Genome-Wide Analysis of Somatic Embryogenesis-Related Transcription Factors in Cultivated Strawberry (Fragaria × ananassa) and Evolutionary Relationships among Rosaceae Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Somatic Embryogenesis-Related Transcription Factors in Fragaria × ananassa and Other Rosaceae Species
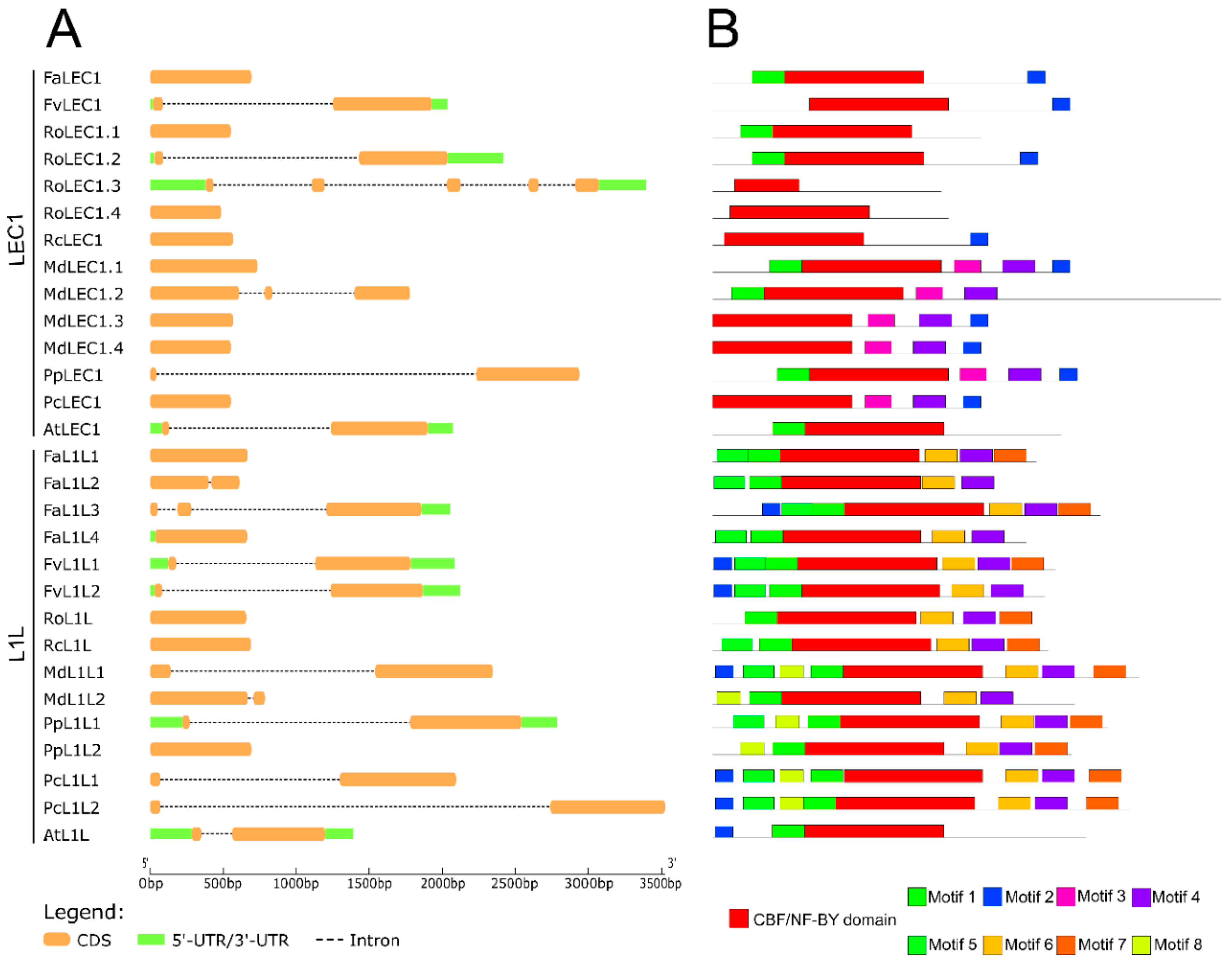
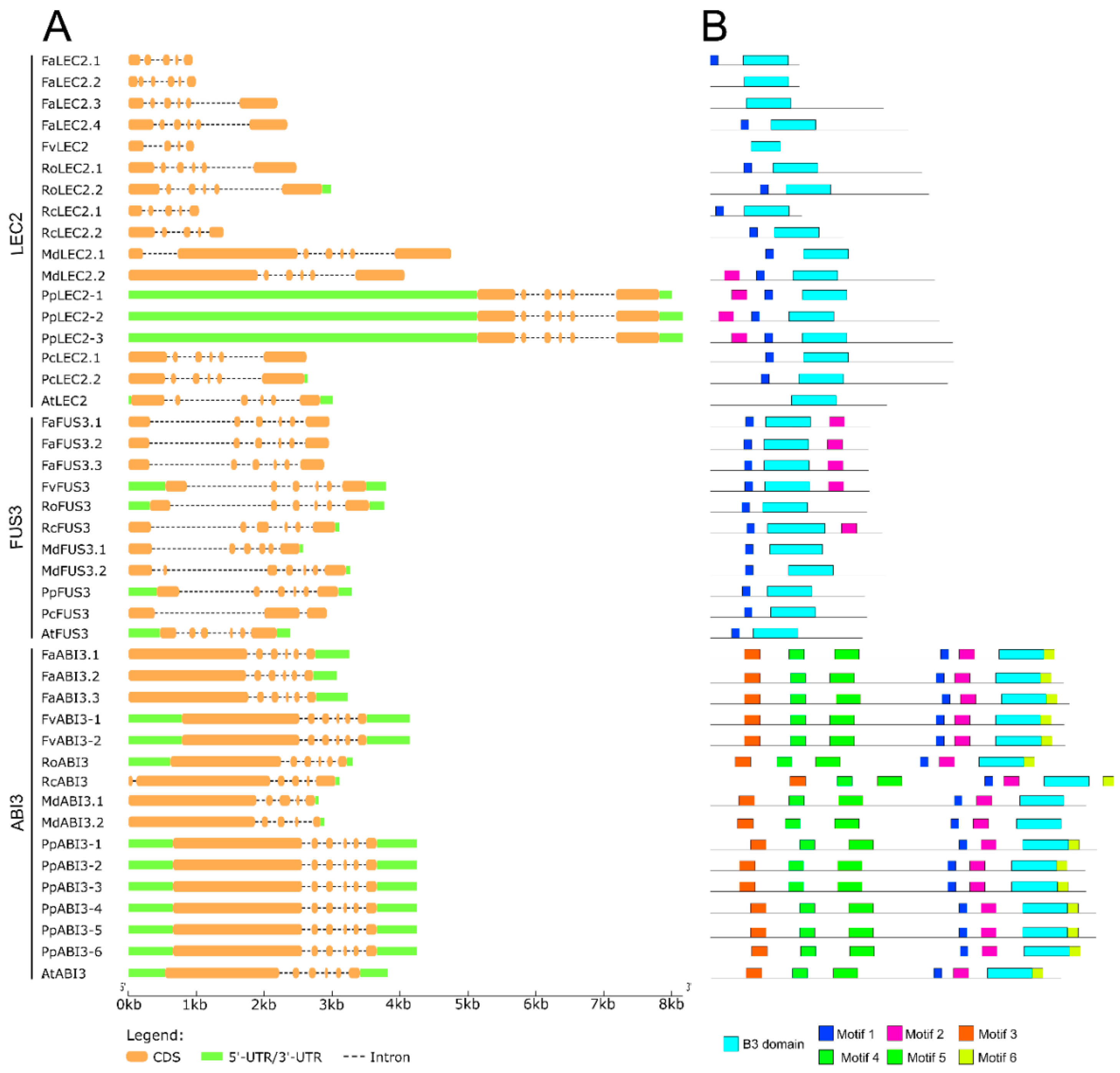
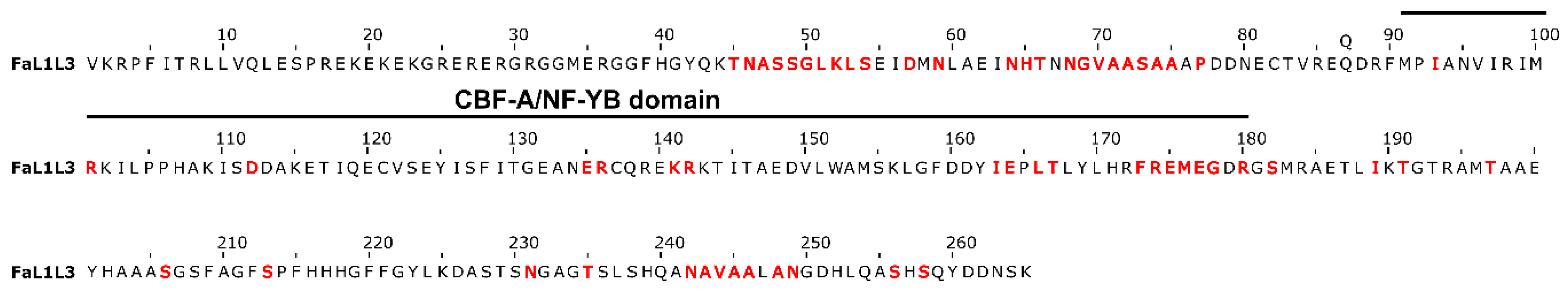
2.2. Characterization of Gene and Protein Structures for Somatic Embryogenesis-Related Transcription Factors in Fragaria × ananassa
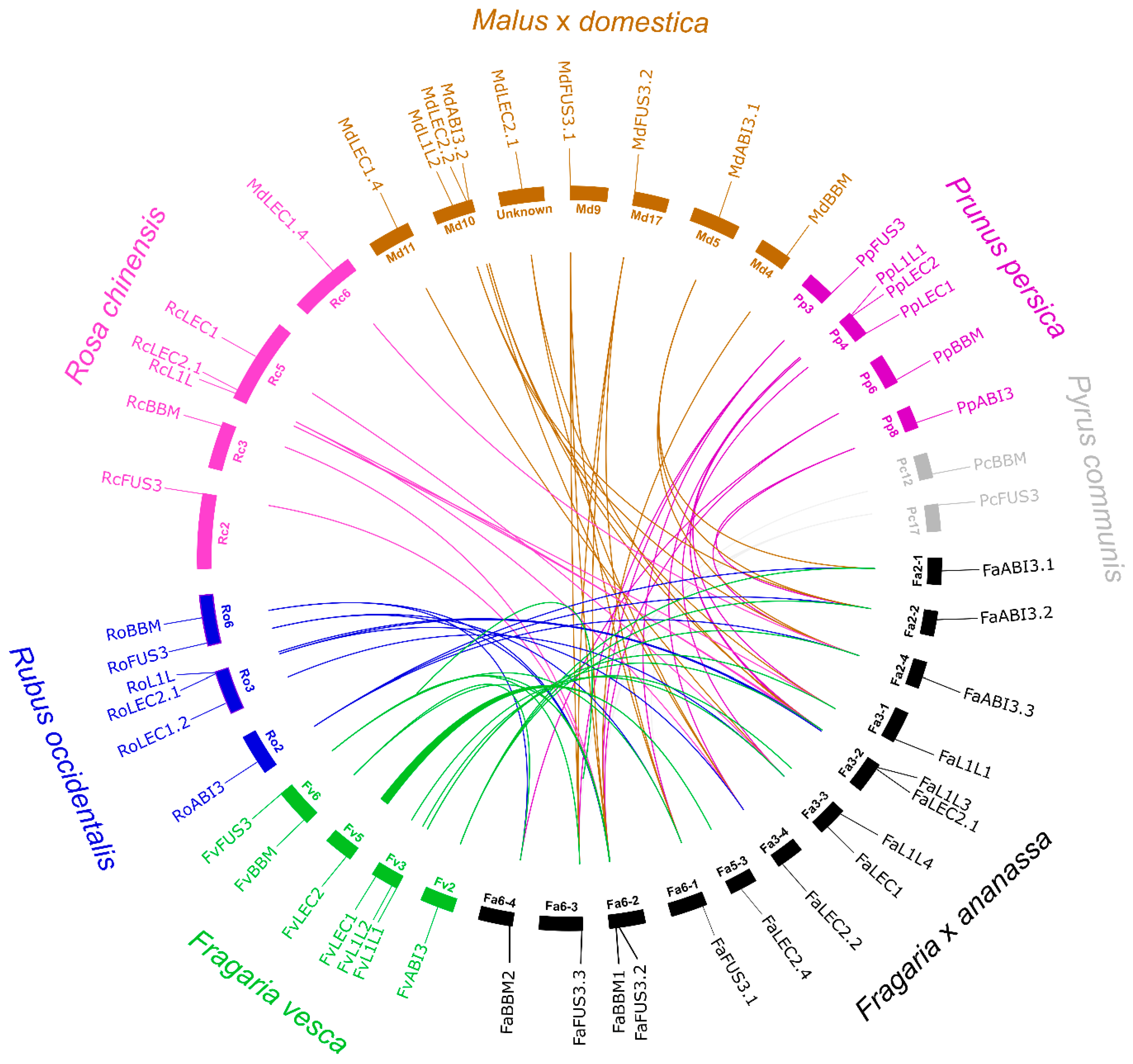
2.3. Synteny Analysis and Molecular Evolutionary Rates between Somatic Embryogenesis-Related Transcription Factors in Fragaria × ananassa and Other Rosaceae Species
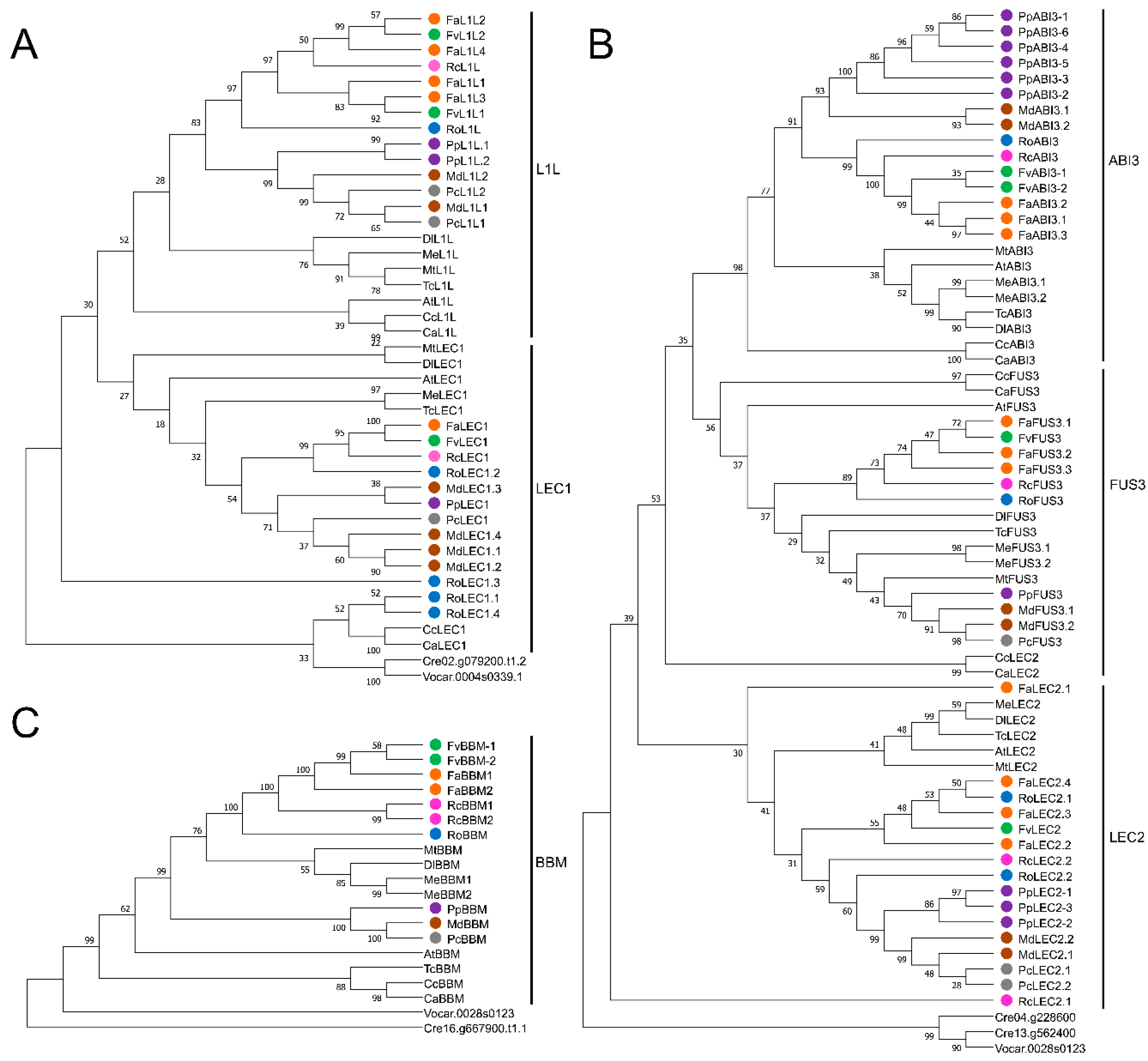
2.4. Phylogenetic Analyses between Somatic Embryogenesis-Related Transcription Factors in Fragaria × ananassa and Other Rosaceae Species
3. Results
3.1. Identification of LEC1, L1L, LEC2, FUS3, ABI3, and BBM Transcription Factors in Fragaria × ananassa and Six Other Rosaceae Species
3.2. Molecular Characterization of LEC1, L1L, LEC2, FUS3, ABI3, and BBM Transcription Factors in Fragaria × ananassa
3.3. Syntenic Relationships and Molecular Evolutionary Analysis of LEC1, L1L, LEC2, FUS3, ABI3, and BBM Genes in Fragaria × ananassa and Six Other Rosaceae Species
3.4. Phylogenetic Relationships of LEC1, L1L, LEC2, FUS3, ABI3, and BBM Transcription Factors in Fragaria × ananassa and Six Other Rosaceae Species
4. Discussion
4.1. Fragaria × ananassa Genome Contains a Variable Loci Number of SE-related TFs
4.2. Genes and Proteins of LAFL-B Network Are Conserved in Fragaria × ananassa and Other Rosaceae Species
4.3. LAFL-B Genes and Proteins of Rosoideae Evolved Independently of Amygdaloideae Subfamily
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahanger, M.A.; Akram, N.A.; Ashraf, M.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Plant Responses to Environmental Stresses—from Gene to Biotechnology. AoB Plants 2017, 9. [Google Scholar] [CrossRef]
- Radoeva, T.; Vaddepalli, P.; Zhang, Z.; Weijers, D. Evolution, Initiation, and Diversity in Early Plant Embryogenesis. Dev. Cell 2019, 50, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Bidabadi, S.S.; Jain, S.M. Cellular, Molecular, and Physiological Aspects of In Vitro Plant Regeneration. Plants 2020, 9, 702. [Google Scholar] [CrossRef] [PubMed]
- Fehér, A. Callus, Dedifferentiation, Totipotency, Somatic Embryogenesis: What These Terms Mean in the Era of Molecular Plant Biology? Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, T.; Dudits, D. Epigenetic Clues to Better Understanding of the Asexual Embryogenesis in Planta and in Vitro. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Fehér, A. Somatic Embryogenesis—Stress-Induced Remodeling of Plant Cell Fate. Biochimica et Biophysica Acta 2015. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.L.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 Is Sufficient to Induce Embryo Development in Vegetative Cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef]
- Stone, S.L.; Braybrook, S.A.; Paula, S.L.; Kwong, L.W.; Meuser, J.; Pelletier, J.; Hsieh, T.-F.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON2 Induces Maturation Traits and Auxin Activity: Implications for Somatic Embryogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3151–3156. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; van Lammeren, A.A.M.; Miki, B.L.A.; et al. Ectopic Expression of BABY BOOM Triggers a Conversion from Vegetative to Embryonic Growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.J. Somatic Embryogenesis in the Medicago Truncatula Model: Cellular and Molecular Mechanisms. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Nic-Can, G.I.; López-Torres, A.; Barredo-Pool, F.; Wrobel, K.; Loyola-Vargas, V.M.; Rojas-Herrera, R.; De-la-Peña, C. New Insights into Somatic Embryogenesis: LEAFY COTYLEDON1, BABY BOOM1 and WUSCHEL-RELATED HOMEOBOX4 Are Epigenetically Regulated in Coffea Canephora. PLoS ONE 2013, 8, e72160. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, B.; Meisel, L.; Gebauer, M.; Garcia-Gonzales, R.; Silva, H. Breeding in Peach, Cherry and Plum: From a Tissue Culture, Genetic, Transcriptomic and Genomic Perspective. Biol. Res. 2013, 46, 219–230. [Google Scholar] [CrossRef]
- Gao, L.M.; Zhang, J.; Hou, Y.; Yao, Y.C.; Ji, Q.L. RNA-Seq Screening of Differentially-Expressed Genes during Somatic Embryogenesis in Fragaria × ananassa Duch. ‘Benihopp’. J. Hort. Sci. Biotech. 2015, 90, 671–681. [Google Scholar] [CrossRef]
- Biswas, M.K.; Islam, R.; Hossain, M. Somatic Embryogenesis in Strawberry (Fragaria Sp.) through Callus Culture. Plant Cell Tiss. Org. 2007, 90, 49–54. [Google Scholar] [CrossRef]
- Husaini, A.M.; Abdin, M.Z. Interactive Effect of Light, Temperature and TDZ on the Regeneration Potential of Leaf Discs of Fragaria × Ananassa Duch. In Vitro Cell. Dev. Biol. Plant 2007, 43, 576–584. [Google Scholar] [CrossRef]
- Gerdakaneh, M.; Mozafari, A.-A.; Sioseh-mardah, A.; Sarabi, B. Effects of Different Amino Acids on Somatic Embryogenesis of Strawberry (Fragaria × ananassa Duch.). Acta Physiologiae Plantarum 2011, 33, 1847–1852. [Google Scholar] [CrossRef]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muino, J.M.; Angenent, G.C.; Boutilier, K. The BABY BOOM Transcription Factor Activates the LEC1-ABI3-FUS3-LEC2 Network to Induce Somatic Embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef]
- Jia, H.; McCarty, D.R.; Suzuki, M. Distinct Roles of LAFL Network Genes in Promoting the Embryonic Seedling Fate in the Absence of VAL Repression. Plant Physiol. 2013, 163, 1293–1305. [Google Scholar] [CrossRef]
- Horstman, A.; Bemer, M.; Boutilier, K. A Transcriptional View on Somatic Embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Kumar, V. BABY BOOM (BBM): A Candidate Transcription Factor Gene in Plant Biotechnology. Biotech. Lett. 2018, 40, 1467–1475. [Google Scholar] [CrossRef]
- Kumar, V.; Jha, P.; Van Staden, J. LEAFY COTYLEDONs (LECs): Master Regulators in Plant Embryo Development. Plant Cell Tiss. Org. 2020, 140, 475–487. [Google Scholar] [CrossRef]
- Kwong, R.W.; Bui, A.Q.; Lee, H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1-LIKE Defines a Class of Regulators Essential for Embryo Development. Plant Cell 2003, 15, 5–18. [Google Scholar] [CrossRef]
- Lee, H.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 Represents a Functionally Specialized Subunit of the CCAAT Binding Transcription Factor. Proc. Natl. Acad. Sci. USA 2003, 100, 2152–2156. [Google Scholar] [CrossRef] [PubMed]
- Ledwoń, A.; Gaj, M.D. LEAFY COTYLEDON1, FUSCA3 Expression and Auxin Treatment in Relation to Somatic Embryogenesis Induction in Arabidopsis. Plant Growth Regul. 2011, 65, 157–167. [Google Scholar] [CrossRef]
- Parcy, F.; Valon, C.; Raynal, M.; Gaubier-Comella, P.; Delseny, M.; Giraudat, J. Regulation of Gene Expression Programs during Arabidopsis Seed Development: Roles of the ABI3 Locus and of Endogenous Abscisic Acid. Plant Cell 1994, 6, 1567–1582. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, K.; Peterson, K.; Jack, T. The Plant B3 Superfamily. Trends Plant Sci. 2008, 13, 647–655. [Google Scholar] [CrossRef] [PubMed]
- El Ouakfaoui, S.; Schnell, J.; Abdeen, A.; Colville, A.; Labbé, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of Somatic Embryogenesis and Embryo Development by AP2 Transcription Factors. Plant Mol. Biol. 2010, 74, 313–326. [Google Scholar] [CrossRef]
- Kumar, R.; Khurana, A.; Sharma, A.K. Role of Plant Hormones and Their Interplay in Development and Ripening of Fleshy Fruits. J. Exp. Bot. 2014, 65, eru277. [Google Scholar] [CrossRef]
- Han, J.-D.; Li, X.; Jiang, C.-K.; Wong, G.K.-S.; Rothfels, C.J.; Rao, G.-Y. Evolutionary Analysis of the LAFL Genes Involved in the Land Plant Seed Maturation Program. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Cagliari, A.; Turchetto-Zolet, A.C.; Korbes, A.P.; Maraschin, F.d.S.; Margis, R.; Margis-Pinheiro, M. New Insights on the Evolution of Leafy Cotyledon1 (LEC1) Type Genes in Vascular Plants. Genomics 2014, 103, 380–387. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.; Yang, Z.; Xiao, Y.; Wang, P.; Wang, Y.; Ge, X.; Zhang, C.; Zhang, X.; Li, F. Genome-Wide Analysis of the NF-YB Gene Family in Gossypium hirsutum L. and Characterization of the Role of GhDNF-YB22 in Embryogenesis. Int. J. Mol. Sci. 2018, 19, 483. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Zhang, S.; Yao, J.; Rahman, M.U.; Hanif, M.; Zhu, Y.; Wang, X. Genomic Organization of the B3-Domain Transcription Factor Family in Grapevine (Vitis vinifera L.) and Expression during Seed Development in Seedless and Seeded Cultivars. Int. J. Mol. Sci. 2019, 20, 4553. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Zhang, Z.; Wang, Y.; Li, S.; Liang, Z. Genome-Wide Identification and Characterization of the NF-Y Gene Family in Grape (Vitis vinifera L.). BMC Genomics 2016, 17, 605. [Google Scholar] [CrossRef]
- Zhang, Y.; Clemens, A.; Maximova, S.N.; Guiltinan, M.J. The Theobroma Cacao B3 Domain Transcription Factor TcLEC2 Plays a Duel Role in Control of Embryo Development and Maturation. BMC Plant Biol. 2014, 14, 106. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, G.; Liu, W.; Dong, X.; Zhang, A. Genome-Wide Analysis of the NF-Y Gene Family in Peach (Prunus persica L.). BMC Genomics 2019, 20, 612. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and Evolution of the Octoploid Strawberry Genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Stevens, P.F. Onwards. Angiosperm Phylogeny Website. Version 12, July 2012 [and More or Less Continuously Updated since]. 2001. Available online: Http://www.Mobot.Org/MOBOT/Research/APweb/ (accessed on 3 February 2021).
- Xiang, Y.; Huang, C.-H.; Hu, Y.; Wen, J.; Li, S.; Yi, T.; Chen, H.; Xiang, J.; Ma, H. Evolution of Rosaceae Fruit Types Based on Nuclear Phylogeny in the Context of Geological Times and Genome Duplication. Mol. Biol. Evol. 2017, 34, 262–281. [Google Scholar] [CrossRef]
- Potter, D.; Gao, F.; Bortiri, P.E.; Oh, S.-H.; Baggett, S. Phylogenetic Relationships in Rosaceae Inferred from Chloroplast Mat K and Trn L-Trn F Nucleotide Sequence Data. Plant Syst. Evol. 2002, 231, 77–89. [Google Scholar] [CrossRef]
- Shulaev, V.; Korban, S.S.; Sosinski, B.; Abbott, A.G.; Aldwinckle, H.S.; Folta, K.M.; Iezzoni, A.; Main, D.; Arús, P.; Dandekar, A.M.; et al. Multiple Models for Rosaceae Genomics. Plant Physiol. 2008, 147, 985–1003. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-59259-890-8. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.; Doron-Faigenboim, A.; Erez, E.; Martz, E.; Bacharach, E.; Pupko, T. Selecton 2007: Advanced Models for Detecting Positive and Purifying Selection Using a Bayesian Inference Approach. Nucleic Acids Res. 2007, 35, W506–W511. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-Molecule Sequencing and Optical Mapping Yields an Improved Genome of Woodland Strawberry (Fragaria vesca) with Chromosome-Scale Contiguity. Gigascience 2018, 7. [Google Scholar] [CrossRef]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated Annotation of the Wild Strawberry Fragaria Vesca V4 Genome. Hort. Res. 2019, 6, 1–9. [Google Scholar] [CrossRef]
- VanBuren, R.; Wai, C.M.; Colle, M.; Wang, J.; Sullivan, S.; Bushakra, J.M.; Liachko, I.; Vining, K.J.; Dossett, M.; Finn, C.E.; et al. A near Complete, Chromosome-Scale Assembly of the Black Raspberry (Rubus occidentalis) Genome. Gigascience 2018, 7. [Google Scholar] [CrossRef]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa Genome Provides New Insights into the Domestication of Modern Roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-Quality de Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 Release: High-Resolution Linkage Mapping and Deep Resequencing Improve Chromosome-Scale Assembly and Contiguity. BMC Genomics 2017, 18, 225. [Google Scholar] [CrossRef]
- Chagné, D.; Crowhurst, R.N.; Pindo, M.; Thrimawithana, A.; Deng, C.; Ireland, H.; Fiers, M.; Dzierzon, H.; Cestaro, A.; Fontana, P.; et al. The Draft Genome Sequence of European Pear (Pyrus communis L. ’Bartlett’). PLoS ONE 2014, 9, e92644. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Bassett, C.; Bielenberg, D.G.; Cheng, C.-H.; Dardick, C.; Main, D.; Meisel, L.; Slovin, J.; Troggio, M.; Schaffer, R.J. A Standard Nomenclature for Gene Designation in the Rosaceae. Tree Genet. Genomes 2015, 11, 108. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Khan, A.; Hashem, A.; Abd Allah, E.F.; Al-Harrasi, A. The Molecular Mass and Isoelectric Point of Plant Proteomes. BMC Genomics 2019, 20, 631. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene Duplication and Evolution in Recurring Polyploidization–Diploidization Cycles in Plants. Genome Biol. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.; Stein, L.; Ware, D. Evolution of Arabidopsis MicroRNA Families through Duplication Events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ortiz, C.; Peña-Garcia, Y.; Natarajan, P.; Bhandari, M.; Abburi, V.; Dutta, S.K.; Yadav, L.; Stommel, J.; Nimmakayala, P.; Reddy, U.K. The Ankyrin Repeat Gene Family in Capsicum Spp: Genome-Wide Survey, Characterization and Gene Expression Profile. Sci. Rep. 2020, 10, 4044. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks Ratio: Diagnosing the Form of Sequence Evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Etienne, H.; Breton, D.; Breitler, J.-C.; Bertrand, B.; Déchamp, E.; Awada, R.; Marraccini, P.; Léran, S.; Alpizar, E.; Campa, C.; et al. Coffee Somatic Embryogenesis: How Did Research, Experience Gained and Innovations Promote the Commercial Propagation of Elite Clones From the Two Cultivated Species? Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Sabooni, N.; Shekafandeh, A. Somatic Embryogenesis and Plant Regeneration of Blackberry Using the Thin Cell Layer Technique. Plant Cell Tiss. Org. 2017, 130, 313–321. [Google Scholar] [CrossRef]
- Ameri, A.; Davarynejad, G.H.; Moshtaghi, N.; Tehranifar, A. The Role of Carbohydrates on The Induction of Somatic Embryogenesis and The Biochemical State of The Embryogenic Callus in Pyrus communis L. Cv. ‘Dar Gazi’. Erwerbs-Obstbau 2020, 62. [Google Scholar] [CrossRef]
- Bao, Y.; Liu, G.; Shi, X.; Xing, W.; Ning, G.; Liu, J.; Bao, M. Primary and Repetitive Secondary Somatic Embryogenesis in Rosa hybrida ‘Samantha’. Plant Cell Tiss. Org. 2012, 109, 411–418. [Google Scholar] [CrossRef]
- Paul, H.; Belaizi, M.; Sangwan-Norreel, B.S. Somatic Embryogenesis in Apple. J. Plant Physiol. 1994, 143, 78–86. [Google Scholar] [CrossRef]
- Pesce, P.G.; Rugini, E. Influence of Plant Growth Regulators, Carbon Sources and Iron on the Cyclic Secondary Somatic Embryogenesis and Plant Regeneration of Transgenic Cherry Rootstock ‘Colt’ (Prunus avium × P. pseudocerasus). Plant Cell Tiss. Org. 2004, 79, 223–232. [Google Scholar] [CrossRef]
- Zakizadeh, H.; Stummann, B.M.; Lütken, H.; Müller, R. Isolation and Characterization of Four Somatic Embryogenesis Receptor-like Kinase (RhSERK) Genes from Miniature Potted Rose (Rosa hybrida Cv. Linda). Plant Cell Tiss. Org. 2010, 101, 331–338. [Google Scholar] [CrossRef]
- Zheng, L.; Ma, J.; Mao, J.; Fan, S.; Zhang, D.; Zhao, C.; An, N.; Han, M. Genome-Wide Identification of SERK Genes in Apple and Analyses of Their Role in Stress Responses and Growth. BMC Genomics 2018, 19, 962. [Google Scholar] [CrossRef]
- Adams, K.L.; Wendel, J.F. Polyploidy and Genome Evolution in Plants. Curr. Op. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Panchy, N.; Wang, P.; Uygun, S.; Shiu, S.-H. Diversity, Expansion, and Evolutionary Novelty of Plant DNA-Binding Transcription Factor Families. Biochimica et Biophysica Acta Gene Regul. Mech. 2017, 1860, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Cheng, T.; Zhang, W.; Liu, Y.; Wu, P.; Yang, X.; Wang, L.; Zhou, S. Molecular Systematics of Rosoideae (Rosaceae). Plant Syst. Evol. 2020, 306, 9. [Google Scholar] [CrossRef]
- Chin, S.-W.; Shaw, J.; Haberle, R.; Wen, J.; Potter, D. Diversification of Almonds, Peaches, Plums and Cherries—Molecular Systematics and Biogeographic History of Prunus (Rosaceae). Mol. Phylogenet. Evol. 2014, 76, 34–48. [Google Scholar] [CrossRef]
- Brand, A.; Quimbaya, M.; Tohme, J.; Chavarriaga-Aguirre, P. Arabidopsis LEC1 and LEC2 Orthologous Genes Are Key Regulators of Somatic Embryogenesis in Cassava. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Davis, T.M. A New Perspective on Polyploid Fragaria (Strawberry) Genome Composition Based on Large-Scale, Multi-Locus Phylogenetic Analysis. Genome Biol. Evol. 2017, 9, 3433–3448. [Google Scholar] [CrossRef] [PubMed]
- Gmitter, F.G.; Ling, X.; Cai, C.; Grosser, J.W. Colchicine-Induced Polyploidy in Citrus Embryogenic Cultures, Somatic Embryos, and Regenerated Plantlets. Plant Sci. 1991, 74, 135–141. [Google Scholar] [CrossRef]
- Ghotbi Ravandi, E.; Rezanejad, F.; Dehghan, E. In Vitro Regeneration Ability of Diploid and Autotetraploid Plants of Cichorium intybus L. Cytol. Genet. 2014, 48, 166–170. [Google Scholar] [CrossRef]
- Jo, B.-S.; Choi, S.S. Introns: The Functional Benefits of Introns in Genomes. Genom. Inform. 2015, 13, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhang, Y.; Zhang, W.; Yang, S.; Chen, J.-Q.; Tian, D. Patterns of Exon-Intron Architecture Variation of Genes in Eukaryotic Genomes. BMC Genom. 2009, 10, 47. [Google Scholar] [CrossRef]
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.N.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative Splicing and Protein Diversity: Plants Versus Animals. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Paterson, A.H.; Chapman, B.A.; Kissinger, J.C.; Bowers, J.E.; Feltus, F.A.; Estill, J.C. Many Gene and Domain Families Have Convergent Fates Following Independent Whole-Genome Duplication Events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 2006, 22, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Xi, X.; Li, S.; Chen, W.; Zhang, B.; Liu, D.; Liu, B.; Wang, D.; Zhang, H. Allelic Variation and Transcriptional Isoforms of Wheat TaMYC1 Gene Regulating Anthocyanin Synthesis in Pericarp. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.F.; Kou, Y.P.; Gao, B.; Soliman, T.M.A.; Xu, K.D.; Ma, N.; Cao, X.; Zhao, L.J. Identification and Functional Analysis of BABY BOOM Genes from Rosa canina. Biologia Plantarum 2014, 58, 427–435. [Google Scholar] [CrossRef]
- Hummer, K.E.; Janick, J. Rosaceae: Taxonomy, Economic Importance, Genomics. In Genetics and Genomics of Rosaceae, 1st ed.; Folta, K.M., Gardiner, S.E., Eds.; Springer: New York, NY, USA, 2009; pp. 1–17. ISBN 978-0-387-77490-9. [Google Scholar]
- Silva, A.T.; Barduche, D.; do Livramento, K.G.; Paiva, L.V. A Putative BABY BOOM-like Gene (CaBBM) Is Expressed in Embryogenic Calli and Embryogenic Cell Suspension Culture of Coffea arabica L. In Vitro Cell. Dev. Biol. Plant 2015, 51, 93–101. [Google Scholar] [CrossRef]
- Maulidiya, A.U.K.; Sugiharto, B.; Dewanti, P.; Handoyo, T. Expression of Somatic Embryogenesis-Related Genes in Sugarcane (Saccharum officinarum L.). J. Crop Sci. Biotech. 2020, 23, 207–214. [Google Scholar] [CrossRef]
- Considine, M.J.; Wan, Y.; D’Antuono, M.F.; Zhou, Q.; Han, M.; Gao, H.; Wang, M. Molecular Genetic Features of Polyploidization and Aneuploidization Reveal Unique Patterns for Genome Duplication in Diploid Malus. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Yamasaki, M.; Wright, S.I.; McMullen, M.D. Genomic Screening for Artificial Selection during Domestication and Improvement in Maize. Ann. Bot. 2007, 100, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.; Liberles, D.A. A Systematic Search for Positive Selection in Higher Plants (Embryophytes). BMC Plant Biol. 2006, 6, 12. [Google Scholar] [CrossRef][Green Version]

| Gene | Accession * | Chr | Coordinates | Gene (bp) | CDS (bp) | Protein (aa) | Molecular Weight (kDa) | pI |
|---|---|---|---|---|---|---|---|---|
| FaLEC1 | augustus_masked-Fvb3-3-processed-gene-175.1 | 3-3 | 17523635-17524327 | 693 | 693 | 230 | 25.46 | 5.42 |
| FaL1L1 | augustus_masked-Fvb3-1-processed-gene-317.10 | 3-1 | 31777224-31777889 | 666 | 666 | 221 | 24.33 | 5.66 |
| FaL1L2 | maker-Fvb3-1-snap-gene-259.54 | 3-1 | 25951232-25951844 | 613 | 594 | 197 | 21.29 | 5.88 |
| FaL1L3 | maker-Fvb3-2-snap-gene-30.79 | 3-2 | 3007308-3009361 | 2054 | 798 | 265 | 29.45 | 6.64 |
| FaL1L4 | snap_masked-Fvb3-3-processed-gene-44.47 | 3-3 | 4402948-4403610 | 663 | 645 | 214 | 23.69 | 6.05 |
| FaLEC2.1 | snap_masked-Fvb3-2-processed-gene-48.18 | 3-2 | 4803028-4803975 | 948 | 552 | 183 | 20.98 | 9.44 |
| FaLEC2.2 | maker-Fvb3-4-snap-gene-246.68 | 3-4 | 24622191-24623184 | 994 | 555 | 184 | 21.16 | 9.03 |
| FaLEC2.3 | maker-Fvb5-1-snap-gene-133.63 | 5-1 | 13346240-13348435 | 2196 | 1074 | 357 | 40.51 | 6.87 |
| FaLEC2.4 | augustus_masked-Fvb5-3-processed-gene-156.8 | 5-3 | 15664339-15666680 | 2342 | 1227 | 408 | 45.90 | 5.82 |
| FaFUS3.1 | maker-Fvb6-1-augustus-gene-7.51 | 6-1 | 758349-761311 | 2963 | 990 | 329 | 37.37 | 5.80 |
| FaFUS3.2 | maker-Fvb6-2-snap-gene-275.28 | 6-2 | 27584481-27587435 | 2955 | 978 | 325 | 36.78 | 5.45 |
| FaFUS3.3 | maker-Fvb6-3-augustus-gene-7.37 | 6-3 | 736263-739146 | 2884 | 981 | 326 | 36.96 | 5.35 |
| FaABI3.1 | maker-Fvb2-1-snap-gene-128.30 | 2-1 | 12810779-12814037 | 3259 | 2205 | 734 | 81.60 | 5.75 |
| FaABI3.2 | maker-Fvb2-2-augustus-gene-81.38 | 2-2 | 8128310-8131378 | 3069 | 2184 | 727 | 80.61 | 5.81 |
| FaABI3.3 | maker-Fvb2-4-augustus-gene-151.48 | 2-4 | 15160217-15163448 | 3232 | 2220 | 739 | 82.15 | 6.00 |
| FaBBM1 | maker-Fvb6-2-augustus-gene-270.50 | 6-2 | 27083071-27087139 | 4069 | 2478 | 825 | 89.76 | 6.11 |
| FaBBM2 | maker-Fvb6-4-augustus-gene-24.29 | 6-4 | 2470827-2474851 | 4025 | 2478 | 825 | 89.80 | 6.10 |
| LEC1 | L1L | LEC2 | FUS3 | ABI3 | BBM | Protein-Coding Genes | Genome Version ** | |
|---|---|---|---|---|---|---|---|---|
| Fragaria × ananassa | 1 | 4 | 4 | 3 | 3 | 2 | 108,087 | v1.0 [36] |
| Fragaria vesca | 1 | 2 | 1 | 1 | 1 (2) * | 1 (2) * | 34,007 | v4.0 [50] |
| Rubus occidentalis | 4 | 1 | 1 | 1 | 1 | 1 | 34,545 | v3.0 [51] |
| Rosa chinensis | 1 | 1 | 2 | 1 | 1 | 2 | 36,377 | v1.0 [52] |
| Malus × domestica | 4 | 2 | 2 | 2 | 2 | 1 | 42,140 | v1.1 [53] |
| Prunus persica | 1 | 2 | 1 (3) * | 1 | 1 (6) * | 1 | 26,873 | v2.0 [54] |
| Pyrus communis | 1 | 2 | 2 | 1 | 0 | 1 | 37,445 | v2.0 [55] |
| Gene A | Gene B | Duplication * | Ka | Ks | Ka/Ks | Selection Pressure |
|---|---|---|---|---|---|---|
| FaL1L1 | FaL1L3 | DSD | 0.026 | 0.038 | 0.684 | Negative |
| FaL1L2 | FaL1L3 | DSD | 0.134 | 0.231 | 0.580 | Negative |
| FvL1L1 | FvL1L2 | DSD | 0.043 | 0.173 | 0.248 | Negative |
| PcL1L1 | PcL1L2 | DSD | 0.039 | 0.155 | 0.252 | Negative |
| FaLEC2.1 | FaLEC2.4 | DSD | 0.130 | 0.236 | 0.551 | Negative |
| FaLEC2.2 | FaLEC2.3 | DSD | 0.385 | 0.629 | 0.612 | Negative |
| RoLEC2.1 | RoLEC2.2 | DSD | 0.338 | 0.654 | 0.517 | Negative |
| RcLEC2.1 | RcLEC2.2 | DSD | 0.327 | 0.802 | 0.408 | Negative |
| MdLEC1.1 | MdLEC1.2 | PD | 0.018 | 0.076 | 0.237 | Negative |
| PcLEC2.1 | PcLEC2.2 | DSD | 0.097 | 0.203 | 0.478 | Negative |
| FaFUS3.1 | FaFUS3.2 | DSD | 0.004 | 0.005 | 0.800 | Negative |
| FaABI3.1 | FaABI3.3 | DSD | 0.015 | 0.037 | 0.405 | Negative |
| FaBBM1 | FaBBM2 | DSD | 0.009 | 0.035 | 0.257 | Negative |
| FaLEC1 | FvLEC1 | 0.063 | 0.131 | 0.481 | Negative | |
| FaL1L3 | FvL1L1 | 0.015 | 0.012 | 1.250 | Positive | |
| FaL1L4 | FvL1L2 | 0.033 | 0.063 | 0.524 | Negative | |
| FaLEC2.1 | FvLEC2 | 0.218 | 0.294 | 0.741 | Negative | |
| FaFUS3.1 | FvFUS3 | 0.003 | 0.009 | 0.333 | Negative | |
| FaABI3.2 | FvABI3 | 0.004 | 0.010 | 0.400 | Negative | |
| FaBBM1 | FvBBM | 0.002 | 0.004 | 0.500 | Negative |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrido-Bigotes, A.; Silva, H.; Hasbún, R. Genome-Wide Analysis of Somatic Embryogenesis-Related Transcription Factors in Cultivated Strawberry (Fragaria × ananassa) and Evolutionary Relationships among Rosaceae Species. Agronomy 2021, 11, 356. https://doi.org/10.3390/agronomy11020356
Garrido-Bigotes A, Silva H, Hasbún R. Genome-Wide Analysis of Somatic Embryogenesis-Related Transcription Factors in Cultivated Strawberry (Fragaria × ananassa) and Evolutionary Relationships among Rosaceae Species. Agronomy. 2021; 11(2):356. https://doi.org/10.3390/agronomy11020356
Chicago/Turabian StyleGarrido-Bigotes, Adrián, Herman Silva, and Rodrigo Hasbún. 2021. "Genome-Wide Analysis of Somatic Embryogenesis-Related Transcription Factors in Cultivated Strawberry (Fragaria × ananassa) and Evolutionary Relationships among Rosaceae Species" Agronomy 11, no. 2: 356. https://doi.org/10.3390/agronomy11020356
APA StyleGarrido-Bigotes, A., Silva, H., & Hasbún, R. (2021). Genome-Wide Analysis of Somatic Embryogenesis-Related Transcription Factors in Cultivated Strawberry (Fragaria × ananassa) and Evolutionary Relationships among Rosaceae Species. Agronomy, 11(2), 356. https://doi.org/10.3390/agronomy11020356






