Phenotypic and Molecular Characterization of Brazilian Capsicum Germplasm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Experimental Conditions, and Phenotyping
2.2. Genotyping
2.3. Data Analysis
3. Results
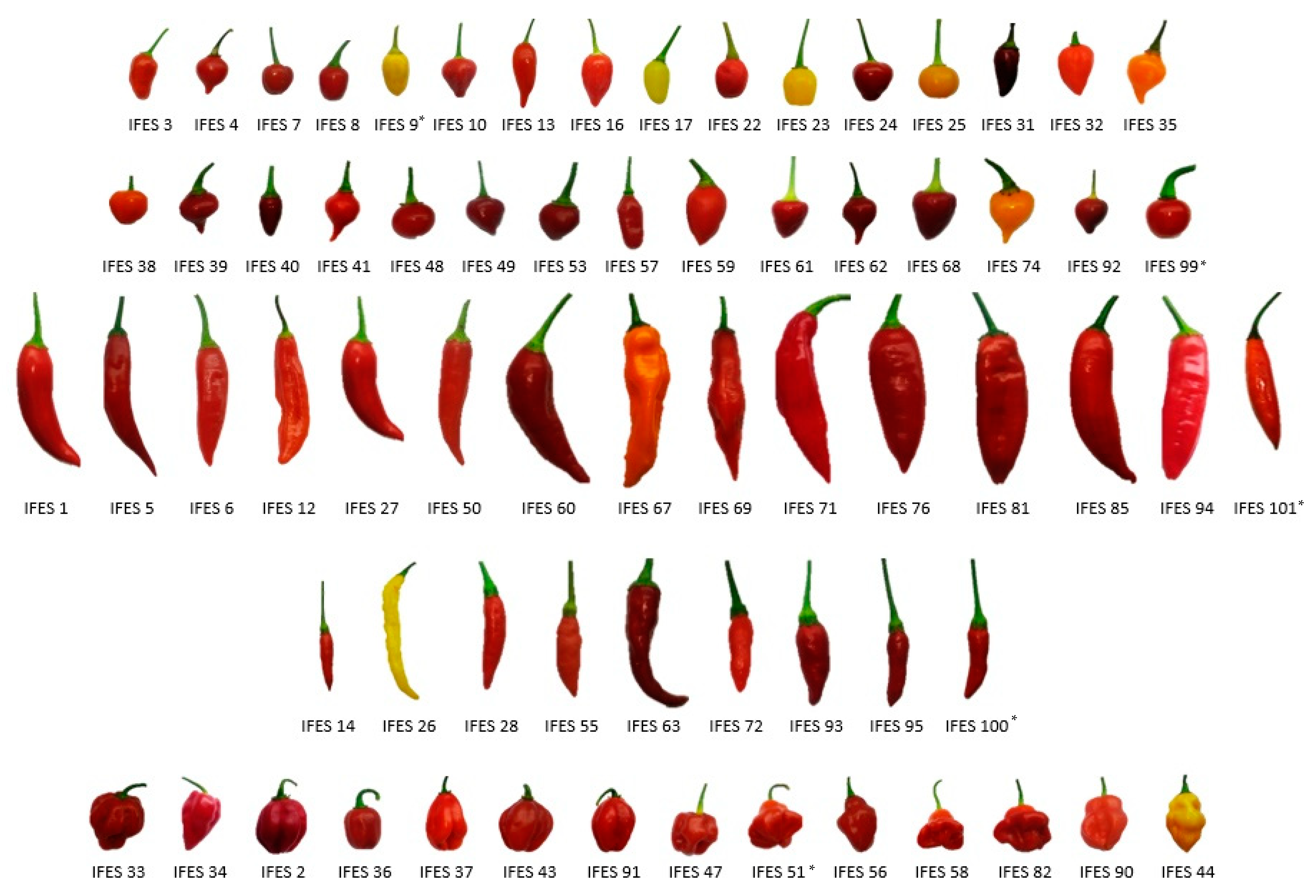
3.1. Morpho-Agronomic Characterization
3.2. Diversity Among Capsicum Accessions and Correlated Traits
3.3. ISSR Characterization and Molecular Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barboza, G.E.; Carrizo García, C.; Leiva González, S.; Scaldaferro, M.; Reyes, X. Four new species of Capsicum (Solanaceae) from the tropical Andes and an update on the phylogeny of the genus. PLoS ONE 2019, 14, e0209792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zonneveld, M.; Ramirez, M.; Williams, D.E.; Petz, M.; Meckelmann, S.; Avila, T.; Bejarano, C.; Ríos, L.; Peña, K.; Jäger, M.; et al. Screening Genetic Resources of Capsicum Peppers in Their Primary Center of Diversity in Bolivia and Peru. PLoS ONE 2015, 10, e0134663. [Google Scholar] [CrossRef] [PubMed]
- Scossa, F.; Roda, F.; Tohge, T.; Georgiev, M.I.; Fernie, A.R. The Hot and the Colorful: Understanding the Metabolism, Genetics and Evolution of Consumer Preferred Metabolic Traits in Pepper and Related Species. CRC. Crit. Rev. Plant Sci. 2019, 38, 339–381. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. FAOSTAT—Food and Agriculture Database. Available online: http://www.fao.org/faostat (accessed on 19 March 2021).
- Neitzke, R.S.; Barbieri, R.L.; Vasconcelos, C.S.; Priori, D.; Fischer, S.Z.; Götzke, M.L.D.; Romano, C.M.; Heiden, G. Diversity in Capsicum Landraces Cultivated in Brazil. Acta Hortic. 2011, 531–536. [Google Scholar] [CrossRef]
- Da Silva Neto, J.J.; do Rêgo, E.R.; Nascimento, M.F.; Silva Filho, V.A.L.; de Almeida Neto, J.X.; do Rêgo, M.M. Variabilidade em população base de pimenteiras ornamentais (Capsicum annuum L.). Rev. Ceres 2014, 61, 84–89. [Google Scholar] [CrossRef]
- Fayos, O.; de Aguiar, A.; Jiménez-Cantizano, A.; Ferreiro-González, M.; Garcés-Claver, A.; Martínez, J.; Mallor, C.; Ruiz-Rodríguez, A.; Palma, M.; Barroso, C.; et al. Ontogenetic Variation of Individual and Total Capsaicinoids in Malagueta Peppers (Capsicum frutescens) during Fruit Maturation. Molecules 2017, 22, 736. [Google Scholar] [CrossRef]
- Rodrigues, R.; Gonçalves, L.S.; dos Bento, C.S.; Sudré, C.P.; Robaina, R.R.; do Amaral Júnior, A.T. Combining ability and heterosis for agronomic traits in chili pepper. Hortic. Bras. 2012, 30, 226–233. [Google Scholar] [CrossRef] [Green Version]
- De Santos, T.O.; Moulin, M.M.; Rangel, L.H.; Pirovani, R.O.L.; Valadares, F.V.; de Almeida, R.N.; Silva, L.O.E. Characterization and Diversity of Peppers (Capsicum spp.) Genotypes Based on Morphological Traits Using Multivariate Analysis. J. Exp. Agric. Int. 2019, 1–10. [Google Scholar] [CrossRef]
- Grafton, R.Q.; Daugbjerg, C.; Qureshi, M.E. Towards food security by 2050. Food Secur. 2015, 7, 179–183. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Dias, L.; Gil-Villar, D.; Castell-Zeising, V.; Quiñones, A.; Calatayud, Á.; Rodríguez-Burruezo, A.; Fita, A. Main Root Adaptations in Pepper Germplasm (Capsicum spp.) to Phosphorus Low-Input Conditions. Agronomy 2020, 10, 637. [Google Scholar] [CrossRef]
- United Nations Transforming Our World: The 2030 Agenda for Sustainable Development; ONU: New York, NY, USA, 2015.
- Moulin, M.M.; Rodrigues, R.; Ramos, H.C.C.; Bento, C.S.; Sudré, C.P.; Gonçalves, L.S.A.; Viana, A.P. Construction of an integrated genetic map for Capsicum baccatum L. Genet. Mol. Res. 2015, 14, 6683–6694. [Google Scholar] [CrossRef]
- Moulin, M.M.; Rodrigues, R.; Bento, C.S.; Gonçalves, L.S.A.; Santos, J.O.; Sudré, C.P.; Viana, A.P. Genetic dissection of agronomic traits in Capsicum baccatum var. pendulum. Genet. Mol. Res. 2015, 14, 2122–2132. [Google Scholar] [CrossRef]
- Baba, V.Y.; Rocha, K.R.; Gomes, G.P.; de Fátima Ruas, C.; Ruas, P.M.; Rodrigues, R.; Gonçalves, L.S.A. Genetic diversity of Capsicum chinense accessions based on fruit morphological characterization and AFLP markers. Genet. Resour. Crop Evol. 2016, 63, 1371–1381. [Google Scholar] [CrossRef]
- Cardoso, R.; Ruas, C.F.; Giacomin, R.M.; Ruas, P.M.; Ruas, E.A.; Barbieri, R.L.; Rodrigues, R.; Gonçalves, L.S.A. Genetic variability in Brazilian Capsicum baccatum germplasm collection assessed by morphological fruit traits and AFLP markers. PLoS ONE 2018, 13, e0196468. [Google Scholar] [CrossRef] [Green Version]
- Kantar, M.B.; Anderson, J.E.; Lucht, S.A.; Mercer, K.; Bernau, V.; Case, K.A.; Le, N.C.; Frederiksen, M.K.; DeKeyser, H.C.; Wong, Z.-Z.; et al. Vitamin Variation in Capsicum Spp. Provides Opportunities to Improve Nutritional Value of Human Diets. PLoS ONE 2016, 11, e0161464. [Google Scholar] [CrossRef] [PubMed]
- Deka, S.D. Taxonomic diversity of cultivated Capsicum genotypes of India. Electron. J. Plant Breed. 2017, 8, 660. [Google Scholar] [CrossRef]
- Dos Pessoa, A.M.S.; do Rêgo, E.R.; dos Santos, C.A.P.; de Carvalho, M.G.; de Mesquita, J.C.P.; do Rêgo, M.M. Inheritance of flower traits in ornamental pepper. Agropecuária Técnica 2018, 39, 50. [Google Scholar] [CrossRef]
- Sharmin, A.; Hoque, M.E.; Haque, M.M.; Khatun, F. Molecular Diversity Analysis of Some Chilli (Capsicum spp.) Genotypes Using SSR Markers. Am. J. Plant Sci. 2018, 9, 368–379. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, P.A.; Silva, L.R.A.; Alencar, A.A.S.; Santos, P.H.A.D.; Pimenta, S.; Sudré, C.P.; Corte, L.E.D.; Gonçalves, L.S.A.; Rodrigues, R. Biomorphological Characterization of Brazilian Capsicum Chinense Jacq. Germplasm. Agronomy 2020, 10, 447. [Google Scholar] [CrossRef] [Green Version]
- Costa, F.R.; Pereira, T.N.S.; Vitória, A.P.; Campos, K.P.; Rodrigues, R.; Silva, D.H.; Pereira, M.G. Genetic diversity among Capsicum accessions using RAPD markers. Crop. Breed. Appl. Biotechnol. 2006, 6, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Thul, S.T.; Darokar, M.P.; Shasany, A.K.; Khanuja, S.P.S. Molecular Profiling for Genetic Variability in Capsicum Species Based on ISSR and RAPD Markers. Mol. Biotechnol. 2012, 51, 137–147. [Google Scholar] [CrossRef]
- Soares de Souza, C.; Schuelter, A.R.; Finger, F.L.; Dias Casali, V.W. Variabilidade genética em acessos de Capsicum chinense por meio de marcadores isoenzimáticos e rapd. Sci. Agrar. 2013, 14. [Google Scholar] [CrossRef] [Green Version]
- Thul, S.T.; Shasany, A.K.; Darokar, M.P.; Khanuja, S.P.S. AFLP Analysis for Genetic Diversity in Capsicum Annuum and Related Species. Nat. Prod. Commun. 2006, 1, 1934578X0600100309. [Google Scholar] [CrossRef]
- Islam, M.A.; Sinha, P.; Sharma, S.S.; Negi, M.S.; Neog, B.; Tripathi, S.B. Analysis of genetic diversity and population structure in Capsicum landraces from North Eastern India using TE-AFLP markers. Plant Mol. Biol. Report. 2016, 34, 869–875. [Google Scholar] [CrossRef]
- Moreira, A.F.P.; Ruas, P.M.; de Ruas, C.F.; Baba, V.Y.; Giordani, W.; Arruda, I.M.; Rodrigues, R.; Gonçalves, L.S.A. Genetic diversity, population structure and genetic parameters of fruit traits in Capsicum chinense. Sci. Hortic. 2018, 236, 1–9. [Google Scholar] [CrossRef]
- Shirasawa, K.; Ishii, K.; Kim, C.; Ban, T.; Suzuki, M.; Ito, T.; Muranaka, T.; Kobayashi, M.; Nagata, N.; Isobe, S.; et al. Development of Capsicum EST–SSR markers for species identification and in silico mapping onto the tomato genome sequence. Mol. Breed. 2013, 31, 101–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, C.; Wei, X.; Zhao, Y.; Yuan, Y.; Yang, S.; Wang, Z.; Zhang, X.; Sun, J.; Zheng, X.; Yao, Q.; et al. Genetic Diversity Analysis of Capsicum Genus by SSR Markers. Mol. Plant Breed. 2017. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, C.D.O.; Rodrigues, R.; Sudré, C.P. Microsatellites for detecting inconsistencies in Capsicum cultivars registration in Brazilian database: More than meets the eye. Hortic. Bras. 2019, 37, 285–293. [Google Scholar] [CrossRef]
- Guzmán, F.A.; Moore, S.; de Vicente, M.C.; Jahn, M.M. Microsatellites to enhance characterization, conservation and breeding value of Capsicum germplasm. Genet. Resour. Crop Evol. 2020, 67, 569–585. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.; da Costa Batista, F.R.; Moulin, M.M. Molecular Markers in Capsicum spp. Breeding. In Production and Breeding of Chilli Peppers (Capsicum spp.); Springer International Publishing: Cham, Switzerland, 2016; pp. 81–95. [Google Scholar] [CrossRef]
- Ou, L.; Zou, X. Inter simple sequence repeat analysis of genetic diversity of five cultivated pepper species. Afr. J. Biotechnol. 2012, 11, 752–757. [Google Scholar] [CrossRef] [Green Version]
- Olantunji, T.L.; Afolayan, A.J. Contributions to the Classification of Capsicum annuum L. and Capsicum frutescens L. in West Africa Using Morphological Traits. Not. Bot. Horti Agrobot. Cluj-Napoca 2018, 47, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Rana, M.; Sharma, R.; Sharma, P.; Bhardwaj, S.V.; Sharma, M. Estimation of Genetic Diversity in Capsicum annuum L. Germplasm Using PCR-Based Molecular Markers. Natl. Acad. Sci. Lett. 2014, 37, 295–301. [Google Scholar] [CrossRef]
- D’Agostino, N.; Tamburino, R.; Cantarella, C.; De Carluccio, V.; Sannino, L.; Cozzolino, S.; Cardi, T.; Scotti, N. The Complete Plastome Sequences of Eleven Capsicum Genotypes: Insights into DNA Variation and Molecular Evolution. Genes 2018, 9, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colonna, V.; D’Agostino, N.; Garrison, E.; Albrechtsen, A.; Meisner, J.; Facchiano, A.; Cardi, T.; Tripodi, P. Genomic diversity and novel genome-wide association with fruit morphology in Capsicum, from 746k polymorphic sites. Sci. Rep. 2019, 9, 10067. [Google Scholar] [CrossRef] [Green Version]
- Olatunji, T.L.; Afolayan, A.J. Evaluation of genetic relationship among varieties of Capsicum annuum L. and Capsicum frutescens L. in West Africa using ISSR markers. Heliyon 2019, 5, e01700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moscone, E.A.; Scaldaferro, M.A.; Grabiele, M.; Cecchini, N.M.; Sánchez García, Y.; Jarret, R.; Daviña, J.R.; Ducasse, D.A.; Barboza, G.E.; Ehrendorfer, F. The evolution of chili peppers (Capsicum—Solanaceae): A cytogenetic perspective. Acta Hortic. 2007, 137–170. [Google Scholar] [CrossRef]
- IPGRI. Descriptors for Capsicum (Capsicum spp.); International Plant Genetic Resources Institute: Rome, Italy, 1995; ISBN 978-92-9043-216-6. [Google Scholar]
- Derera, F. Condiment Paprika: Breeding, Harvesting and Commercialisation: A Report for the Rural Industries Research and Development Corporation; RIRDC: New South Wales, Australia, 2000; p. 33. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1987, 12, 39–40. [Google Scholar]
- Daher, R.F.; Pereira, M.G.; Tupinambá, E.A.; Amaral Junior, A.T.; Aragão, W.M.; Ribeiro, F.E.; Oliveira, L.O.; Sakiyama, N.S. Assessment of coconut tree genetic divergence by compound sample RAPD marker analysis. Crop. Breed. Appl. Biotechnol. 2002, 2, 431–438. [Google Scholar] [CrossRef] [Green Version]
- Souza, D.C.L.; Gomes, L.J.; Blank, A.F.; Gois, I.B.; Pereira, G.S.; Alves, P.B.; Silva-Mann, R. Characterization of wild genotypes of Aroeira: Subsidy for plant breeding. J. Agric. Biotechnol. Sustain. Dev. 2014, 6, 39–49. [Google Scholar] [CrossRef]
- jiang, Y.; Liu, J.-P. Analysis of genetic diversity of Piper spp. in Hainan Island (China) using inter-simple sequence repeat ISSR markers. AFRICAN J. Biotechnol. 2011, 10, 16763–16768. [Google Scholar] [CrossRef]
- Yao, H.; Zhao, Y.; Chen, D.F.; Chen, J.K.; Zhou, T.S. ISSR primer screening and preliminary evaluation of genetic diversity in wild populations of Gycyrrhiza uralensis. Biol. Plant. 2008, 52, 117–120. [Google Scholar] [CrossRef]
- Refaat, M.H.; Elgarhy, H.A.S. Relationship between hybrid performance and genetic diversity based on ISSR-PCR markers in Pepper (Capsicum annuum. L.). Ann. Agric. Sci. 2007, 45, 1565–1579. [Google Scholar]
- Singh, D. The relative importance of characters affecting genetic divergence. Indian J. Genet. Plant Breed. 1981, 41, 237–245. [Google Scholar]
- R Core Team. R: A Language and ENVIRONMENT for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2021. Available online: http://www.R-project.org/ (accessed on 26 April 2021).
- Cruz, C.D..; Regazzi, I.A.J.; Carneiro, P.C.S. Modelos Biométricos Aplicados ao Melhoramento Genético; Editora UFV: Viçosa, MG, Brasil, 2012; Volume 1. [Google Scholar]
- Cruz, C.D. GENES—A software package for analysis in experimental statistics and quantitative genetics. Acta Sci. Agron. 2013, 35, 271–276. [Google Scholar] [CrossRef]
- Galili, T. dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef] [Green Version]
- Lima, M.F.; Carvalho, S.I.C.; Ragassi, C.F.; Bianchetti, L.B.; Faleiro, F.G.; Reifschneider, F.J.B. Characterization of a pepper collection (Capsicum frutescens L.) from Brazil. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef]
- Martinez, A.L.A.; Araújo, J.S.P.; Ragassi, C.F.; Buso, G.S.C.; Reifschneider, F.J.B. Variability among Capsicum baccatum accessions from Goiás, Brazil, assessed by morphological traits and molecular markers. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef]
- Nankar, A.N.; Todorova, V.; Tringovska, I.; Pasev, G.; Radeva-Ivanova, V.; Ivanova, V.; Kostova, D. A step towards Balkan Capsicum annuum L. core collection: Phenotypic and biochemical characterization of 180 accessions for agronomic, fruit quality, and virus resistance traits. PLoS ONE 2020, 15, e0237741. [Google Scholar] [CrossRef]
- Bianchi, P.A.; Dutra, I.P.; Moulin, M.M.; Santos, J.O.; Santos Júnior, A.C. Morphological characterization and analysis of genetic variability among pepper accessions. Ciência Rural 2016, 46, 7. [Google Scholar] [CrossRef]
- González-López, J.; Rodríguez-Moar, S.; Silvar, C. Correlation Analysis of High-Throughput Fruit Phenomics and Biochemical Profiles in Native Peppers (Capsicum spp.) from the Primary Center of Diversification. Agronomy 2021, 11, 262. [Google Scholar] [CrossRef]
- Carvalho, S.I.C.; Bianchetti, L.B.; Ragassi, C.F.; Ribeiro, C.S.C.; Reifschneider, F.J.B.; Buso, G.S.C.; Faleiro, F.G. Genetic variability of a Brazilian Capsicum frutescens germplasm collection using morphological characteristics and SSR markers. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef] [PubMed]
- Baral, J.B.; Bosland, P.W. Unraveling the Species Dilemma in Capsicum frutescens and C. chinense (Solanaceae): A Multiple Evidence Approach Using Morphology, Molecular Analysis, and Sexual Compatibility. J. Am. Soc. Hortic. Sci. 2004, 129, 826–832. [Google Scholar] [CrossRef] [Green Version]
- Leite, W.S.; Pavan, B.E.; Alcântara Neto, F.; Matos Filho, C.H.A.; Feitosa, F.S.; Oliveira, C.B. Multivariate Exploratory Approach and Influence of Six Agronomic Traits on Soybean Genotypes Selection. Nativa 2016, 4, 206–210. [Google Scholar] [CrossRef] [Green Version]
- Abud, H.F.; Araujo, R.F.; Pinto, C.M.F.; Araujo, E.F.; Araujo, A.V.; Santos, J.A. Caraterização morfométrica dos frutos de pimentas malagueta e biquinho. Rev. Bras. Agropecuária Sustentável 2018, 8. [Google Scholar] [CrossRef]
- Rêgo, M.M.; Sapucay, M.J.L.C.; Rêgo, E.R.; Araújo, E.R. Analysis of divergence and correlation of quantitative traits in ornamental pepper (Capsicum SPP.). Acta Hortic. 2015, 389–394. [Google Scholar] [CrossRef]
- Paran, I.; van der Knaap, E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 2007, 58, 3841–3852. [Google Scholar] [CrossRef] [Green Version]
- Naegele, R.P.; Mitchell, J.; Hausbeck, M.K. Genetic Diversity, Population Structure, and Heritability of Fruit Traits in Capsicum annuum. PLoS ONE 2016, 11, e0156969. [Google Scholar] [CrossRef] [PubMed]
- Leite, P.S.S.; Rodrigues, R.; Silva, R.N.O.; Pimenta, S.; Medeiros, A.M.; Bento, C.S.; Gonçalves, L.S.A. Molecular and agronomic analysis of intraspecific variability in Capsicum baccatum var. pendulum accessions. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Fortunato, F.L.G.; Do Rêgo, E.R.; De Carvalho, M.G.; Dos Santos, C.A.P.; Do Rêgo, M.M. Genetic diversity in ornamental pepper plants. Comun. Sci. 2019, 10, 364–375. [Google Scholar] [CrossRef] [Green Version]
- Rohlf, F.J.; Fisher, D.R. Tests for Hierarchical Structure in Random Data Sets. Syst. Zool. 1968, 17, 407. [Google Scholar] [CrossRef]
- Sokal, R.R.; Rohlf, F.J. The comparison of dendrograms by objective methods. Taxon 1962, 11, 33–40. [Google Scholar] [CrossRef]
- Finger, F.L.; Lannes, S.D.; Schuelter, A.R.; Doege, J.; Comerlatto, A.P.; Gonçalves, L.S.A.; Ferreira, F.R.A.; Clovis, L.R.; Scapim, C.A. Genetic diversity of Capsicum chinensis (Solanaceae) accessions based on molecular markers and morphological and agronomic traits. Genet. Mol. Res. 2010, 9, 1852–1864. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Datta, A.K.; Datta, S.; Gupta, S. Genetic assessment of eight Corchorus spp. (Tiliaceae) using RAPD and ISSR markers. Nucleus 2013, 56, 23–30. [Google Scholar] [CrossRef]
- Jia, X.-D.; Wang, T.; Zhai, M.; Li, Y.-R.; Guo, Z.-R. Genetic Diversity and Identification of Chinese-Grown Pecan Using ISSR and SSR Markers. Molecules 2011, 16, 10078–10092. [Google Scholar] [CrossRef] [Green Version]
- Sunar, S.; Yildirim, N.; Sengul, M.; Agar, G. Genetic diversity and relationships detected by ISSR and RAPD analysis among Aethionema species growing in Eastern Anatolia (Turkey). C. R. Biol. 2016, 339, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, F.R.; Pereira, T.N.S.; Sudré, C.P.; Rodrigues, R. Marcadores RAPD e caracteres morfoagronômicos na determinação da diversidade genética entre acessos de pimentas e pimentões. Ciência Rural 2008, 39, 696–704. [Google Scholar] [CrossRef] [Green Version]


| SV | DF | Capsicum chinense—Mean Squares | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | CD | SD | LL | LW | PL | FL | FD | FFM | PT | FLD | ||
| Block | 2 | 1741.84 | 2440.50 | 56.60 | 11.81 | 0.10 | 1.04 | 2.69 | 24.61 | 24.94 | 0.17 | 7.62 |
| Accessions | 36 | 513.35 ** | 910.36 ** | 50.97 ** | 10.58 ** | 2.61 ** | 1.30 ** | 90.08 ** | 266.59 ** | 113.76 ** | 1.36 ** | 8.33 ** |
| Error | 72 | 144.24 | 363.72 | 24.90 | 3.99 | 0.70 | 0.30 | 0.46 | 20.89 | 5.04 | 0.09 | 3.00 |
| CV% | 24.49 | 30.94 | 30.04 | 24.60 | 22.36 | 20.32 | 17.19 | 22.63 | 42.31 | 15.83 | 18.67 | |
| SV | DF | Capsicum baccatum var. pendulum—Mean Squares | ||||||||||
| PH | CD | SD | LL | LW | PL | FL | FD | FFM | PT | FLD | ||
| Block | 2 | 504.13 | 639.35 | 17.26 | 0.34 | 0.66 | 0.14 | 0.06 | 31.47 | 124,497.00 | 0.04 | 2.23 |
| Accessions | 22 | 571.64 ** | 1062.58 ** | 42.51 * | 7.43 * | 1.82 ** | 2.65 ** | 21.07 ** | 427.41 ** | 119.03 ** | 1.08 ** | 12.96 ** |
| Error | 44 | 150.19 | 275.89 | 18.92 | 3.89 | 0.67 | 0.42 | 1.00 | 28.51 | 7.88 | 0.05 | 5.63 |
| CV% | 22.42 | 25.42 | 24.94 | 20.93 | 19.30 | 14.93 | 16.11 | 23.15 | 29.46 | 9.68 | 19.55 | |
| SV | DF | Capsicum frutescens—Mean Squares | ||||||||||
| PH | CD | SD | LL | LW | PL | FL | FD | FFM | PT | FLD | ||
| Block | 2 | 701.29 | 1658.36 | 53.20 | 16.90 | 3.03 | 0.32 | 0.29 | 3.42 | 0.25 | 2.04 | 5.03 |
| Accessions | 6 | 842.05 ns | 1088.96 * | 79.79 ns | 9.50 ns | 2.06 * | 0.59 ns | 1.44 * | 5.22 * | 0.33 ns | 1.71 ns | 6.95 ns |
| Error | 12 | 313.12 | 349.92 | 33.44 | 5.08 | 0.68 | 0.36 | 0.46 | 1.63 | 0.27 | 1.93 | 3.78 |
| CV% | 23.37 | 24.18 | 25.06 | 23.00 | 18.20 | 21.01 | 23.07 | 22.00 | 73.89 | 161.07 | 22.77 | |
| SV | DF | Capsicum annuum var. annuum and Capsicum annuum var. glabriusculum—Mean Squares | ||||||||||
| PH | CD | SD | LL | LW | PL | FL | FD | FFM | PT | FLD | ||
| Block | 2 | 63.17 | 376.54 | 34.39 | 108.79 | 10.09 | 0.17 | 0.42 | 3.42 | 1.17 | 0.21 | 2.08 |
| Accessions | 1 | 620.17 * | 84.38 ns | 183.15 ns | 38.86 ns | 1.34 ns | 0.27 ns | 3.38 ns | 0.18 ns | 0.81 ns | 0.08 ns | 7.09 * |
| Error | 2 | 28.17 | 58.88 | 11.14 | 69.64 | 5.34 | 0.19 | 0.34 | 2.18 | 0.09 | 0.11 | 0.29 |
| CV% | 17.79 | 29.61 | 27.12 | 87.06 | 73.63 | 21.74 | 16.63 | 12.45 | 12.93 | 24.45 | 4.66 | |
| Trait | Relative Contribution (%) |
|---|---|
| Fruit length (cm) | 60.72% |
| Fruit diameter (mm) | 12.71% |
| Petiole length (cm) | 8.52% |
| Pericarp thickness (mm) | 5.49% |
| Fruit fresh mass (g) | 3.26% |
| Plant height (cm) | 3.07% |
| Flower diameter (mm) | 2.31% |
| Leaf length (cm) | 1.92% |
| Stem diameter (mm) | 1.09% |
| Canopy diameter (cm) | 0.75% |
| Leaf width (cm) | 0.17% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brilhante, B.D.G.; Santos, T.d.O.; Santos, P.H.A.D.; Kamphorst, S.H.; Neto, J.D.S.; Rangel, L.H.; Valadares, F.V.; de Almeida, R.N.; Rodrigues, R.; Júnior, A.C.S.; et al. Phenotypic and Molecular Characterization of Brazilian Capsicum Germplasm. Agronomy 2021, 11, 854. https://doi.org/10.3390/agronomy11050854
Brilhante BDG, Santos TdO, Santos PHAD, Kamphorst SH, Neto JDS, Rangel LH, Valadares FV, de Almeida RN, Rodrigues R, Júnior ACS, et al. Phenotypic and Molecular Characterization of Brazilian Capsicum Germplasm. Agronomy. 2021; 11(5):854. https://doi.org/10.3390/agronomy11050854
Chicago/Turabian StyleBrilhante, Bruna Dias Gomes, Talles de Oliveira Santos, Pedro Henrique Araújo Diniz Santos, Samuel Henrique Kamphorst, José Dias Souza Neto, Leandro Heitor Rangel, Fernanda Vargas Valadares, Rafael Nunes de Almeida, Rosana Rodrigues, Alexandre Cristiano Santos Júnior, and et al. 2021. "Phenotypic and Molecular Characterization of Brazilian Capsicum Germplasm" Agronomy 11, no. 5: 854. https://doi.org/10.3390/agronomy11050854
APA StyleBrilhante, B. D. G., Santos, T. d. O., Santos, P. H. A. D., Kamphorst, S. H., Neto, J. D. S., Rangel, L. H., Valadares, F. V., de Almeida, R. N., Rodrigues, R., Júnior, A. C. S., & Moulin, M. M. (2021). Phenotypic and Molecular Characterization of Brazilian Capsicum Germplasm. Agronomy, 11(5), 854. https://doi.org/10.3390/agronomy11050854








