S.; Steber, C.M. Lifting della repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling. Plant Physiol. 2013, 162, 2125–2139. [Google Scholar] [CrossRef]
- Wen, C.K.; Chang, C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 2002, 14, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Tyler, L.; Thomas, S.G.; Hu, J.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef]
- Fu, X.; Richards, D.E.; Fleck, B.; Xie, D.; Burton, N.; Harberd, N.P. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 2004, 16, 1406–1418. [Google Scholar] [CrossRef]
- Gomi, K.; Sasaki, A.; Itoh, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Kitano, H.; Matsuoka, M. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 2004, 37, 626–634. [Google Scholar] [CrossRef]
- Xing, L.; Li, Z.; Khalil, R.; Ren, Z.; Yang, Y. Functional identification of a novel F-box/FBA gene in tomato. Physiol. Plant. 2012, 144, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genom. Hum. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Zhao, Y.; Ma, Z.; Shi, G.; Yang, Y.; Pichersky, E.; Chen, H.; Liu, M.; Chen, Z.; et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005, 17, 2693–2704. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Gene Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Song, J.B.; Huang, S.Q.; Dalmay, T.; Yang, Z.M. Regulation of leaf morphology by MicroRNA394 and its target LEAF CURLING RESPONSIVENESS. Plant Cell Physiol. 2012, 53, 1283–1294. [Google Scholar] [CrossRef]
- Hu, Z.; Keceli, M.A.; Piisilä, M.; Li, J.; Survila, M.; Heino, P.; Brader, G.; Palva, E.T.; Li, J. F-box protein AFB4 plays a crucial role in plant growth, development and innate immunity. Cell Res. 2012, 22, 777–781. [Google Scholar] [CrossRef]
- Han, B.; Chen, L.; Wang, J.; Wu, Z.; Yan, L.; Hou, S. Constitutive expresser of pathogenesis related genes 1 Is required for pavement cell morphogenesis in Arabidopsis. PLoS ONE 2015, 10, e0133249. [Google Scholar] [CrossRef]
- Qiao, H.; Chang, K.N.; Yazaki, J.; Ecker, J.R. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009, 23, 512–521. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Cui, M.H.; Noh, M.S.; Jung, K.W.; Shin, J.S. The FBA motif-containing protein AFBA1 acts as a novel positive regulator of ABA response in Arabidopsis. Plant Cell Physiol. 2017, 58, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Gou, M.; Su, N.; Zheng, J.; Huai, J.; Wu, G.; Zhao, J.; He, J.; Tang, D.; Yang, S.; Wang, G. An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 2009, 60, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Delaney, T.P. Arabidopsis SON1 is an F-Box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 2002, 14, 1469–1482. [Google Scholar] [CrossRef] [PubMed]
- Gou, M.; Shi, Z.; Zhu, Y.; Bao, Z.; Wang, G.; Hua, J. The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 2012, 69, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Grappin, P.; Bouinot, D.; Sotta, B.; Miginiac, E.; Jullien, M. Control of seed dormancy in Nicotiana plumbaginifolia: Post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta 2000, 210, 279–285. [Google Scholar] [CrossRef]
- Dang, J.; Wang, J.; Yang, Y.; Shang, W.; Guo, Q.; Liang, G. Resistance of Nicotiana tabacum to Phytophthora parasitica var. nicotianae race 0 is enhanced by the addition of N. plumbaginifolia chromosome 9 with a slight effect on host genomic expression. Crop Sci. 2019, 59, 2667–2678. [Google Scholar] [CrossRef]
- Hoekema, A.; Hirsch, P.R.; Hooykaas, P.J.J.; Schilperoort, R.A. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 1983, 303, 179–180. [Google Scholar] [CrossRef]
- Wen, G.; Dang, J.; Xie, Z.; Wang, J.; Jiang, P.; Guo, Q.; Liang, G. Molecular karyotypes of loquat (Eriobotrya japonica) aneuploids can be detected by using SSR markers combined with quantitative PCR irrespective of heterozygosity. Plant Methods 2020, 16, 22. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef]
- Dang, J.; Wu, T.; Liang, G.; Wu, D.; He, Q.; Guo, Q. Identification and characterization of a loquat aneuploid with novel leaf phenotypes. HortScience 2019, 54, 1–5. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Q.; Tang, Y.; Ding, W. NtPR1a regulates resistance to Ralstonia solanacearum in Nicotiana tabacum via activating the defense-related genes. Biochem. Biophys. Res. Commun. 2019, 508, 940–945. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Kuang, J.; Wang, F.; Chen, L.; Hong, K.; Xiao, Y.; Xie, H.; Lu, W.; Chen, J. Molecular characterization of PR and WRKY genes during SA- and MeJA-induced resistance against Colletotrichum musae in banana fruit. Postharv. Biol. Technol. 2013, 79, 62–68. [Google Scholar] [CrossRef]
- Li, Q.; Wang, S.; Liu, G.; Zhou, P.; Zheng, X.; Ceng, W.; Chen, Z.; Chen, A. Response of tobacco leaf shape index and auxin to low temperature stress and growth recovery. Jiangsu Agric. Sci. 2019, 47, 60–65. [Google Scholar] [CrossRef]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Kelley, D.R.; Arreola, A.; Gallagher, T.L.; Gasser, C.S. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 2012, 139, 1105–1109. [Google Scholar] [CrossRef]
- Sonoda, Y.; Sako, K.; Maki, Y.; Yamazaki, N.; Yamamoto, H.; Ikeda, A.; Yamaguchi, J. Regulation of leaf organ size by the Arabidopsis RPT2a 19S proteasome subunit. Plant J. 2009, 60, 68–78. [Google Scholar] [CrossRef]
- Fu, X.; Richards, D.E.; Ait-ali, T.; Hynes, L.W.; Ougham, H.; Peng, J.; Harberd, N.P. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 2002, 14, 3191–3200. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, J.M.; Chapple, C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002, 5, 224–229. [Google Scholar] [CrossRef]
- Li, G.; Zhang, K.; Liu, F.; Liu, D.; Wan, Y. Morphological and physiological traits of leaf in different drought resistant peanut cultivars. Sci. Agric. Sin. 2014, 47, 644–654. [Google Scholar] [CrossRef]
- Feng, X.; Li, X.; Yang, X.; Zhu, P. Fine mapping and identification of the leaf shape gene BoFL in ornamental kale. Theor. Appl. Genet. 2020, 133, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Cheng, S.; Dou, Y.; Xie, Y.; Liu, G.; Liu, X.; Du, Y. Relationship between leaf shape and inner quality of Flue-cured Tobacco in Xuanwei County of Yunnan Province. Chin. Tob. Sci. 2011, 32, 6–13. [Google Scholar] [CrossRef]
- Robles, P.; Fleury, D.; Candela, H.; Cnops, G.; Alonso-Peral, M.M.; Anami, S.; Falcone, A.; Caldana, C.; Willmitzer, L.; Ponce, M.R.; et al. The RON1/FRY1/SAL1 gene is required for leaf morphogenesis and venation patterning in Arabidopsis. Plant Physiol. 2010, 152, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dan, Z.; Gao, F.; Chen, P.; Fan, F.; Li, S. Rice GROWTH-REGULATING FACTOR7 modulates plant architecture through regulating GA and Indole-3-Acetic acid metabolism. Plant Physiol. 2020, 184, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Pi, L.; Liang, W.; Xu, B.; Wang, H.; Cai, R.; Huang, H. The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 2006, 18, 2479–2492. [Google Scholar] [CrossRef][Green Version]
- Zhang, Z.; Runions, A.; Mentink, R.A.; Kierzkowski, D.; Karady, M.; Hashemi, B.; Huijser, P.; Strauss, S.; Gan, X.; Ljung, K.; et al. A WOX/Auxin biosynthesis module controls growth to shape leaf form. Curr. Biol. 2020, 30, 4857–4868.e6. [Google Scholar] [CrossRef]
- Paquis, S.; Mazeyrat-Gourbeyre, F.; Fernandez, O.; Crouzet, J.; Clément, C.; Baillieul, F.; Dorey, S. Characterization of a F-box gene up-regulated by phytohormones and upon biotic and abiotic stresses in grapevine. Mol. Biol. Rep. 2011, 38, 3327–3337. [Google Scholar] [CrossRef]
- Meng, Y.; Wei, C.; Fan, R.; Yu, X.; Wang, X.; Zhao, W.; Wei, X.; Kang, Z.; Liu, D. TaPP2-A13 gene shows induced expression pattern in wheat responses to stresses and interacts with adaptor protein SKP1 from SCF complex. Acta Agron. Sin. 2021, 47, 224–236. [Google Scholar] [CrossRef]
- Wei, C.; Meng, Y.; Fan, R.; Zhao, M.; Yu, X.; Zhao, W.; Kang, Z.; Liu, D. Stress-related expression profile of F-box/Kelch gene TaFKOR23 in wheat and molecular characterization of the interacting target protein. J. Plant Genet. Resour. 2020, 21, 695–705. [Google Scholar] [CrossRef]
- Holle, S.V.; Smagghe, G.; Damme, E.J.M.V. Overexpression of Nictaba-Like Lectin genes from Glycine max confers tolerance toward Pseudomonas syringae infection, aphid infestation and salt stress in transgenic Arabidopsis plants. Front Plant Sci. 2016, 7, 1590. [Google Scholar] [CrossRef]
- Hou, J.; Zhao, F.; Yang, X.; Li, W.; Xie, D.; Tang, Z.; Lv, S.; Nie, L.; Sun, Y.; Wang, M.; et al. Lignin synthesis related genes with potential significance in the response of upland cotton to fusarium wilt identified by transcriptome profiling. Trop. Plant Biol. 2021, 14, 106–119. [Google Scholar] [CrossRef]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-Box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef] [PubMed]
- Grones, P.; Friml, J. Auxin transporters and binding proteins at a glance. J. Cell Sci. 2015, 128, 1–7. [Google Scholar] [CrossRef] [PubMed]




| Materials | Leaf Length (cm) | Leaf Width (cm) | Leaf Shape Index (Length/Width) |
|---|---|---|---|
| WT | 19.33 ± 2.35 | 6.90 ± 0.63 | 2.81 ± 0.03 |
| NpFBA1#2 | 16.00 ± 1.75 * | 8.10 ± 0.58 | 2.02 ± 0.01 * |
| NpFBA1#9 | 17.80 ± 0.28 | 7.90 ± 0.13 | 2.29 ± 0.02 * |
| NpFBA1#17 | 17.16 ± 0.58 | 8.00 ± 0.25 | 2.15 ± 0.01 * |
| Materials | Leaf Thickness (µm) | Spongy Tissue Thickness (µm) | Upper Epidermis Thickness (µm) | CTR% | Number of Cells per 1 mm Palisade Tissue |
|---|---|---|---|---|---|
| WT | 177.25 ± 5.27 | 89.09 ± 1.67 | 10.30 ± 1.78 | 33.87 ± 3.03 | 44.33 ± 5.13 |
| NpFBA1#2 | 210.17 ± 18.25 * | 142.66 ± 10.64 ** | 25.47 ± 2.79 ** | 24.10 ± 4.81 * | 57.00 ± 1.73 ** |
| NpFBA1#9 | 210.83 ± 10.57 ** | 97.55 ± 3.21 * | 24.14 ± 1.80 ** | 22.72 ± 4.99 * | 54.67 ± 2.08 * |
| NpFBA1#17 | 193.16 ± 4.05 * | 128.96 ± 8.61 ** | 22.64 ± 2.95 ** | 19.60 ± 0.67 ** | 70.33 ± 4.16 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Z.; Wen, G.; Yang, Y.; Wang, H.; Wang, J.; Lei, C.; Guo, Q.; Dang, J.; Liang, G. The FBA Motif-Containing Protein NpFBA1 Causes Leaf Curling and Reduces Resistance to Black Shank Disease in Tobacco. Agronomy 2021, 11, 2478. https://doi.org/10.3390/agronomy11122478
Xie Z, Wen G, Yang Y, Wang H, Wang J, Lei C, Guo Q, Dang J, Liang G. The FBA Motif-Containing Protein NpFBA1 Causes Leaf Curling and Reduces Resistance to Black Shank Disease in Tobacco. Agronomy. 2021; 11(12):2478. https://doi.org/10.3390/agronomy11122478
Chicago/Turabian StyleXie, Zhongyi, Guo Wen, Yao Yang, Haiyan Wang, Jinying Wang, Chenggong Lei, Qigao Guo, Jiangbo Dang, and Guolu Liang. 2021. "The FBA Motif-Containing Protein NpFBA1 Causes Leaf Curling and Reduces Resistance to Black Shank Disease in Tobacco" Agronomy 11, no. 12: 2478. https://doi.org/10.3390/agronomy11122478
APA StyleXie, Z., Wen, G., Yang, Y., Wang, H., Wang, J., Lei, C., Guo, Q., Dang, J., & Liang, G. (2021). The FBA Motif-Containing Protein NpFBA1 Causes Leaf Curling and Reduces Resistance to Black Shank Disease in Tobacco. Agronomy, 11(12), 2478. https://doi.org/10.3390/agronomy11122478






