Using Principal Component Analysis and RNA-Seq to Identify Candidate Genes That Control Salt Tolerance in Garlic (Allium sativum L.)
Abstract
1. Introduction
2. Materials and Methods
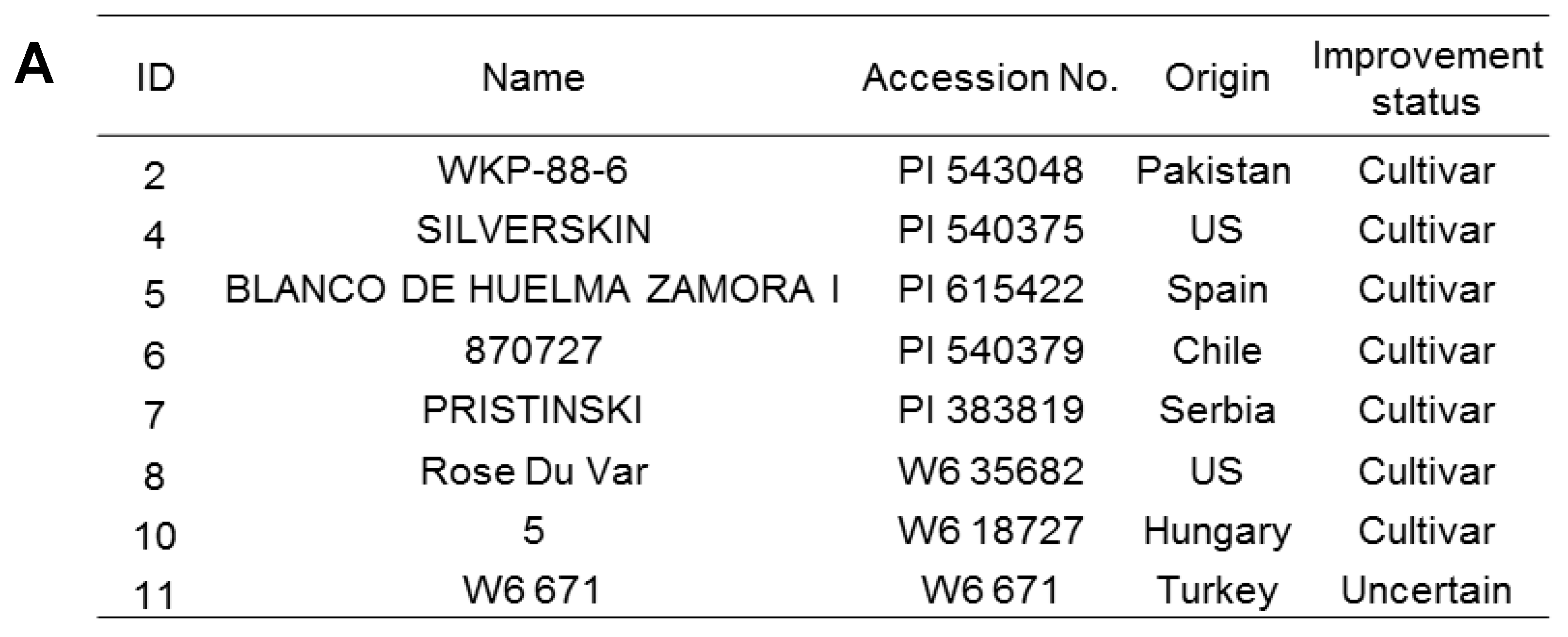
2.1. Plant Materials and Growth Conditions
2.2. Salt Stress Treatment and Physiological Measurements
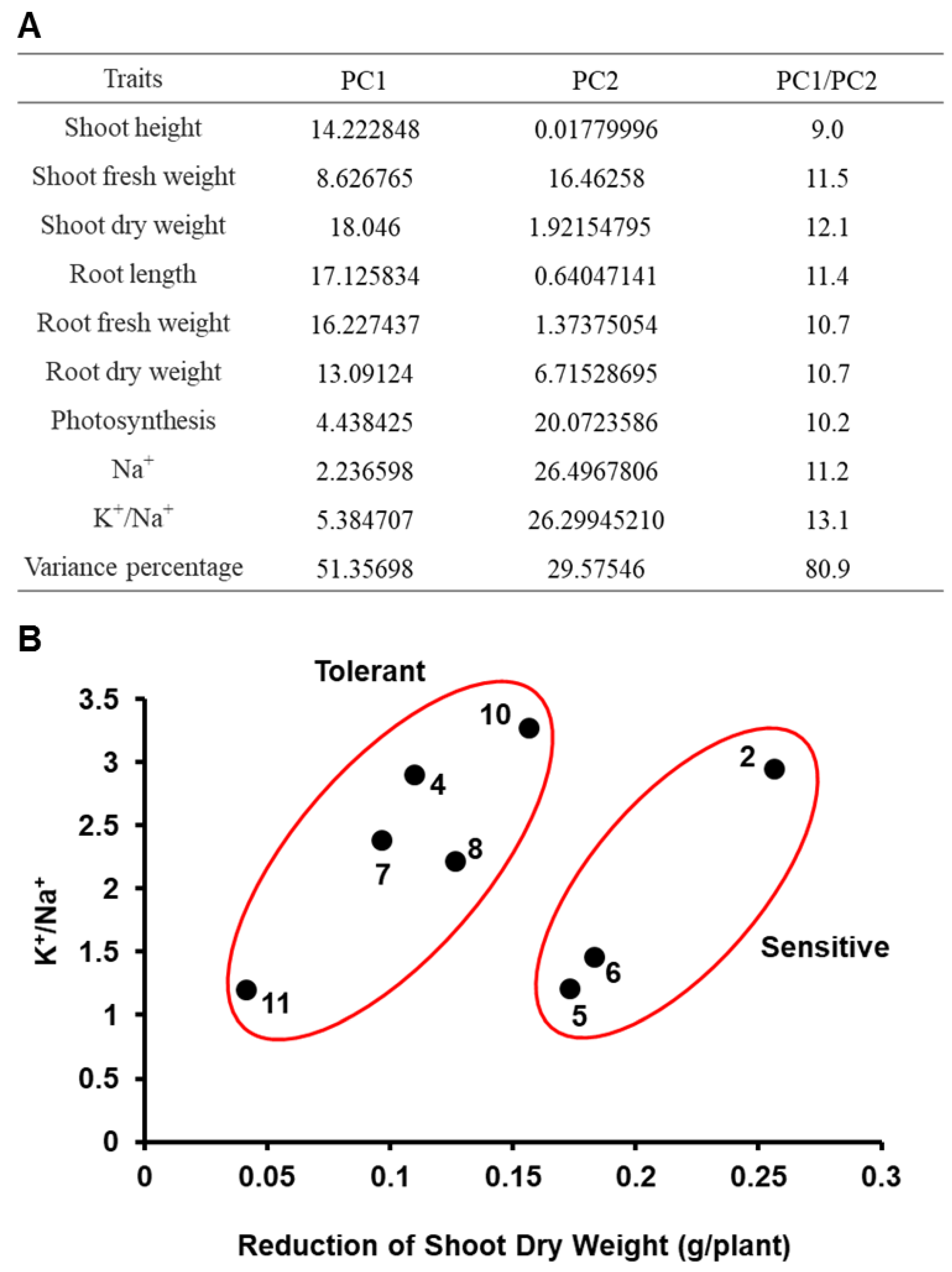
2.3. Principal Component Analysis (PCA)
2.4. RNA Extraction, Library Construction and Quantitative Reverse Transcription PCR (qRT-PCR)
2.5. RNA-Seq Data Analysis and Functional Annotation
3. Results
3.1. Physiological Response of Garlic Cultivars to Salt Stress
3.2. PCA and Separation of Garlic Accessions
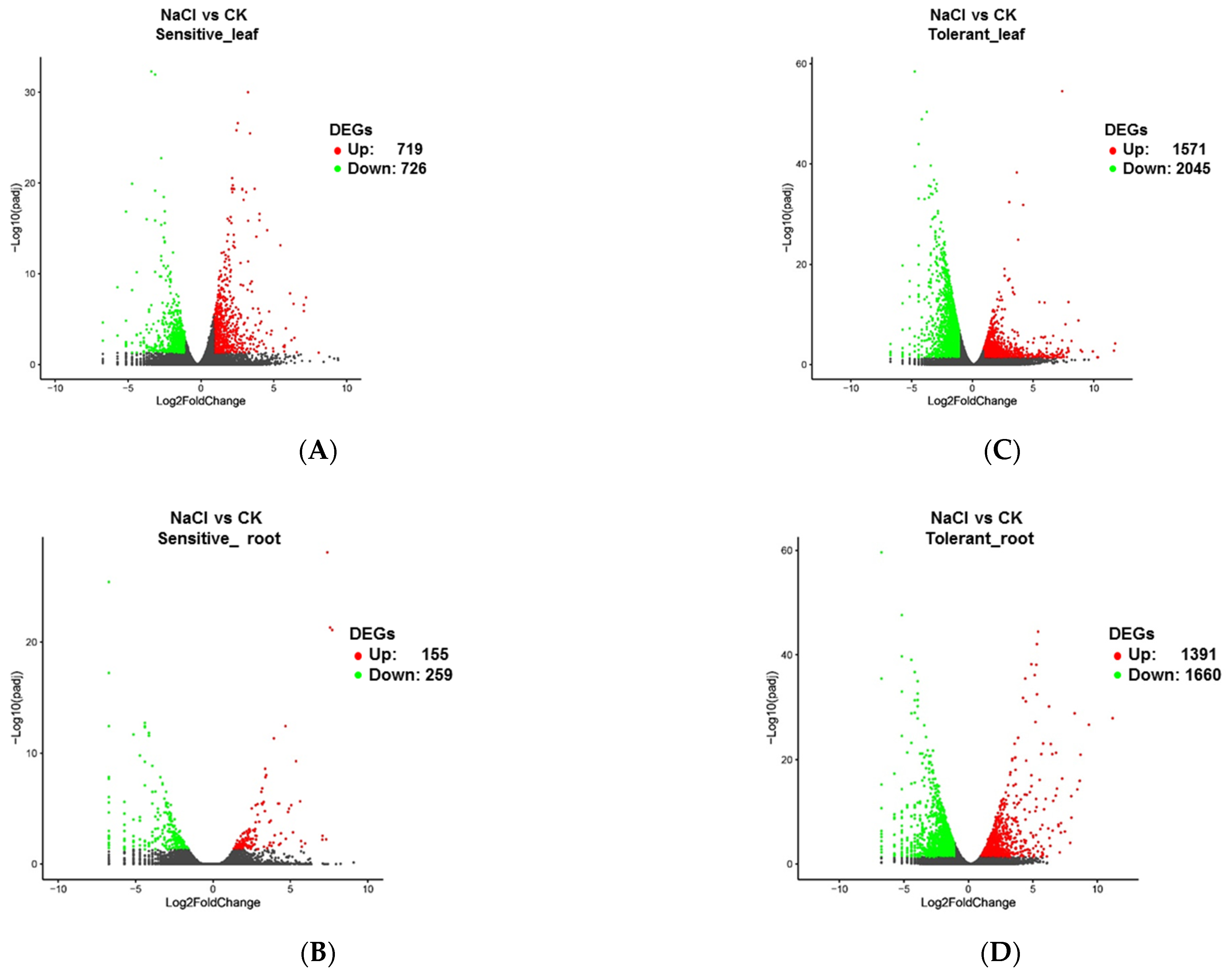
3.3. Comparative Transcriptome in the Salinity-Tolerant and Sensitive Accessions by RNA-Seq
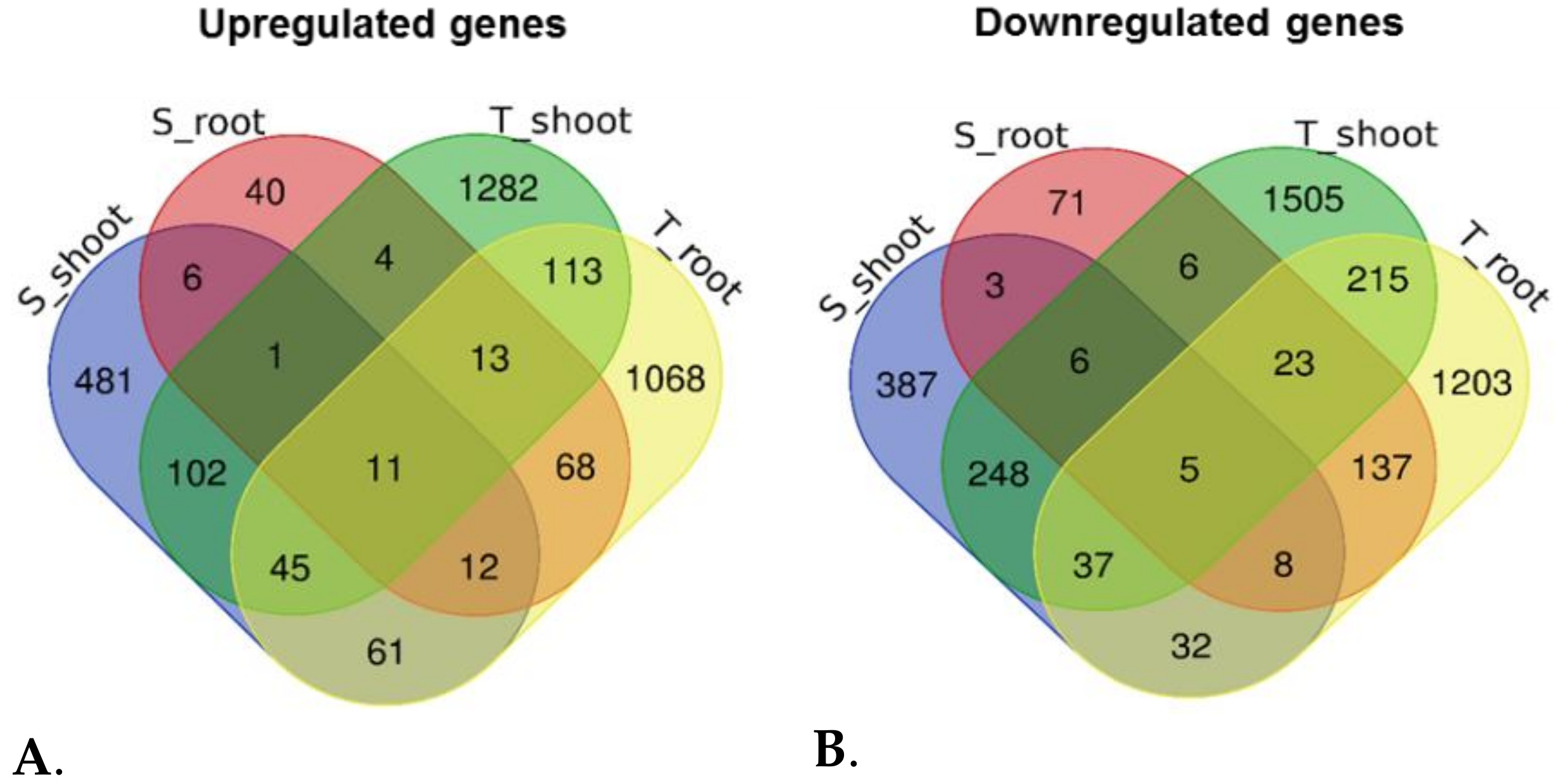
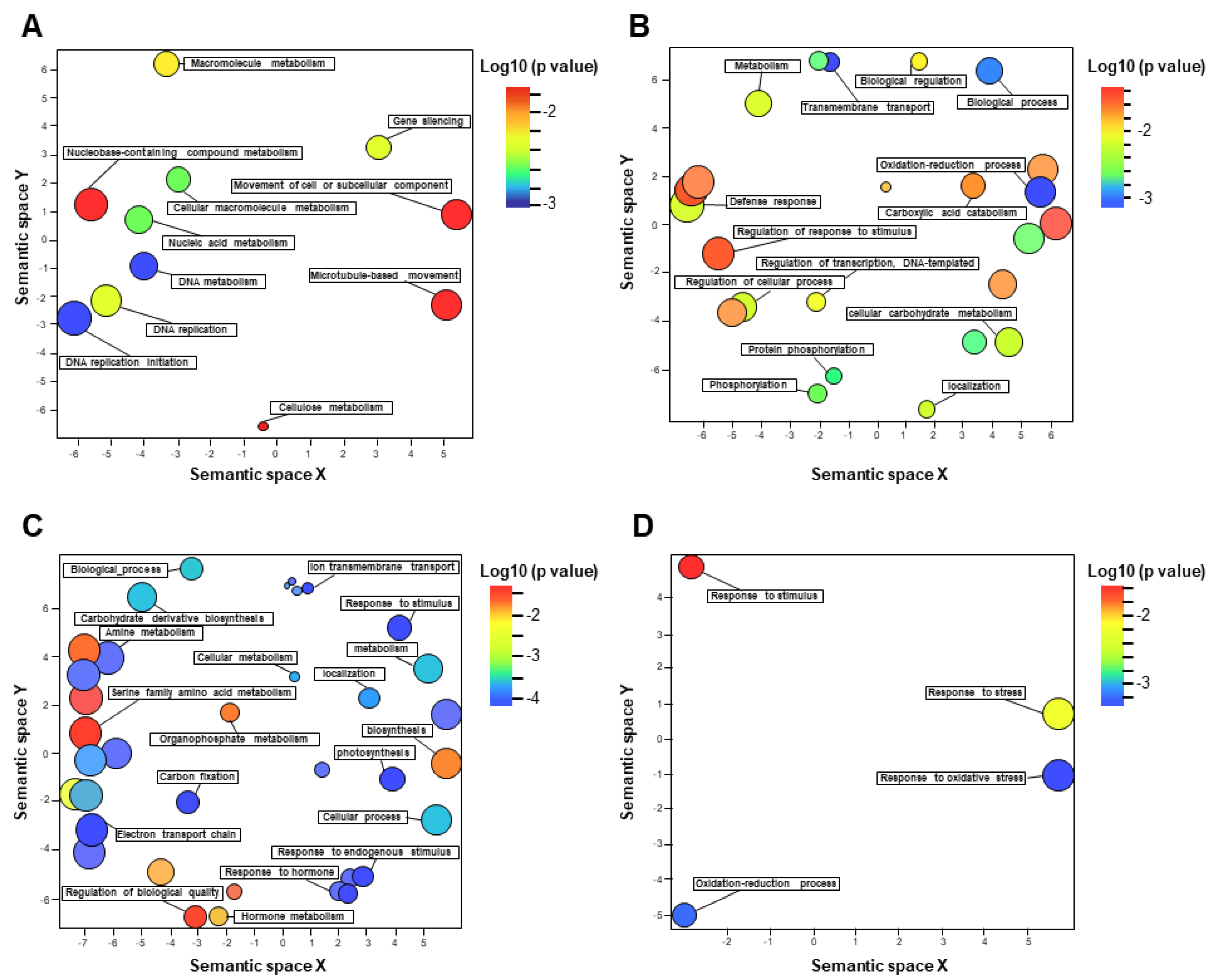
3.4. Functional Categorization of DEGs Identified in the Tolerant Leaves and Roots
3.5. Potassium Transporters were Significantly Upregulated in the Tolerant Roots
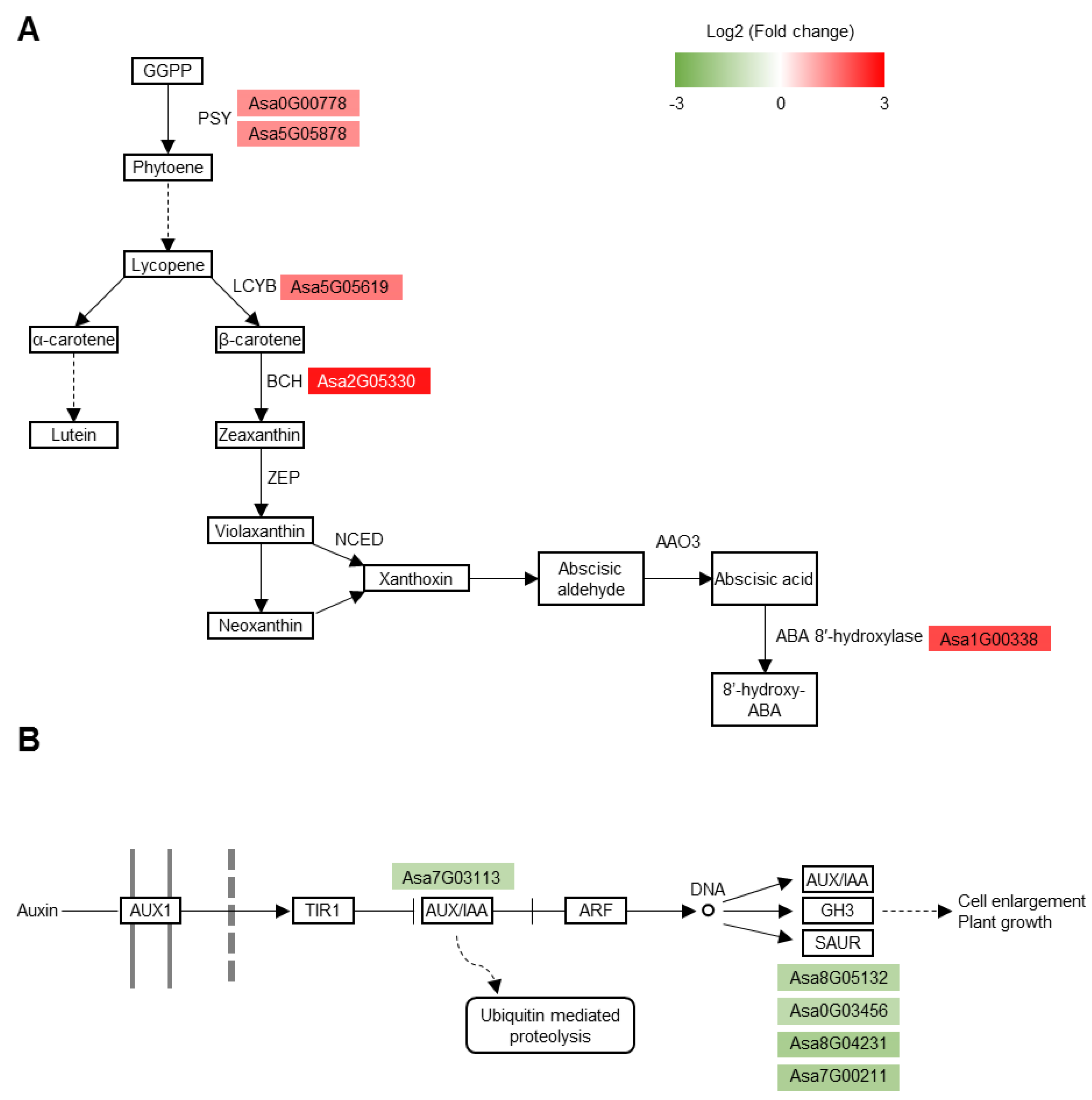
3.6. Carotenoid Biosynthesis Pathway and Auxin Signaling Pathway Are Substantially Altered in the Tolerant Roots
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rengasamy, P. Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 2010, 37, 613–620. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Tsugane, K.; Kobayashi, K.; Niwa, Y.; Ohba, Y.; Wada, K.; Kobayashi, H. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 1999, 11, 1195–1206. [Google Scholar] [CrossRef]
- Hernández, J.A.; Ferrer, M.A.; Jiménez, A.; Barceló, A.R.; Sevilla, F. Antioxidant systems and O2.−/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 2001, 127, 817–831. [Google Scholar] [CrossRef]
- Isayenkov, S.V. Physiological and molecular aspects of salt stress in plants. Cytol. Genet. 2012, 46, 302–318. [Google Scholar] [CrossRef]
- Zhang, H.-H.; Xu, N.; Li, X.; Jin, W.-W.; Tian, Q.; Gu, S.-Y.; Sun, G.-Y. Overexpression of 2-Cys Prx increased salt tolerance of photosystem II in tobacco. Int. J. Agric. Biol. 2017, 19, 735–745. [Google Scholar] [CrossRef]
- Niu, X.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995, 109, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.S.; Gilliham, M.; Jha, D.; Mayo, G.M.; Roy, S.J.; Coates, J.C.; Haseloff, J.; Tester, M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type–specific alteration of Na+ transport in Arabidopsis. Plant Cell 2009, 21, 2163–2178. [Google Scholar] [CrossRef]
- Manishankar, P.; Wang, N.; Köster, P.; Alatar, A.A.; Kudla, J. Calcium signaling during salt stress and in the regulation of ion homeostasis. J. Exp. Bot. 2018, 69, 4215–4226. [Google Scholar] [CrossRef]
- Wang, Y.; Li, K.; Li, X. Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. J. Plant Physiol. 2009, 166, 1637–1645. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, Y.; Chen, S.; Ning, N.; Hu, H. Arabidopsis IAR4 modulates primary root growth under salt stress through ROS-mediated modulation of auxin distribution. Front. Plant Sci. 2019, 10, 522. [Google Scholar] [CrossRef]
- Ding, Z.; De Smet, I. Localised ABA signalling mediates root growth plasticity. Trends Plant Sci. 2013, 18, 533–535. [Google Scholar] [CrossRef]
- Geng, Y.; Wu, R.; Wee, C.W.; Xie, F.; Wei, X.; Chan, P.M.Y.; Tham, C.; Duan, L.; Dinneny, J.R. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 2013, 25, 2132–2154. [Google Scholar] [CrossRef] [PubMed]
- Barrero, J.M.; Rodríguez, P.L.; Quesada, V.; Piqueras, P.; Ponce, M.R.; Micol, J.L. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006, 29, 2000–2008. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sola, M.Á.; Arbona, V.; Gómez-Cadenas, A.; Rodríguez-Concepción, M.; Rodríguez-Villalón, A. A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in Arabidopsis. PLoS ONE 2014, 9, e90765. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.-K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J.; Ding, L.; Zhu, J.K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 1996, 8, 617–627. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, J.-K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef]
- Halfter, U.; Ishitani, M.; Zhu, J.-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Purty, R.S.; Sharma, M.P.; Singla-Pareek, S.L.; Pareek, A. Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J. Plant Physiol. 2009, 166, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Francois, L.E. Yield and quality response of salt-stressed garlic. HortScience 1994, 29, 1314–1317. [Google Scholar] [CrossRef]
- Arumuganathan, K.; Earle, E.D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Egea, L.A.; Mérida-García, R.; Kilian, A.; Hernandez, P.; Dorado, G. Assessment of genetic diversity and structure of large garlic (Allium sativum) germplasm bank, by diversity arrays technology “genotyping-by-sequencing” platform (DArTseq). Front. Genet. 2017, 8, 98. [Google Scholar] [CrossRef]
- Sun, X.; Zhu, S.; Li, N.; Cheng, Y.; Zhao, J.; Qiao, X.; Lu, L.; Liu, S.; Wang, Y.; Liu, C.; et al. A chromosome-level genome assembly of garlic (Allium sativum) provides insights into genome evolution and allicin biosynthesis. Mol. Plant 2020, 13, 1328–1339. [Google Scholar] [CrossRef]
- Chen, X.; Liu, X.; Zhu, S.; Tang, S.; Mei, S.; Chen, J.; Li, S.; Liu, M.; Gu, Y.; Dai, Q.; et al. Transcriptome-referenced association study of clove shape traits in garlic. DNA Res. 2018, 25, 587–596. [Google Scholar] [CrossRef]
- Li, N.; Qiu, Z.; Lu, X.; Shi, B.; Sun, X.; Tang, X.; Qiao, X. Comparative transcriptome analysis of temperature-induced green discoloration in garlic. Int. J. Genomics 2018, 2018, 6725728. [Google Scholar] [CrossRef]
- Sun, X.; Zhou, S.; Meng, F.; Liu, S. De novo assembly and characterization of the garlic (Allium sativum) bud transcriptome by Illumina sequencing. Plant Cell Rep. 2012, 31, 1823–1828. [Google Scholar] [CrossRef]
- Mehra, R.; Jasrotia, R.S.; Mahajan, A.; Sharma, D.; Iquebal, M.A.; Kaul, S.; Dhar, M.K. Transcriptome analysis of Snow Mountain garlic for unraveling the organosulfur metabolic pathway. Genomics 2020, 112, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Ren, X.Q.; Liu, J.X.; Yang, F.; Wang, Y.P.; Xiong, A.S. Transcript profiling reveals an important role of cell wall remodeling and hormone signaling under salt stress in garlic. Plant Physiol. Biochem. 2019, 135, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Shemesh-Mayer, E.; Ben-Michael, T.; Rotem, N.; Rabinowitch, H.D.; Doron-Faigenboim, A.; Kosmala, A.; Perlikowski, D.; Sherman, A.; Kamenetsky, R. Garlic (Allium sativum L.) fertility: Transcriptome and proteome analyses provide insight into flower and pollen development. Front. Plant Sci. 2015, 6, 271. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Langmead, B. Aligning short sequencing reads with bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.7.1–11.7.14. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.M. Sodium in plants: Perception, signalling, and regulation of sodium fluxes. J. Exp. Bot. 2014, 65, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yu, X.; Luo, N.A.; Xiao, F.; Camberato, J.J.; Jiang, Y. Natural variation of salinity response, population structure and candidate genes associated with salinity tolerance in perennial ryegrass accessions. Plant Cell Environ. 2013, 36, 2021–2033. [Google Scholar] [CrossRef] [PubMed]
- Ringnér, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Xin, S.; Suo-min, W.; Yiwei, J. Genotypic variations in plant growth and nutritional elements of perennial ryegrass accessions under salinity stress. J. Amer. Soc. Hortic. Sci. 2017, 142, 476–483. [Google Scholar] [CrossRef]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A mechanism for sustained cellulose synthesis during salt stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.Q.; Tran, N.Q.; Gill, S.S.; Tuteja, R.; Tuteja, N. A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol. Biol. 2011, 76, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.; Dreyer, I.; Riedelsberger, J. The role of K+ channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front. Plant Sci. 2013, 4, 224. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Welsch, R.; Beyer, P.; Hugueney, P.; Kleinig, H.; von Lintig, J. Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 2000, 211, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.X., Jr.; Chamovitz, D.; Misawa, N.; Gantt, E.; Hirschberg, J. Cloning and functional expression in Escherichia coli of a cyanobacterial gene for lycopene cyclase, the enzyme that catalyzes the biosynthesis of beta-carotene. FEBS Lett. 1993, 328, 130–138. [Google Scholar] [CrossRef]
- Davison, P.A.; Hunter, C.N.; Horton, P. Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 2002, 418, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Magallanes-Lundback, M.; Musetti, V.; DellaPenna, D. Functional analysis of beta- and epsilon-ring carotenoid hydroxylases in Arabidopsis. Plant Cell 2003, 15, 1320–1332. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 2004, 16, 533–543. [Google Scholar] [CrossRef]
- Ren, H.; Gray, W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef]
- Li, Z.-G.; Chen, H.-W.; Li, Q.-T.; Tao, J.-J.; Bian, X.-H.; Ma, B.; Zhang, W.-K.; Chen, S.-Y.; Zhang, J.-S. Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci. Rep. 2015, 5, 12477. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.-M.; Tan, Y.; Chu, H.-J.; Sun, M.-X.; Xing, J.-C. Transcriptome sequencing revealed molecular mechanisms underlying tolerance of Suaeda salsa to saline stress. PLoS ONE 2019, 14, e0219979. [Google Scholar] [CrossRef]
- Zhang, M.; Hong, L.-Z.; Gu, M.-F.; Wu, C.-D.; Zhang, G. Transcriptome analyses revealed molecular responses of Cynanchum auriculatum leaves to saline stress. Sci. Rep. 2020, 10, 449. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA gene family in plants: Molecular structure, regulation, and function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed]
- Kepinski, S.; Leyser, O. Ubiquitination and auxin signaling: A degrading story. Plant Cell 2002, 14 (Suppl. 1), S81–S95. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Kim, Y.-S.; Yoon, H.-K.; Park, C.-M. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007, 172, 150–157. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zhu, Y.; Gao, C.; She, W.; Lin, W.; Chen, Y.; Han, N.; Bian, H.; Zhu, M.; Wang, J. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root Meristem patterning in Arabidopsis. Plant Cell Physiol. 2013, 54, 609–621. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, Q.; Hu, Z.; Sun, X.; Fan, S.; Zhang, H. Function of the auxin-responsive gene TaSAUR75 under salt and drought stress. Crop J. 2018, 6, 181–190. [Google Scholar] [CrossRef]


| Traits | Control | NaCl 200 mM | ||
|---|---|---|---|---|
| Range | Mean | Range | Mean | |
| SH (cm) | 16.5–41.1 | 28.3 | 13.2–29.8 | 19.9 |
| SFW (g/plant) | 1.88–8.59 | 4.07 | 0.61–5.4 | 2.16 |
| SDW (g/plant) | 0.30–0.76 | 0.47 | 0.15–0.58 | 0.33 |
| RL (cm) | 9.6–33.1 | 26.2 | 9.2–21.1 | 15.1 |
| RFW (g/plant) | 1.13–5.21 | 3.0 | 0.4–3.68 | 1.78 |
| RDW (g/plant) | 0.11–0.37 | 0.25 | 0.04–0.27 | 0.16 |
| Photo (μmol m−2 s−1) | 4.42–18.03 | 11.87 | 1.20–4.68 | 2.65 |
| Na+(mg/g) | ND | ND | 14.0–28.2 | 20.2 |
| K+(mg/g) | ND | ND | 29.3–46.0 | 37.8 |
| Gene ID | Annotation | Fold Change | Adjusted p Value | qRT-PCR |
|---|---|---|---|---|
| Asa2G07164 | Potassium channel like | 4.91 | 0.038099708 | 6.1 |
| Asa7G06244 | Potassium transporter | 2.72 | 0.008903446 | 3.7 |
| Asa7G02557 | Two-pore potassium channel like | 2.69 | 0.028014856 | 7.7 |
| Asa8G02052 | Potassium transporter | 2.36 | 0.039404635 | 4.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Dou, Y.; Huang, X.; Li, G.; Zhang, H.; Jiang, D.; Fan, J.; Condori-Apfata, J.A.; Liu, X.; Condori-Pacsi, S.J.; et al. Using Principal Component Analysis and RNA-Seq to Identify Candidate Genes That Control Salt Tolerance in Garlic (Allium sativum L.). Agronomy 2021, 11, 691. https://doi.org/10.3390/agronomy11040691
Zhou X, Dou Y, Huang X, Li G, Zhang H, Jiang D, Fan J, Condori-Apfata JA, Liu X, Condori-Pacsi SJ, et al. Using Principal Component Analysis and RNA-Seq to Identify Candidate Genes That Control Salt Tolerance in Garlic (Allium sativum L.). Agronomy. 2021; 11(4):691. https://doi.org/10.3390/agronomy11040691
Chicago/Turabian StyleZhou, Xiangjun, Yanxia Dou, Xiaoxia Huang, Gang Li, Hongrui Zhang, Dagang Jiang, Jinping Fan, Jorge Alberto Condori-Apfata, Xiaoqin Liu, Sandro Jhonatan Condori-Pacsi, and et al. 2021. "Using Principal Component Analysis and RNA-Seq to Identify Candidate Genes That Control Salt Tolerance in Garlic (Allium sativum L.)" Agronomy 11, no. 4: 691. https://doi.org/10.3390/agronomy11040691
APA StyleZhou, X., Dou, Y., Huang, X., Li, G., Zhang, H., Jiang, D., Fan, J., Condori-Apfata, J. A., Liu, X., Condori-Pacsi, S. J., Bardales, R. M., Anco, M. E. M., Lazo, H. O., Salazar, H. A. D., Valencia, M. V., & Zhang, C. (2021). Using Principal Component Analysis and RNA-Seq to Identify Candidate Genes That Control Salt Tolerance in Garlic (Allium sativum L.). Agronomy, 11(4), 691. https://doi.org/10.3390/agronomy11040691








